[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV642469 HCL052d06_r
(508 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
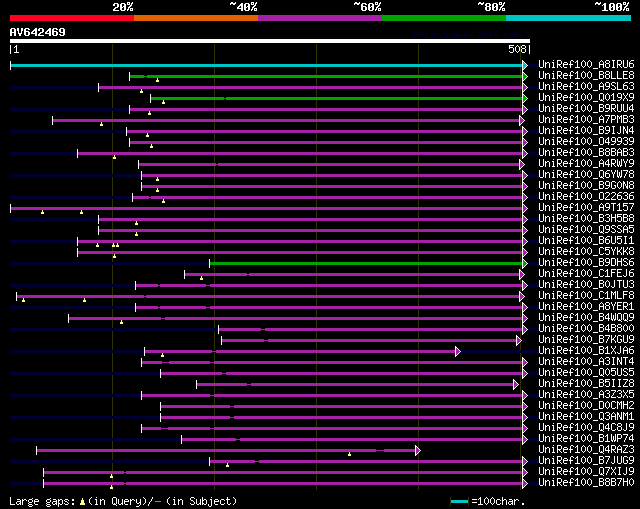
significant alignments:[graphical|details]
UniRef100_A8IRU6 Peptidyl-prolyl cis-trans isomerase, cyclophili... 320 3e-86
UniRef100_B8LLE8 Putative uncharacterized protein n=1 Tax=Picea ... 105 2e-21
UniRef100_A9SL63 Predicted protein n=1 Tax=Physcomitrella patens... 94 4e-18
UniRef100_Q019X9 Cyclophilin type, U box-containing peptidyl-pro... 94 5e-18
UniRef100_B9RUU4 Peptidyl-prolyl cis-trans isomerase, putative n... 92 2e-17
UniRef100_A7PMB3 Chromosome chr14 scaffold_21, whole genome shot... 92 3e-17
UniRef100_B9IJN4 Predicted protein n=1 Tax=Populus trichocarpa R... 91 6e-17
UniRef100_O49939 Peptidyl-prolyl cis-trans isomerase, chloroplas... 90 8e-17
UniRef100_B8BAB3 Putative uncharacterized protein n=1 Tax=Oryza ... 89 2e-16
UniRef100_A4RWY9 Predicted protein n=1 Tax=Ostreococcus lucimari... 89 2e-16
UniRef100_Q6YW78 Os08g0382400 protein n=1 Tax=Oryza sativa Japon... 87 6e-16
UniRef100_B9G0N8 Putative uncharacterized protein n=1 Tax=Oryza ... 87 6e-16
UniRef100_O22636 Poly(A) polymerase n=1 Tax=Pisum sativum RepID=... 86 1e-15
UniRef100_A9T157 Predicted protein n=1 Tax=Physcomitrella patens... 85 2e-15
UniRef100_B3H5B8 Uncharacterized protein At3g01480.2 n=1 Tax=Ara... 84 4e-15
UniRef100_Q9SSA5 Peptidyl-prolyl cis-trans isomerase CYP38, chlo... 84 4e-15
UniRef100_B6U5I1 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Zea... 84 5e-15
UniRef100_C5YKK8 Putative uncharacterized protein Sb07g019320 n=... 83 1e-14
UniRef100_B9DHS6 AT3G01480 protein (Fragment) n=1 Tax=Arabidopsi... 82 2e-14
UniRef100_C1FEJ6 Peptidyl-prolyl cis-trans isomerase chloroplast... 77 7e-13
UniRef100_B0JTU3 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Mic... 75 3e-12
UniRef100_C1MLF8 Peptidyl-prolyl cis-trans isomerase chloroplast... 73 1e-11
UniRef100_A8YER1 Genome sequencing data, contig C300 n=1 Tax=Mic... 72 2e-11
UniRef100_B4WQQ9 Peptidyl-prolyl cis-trans isomerase, cyclophili... 70 6e-11
UniRef100_B4B800 Peptidyl-prolyl cis-trans isomerase cyclophilin... 66 2e-09
UniRef100_B7KGU9 Peptidyl-prolyl cis-trans isomerase cyclophilin... 62 3e-08
UniRef100_B1XJA6 Putative peptidyl-prolyl cis-trans isomerase n=... 61 4e-08
UniRef100_A3INT4 Putative uncharacterized protein n=1 Tax=Cyanot... 59 1e-07
UniRef100_Q05US5 Putative cyclophilin-type peptidyl-prolyl cis-t... 59 2e-07
UniRef100_B5IIZ8 Peptidyl-prolyl cis-trans isomerase, cyclophili... 59 2e-07
UniRef100_A3Z3X5 Putative cyclophilin-type peptidyl-prolyl cis-t... 58 4e-07
UniRef100_D0CMH2 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Syn... 57 7e-07
UniRef100_Q3ANM1 Putative cyclophilin-type peptidyl-prolyl cis-t... 57 9e-07
UniRef100_Q4C8J9 Putative uncharacterized protein n=1 Tax=Crocos... 57 9e-07
UniRef100_B1WP74 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Cya... 56 2e-06
UniRef100_Q4RAZ3 Chromosome undetermined SCAF22622, whole genome... 55 2e-06
UniRef100_B7JUG9 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Cya... 55 3e-06
UniRef100_Q7XIJ9 Os07g0565600 protein n=1 Tax=Oryza sativa Japon... 55 3e-06
UniRef100_B8B7H0 Putative uncharacterized protein n=1 Tax=Oryza ... 55 3e-06
UniRef100_C7QUU8 Peptidylprolyl isomerase n=1 Tax=Cyanothece sp.... 55 4e-06