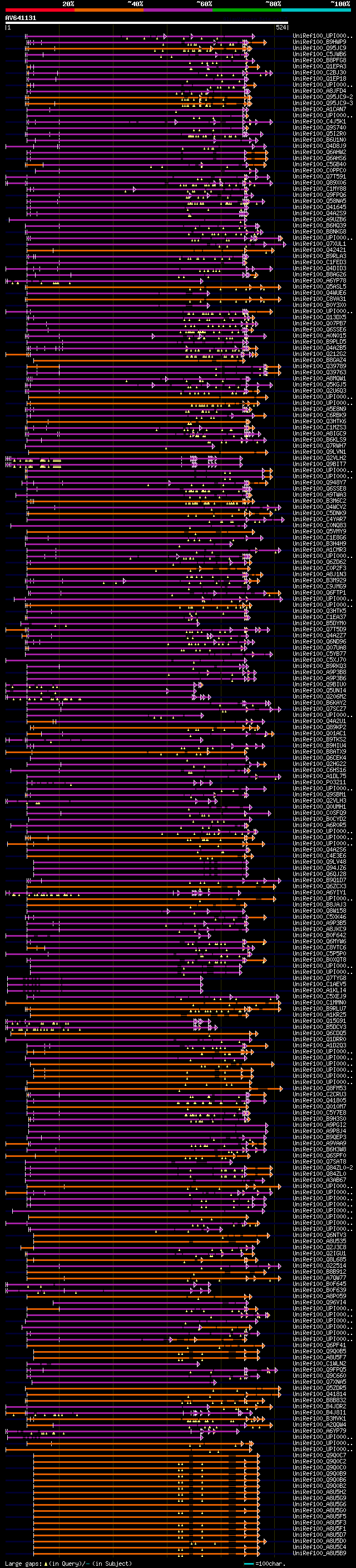
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV641131 HCL028c05_r
(524 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_UPI0001B55E13 putative chaplin n=1 Tax=Streptomyces sp... 79 1e-13
UniRef100_B9HWP9 Predicted protein (Fragment) n=1 Tax=Populus tr... 78 4e-13
UniRef100_Q95JC9 Parotid hormone n=1 Tax=Sus scrofa RepID=PRP_PIG 77 7e-13
UniRef100_C5JWB6 Putative uncharacterized protein n=1 Tax=Ajello... 75 3e-12
UniRef100_B8PFG8 Predicted protein n=1 Tax=Postia placenta Mad-6... 75 3e-12
UniRef100_Q1EPA3 Protein kinase family protein n=1 Tax=Musa acum... 75 4e-12
UniRef100_C2BJ30 Fe-S oxidoreductase n=1 Tax=Corynebacterium pse... 74 6e-12
UniRef100_Q1EP18 Protein kinase family protein n=1 Tax=Musa balb... 74 6e-12
UniRef100_UPI0001982D5B PREDICTED: hypothetical protein n=1 Tax=... 74 8e-12
UniRef100_A8JFD4 Glyoxal or galactose oxidase (Fragment) n=1 Tax... 74 8e-12
UniRef100_Q95JC9-2 Isoform 2 of Basic proline-rich protein n=1 T... 74 8e-12
UniRef100_Q95JC9-3 Isoform 3 of Basic proline-rich protein n=1 T... 74 8e-12
UniRef100_A1CAN7 Cell wall protein, putative n=1 Tax=Aspergillus... 73 1e-11
UniRef100_UPI00017390C7 AGP19 (ARABINOGALACTAN-PROTEIN 19) n=1 T... 73 1e-11
UniRef100_C4J5K1 Putative uncharacterized protein n=1 Tax=Zea ma... 73 1e-11
UniRef100_Q9S740 Lysine-rich arabinogalactan protein 19 n=1 Tax=... 73 1e-11
UniRef100_Q5I2R0 Minus agglutinin n=1 Tax=Chlamydomonas incerta ... 72 2e-11
UniRef100_B6U1N0 Putative uncharacterized protein n=1 Tax=Zea ma... 72 2e-11
UniRef100_Q4D8J9 Putative uncharacterized protein n=1 Tax=Trypan... 72 2e-11
UniRef100_Q6AHW2 Protease-1 (PRT1),, putative (Fragment) n=1 Tax... 72 2e-11
UniRef100_Q6AHS6 Protease-1 (PRT1) protein, putative n=1 Tax=Pne... 72 2e-11
UniRef100_C5GB40 Actin associated protein Wsp1 n=1 Tax=Ajellomyc... 72 2e-11
UniRef100_C0PPC0 Putative uncharacterized protein n=1 Tax=Zea ma... 72 2e-11
UniRef100_Q7T591 Large tegument protein n=1 Tax=Macacine herpesv... 72 3e-11
UniRef100_Q89X06 Blr0521 protein n=1 Tax=Bradyrhizobium japonicu... 72 3e-11
UniRef100_C1MY88 Predicted protein n=1 Tax=Micromonas pusilla CC... 72 3e-11
UniRef100_Q9FPQ6 Vegetative cell wall protein gp1 n=1 Tax=Chlamy... 72 3e-11
UniRef100_Q58NA5 Plus agglutinin (Fragment) n=1 Tax=Chlamydomona... 71 4e-11
UniRef100_Q41645 Extensin (Fragment) n=1 Tax=Volvox carteri RepI... 71 4e-11
UniRef100_Q4A2S9 Putative membrane protein n=2 Tax=Emiliania hux... 71 5e-11
UniRef100_A9UZB6 Predicted protein n=1 Tax=Monosiga brevicollis ... 71 5e-11
UniRef100_B6HQ39 Pc22g23920 protein n=1 Tax=Penicillium chrysoge... 71 5e-11
UniRef100_B8NKG8 Actin associated protein Wsp1, putative n=1 Tax... 70 7e-11
UniRef100_UPI0001925A99 PREDICTED: hypothetical protein, partial... 70 9e-11
UniRef100_Q7XUL1 Os04g0498700 protein n=1 Tax=Oryza sativa Japon... 70 9e-11
UniRef100_Q42421 Chitinase n=1 Tax=Beta vulgaris subsp. vulgaris... 70 9e-11
UniRef100_B9RLA3 ATP binding protein, putative n=1 Tax=Ricinus c... 70 9e-11
UniRef100_C1FED3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 70 1e-10
UniRef100_Q4DID3 Putative uncharacterized protein n=1 Tax=Trypan... 70 1e-10
UniRef100_B8AG26 Putative uncharacterized protein n=1 Tax=Oryza ... 69 2e-10
UniRef100_A6YP78 Major ampullate spidroin-like protein (Fragment... 69 2e-10
UniRef100_Q5ASL5 Putative uncharacterized protein n=1 Tax=Emeric... 69 2e-10
UniRef100_Q4WUE6 Cell wall protein, putative n=1 Tax=Aspergillus... 69 2e-10
UniRef100_C8VA31 Putative uncharacterized protein n=1 Tax=Asperg... 69 2e-10
UniRef100_B0Y3X0 Cell wall protein, putative n=1 Tax=Aspergillus... 69 2e-10
UniRef100_UPI0001925155 PREDICTED: hypothetical protein, partial... 69 2e-10
UniRef100_Q13DX5 OmpA/MotB n=1 Tax=Rhodopseudomonas palustris Bi... 69 2e-10
UniRef100_Q07PB7 Peptidase C14, caspase catalytic subunit p20 n=... 69 2e-10
UniRef100_Q6SSE6 Plus agglutinin n=1 Tax=Chlamydomonas reinhardt... 69 2e-10
UniRef100_A0N015 Vegetative cell wall protein n=1 Tax=Chlamydomo... 69 2e-10
UniRef100_B9PLD5 Putative uncharacterized protein n=1 Tax=Toxopl... 69 2e-10
UniRef100_Q4A2B5 Putative membrane protein n=1 Tax=Emiliania hux... 69 3e-10
UniRef100_Q212G2 Peptidase C14, caspase catalytic subunit p20 n=... 69 3e-10
UniRef100_B8GAZ4 Putative uncharacterized protein n=1 Tax=Chloro... 69 3e-10
UniRef100_Q39789 Proline-rich cell wall protein n=1 Tax=Gossypiu... 69 3e-10
UniRef100_Q39763 Proline-rich cell wall protein n=1 Tax=Gossypiu... 69 3e-10
UniRef100_A8MQW1 Uncharacterized protein At1g44191.1 n=1 Tax=Ara... 69 3e-10
UniRef100_Q5KGJ5 Putative uncharacterized protein n=1 Tax=Filoba... 69 3e-10
UniRef100_Q2U6Q3 Actin regulatory protein n=1 Tax=Aspergillus or... 69 3e-10
UniRef100_UPI0001A7B186 actin binding n=1 Tax=Arabidopsis thalia... 68 3e-10
UniRef100_UPI00015B4CAB PREDICTED: hypothetical protein n=1 Tax=... 68 3e-10
UniRef100_A5E8N9 Putative uncharacterized protein n=1 Tax=Bradyr... 68 3e-10
UniRef100_C6RBK9 Iron-sulfur cluster-binding protein n=1 Tax=Cor... 68 3e-10
UniRef100_Q3HTK6 Pherophorin-C1 protein n=1 Tax=Chlamydomonas re... 68 3e-10
UniRef100_C1MZS3 Predicted protein n=1 Tax=Micromonas pusilla CC... 68 3e-10
UniRef100_A8IGC9 Fibrocystin-L-like protein n=1 Tax=Chlamydomona... 68 3e-10
UniRef100_B6KLS9 Voltage gated chloride channel domain-containin... 68 3e-10
UniRef100_Q7RWH7 Cytokinesis protein sepA n=1 Tax=Neurospora cra... 68 3e-10
UniRef100_Q9LVN1 Formin-like protein 13 n=1 Tax=Arabidopsis thal... 68 3e-10
UniRef100_Q2VLH2 Major ampullate spidroin 2-like (Fragment) n=1 ... 63 4e-10
UniRef100_Q9BIT7 Major ampullate spidroin 2-like protein (Fragme... 63 4e-10
UniRef100_UPI0001A5EE3F PREDICTED: hypothetical protein FLJ22184... 68 4e-10
UniRef100_UPI0001A5EAAB PREDICTED: hypothetical protein LOC80164... 68 4e-10
UniRef100_Q948Y7 VMP3 protein n=1 Tax=Volvox carteri f. nagarien... 68 4e-10
UniRef100_Q6SSE8 Minus agglutinin n=1 Tax=Chlamydomonas reinhard... 68 4e-10
UniRef100_A9TWA3 Predicted protein n=1 Tax=Physcomitrella patens... 68 4e-10
UniRef100_B3M6C2 GF24314 n=1 Tax=Drosophila ananassae RepID=B3M6... 68 4e-10
UniRef100_Q4WCV2 Actin associated protein Wsp1, putative n=1 Tax... 68 4e-10
UniRef100_C5DNK9 KLTH0G17886p n=1 Tax=Lachancea thermotolerans C... 68 4e-10
UniRef100_C4YAR7 Putative uncharacterized protein n=1 Tax=Clavis... 68 4e-10
UniRef100_C0NQ83 Putative uncharacterized protein n=1 Tax=Ajello... 68 4e-10
UniRef100_Q5VMY9 Hydroxyproline-rich glycoprotein-like n=1 Tax=O... 67 6e-10
UniRef100_C1E8G6 Predicted protein n=1 Tax=Micromonas sp. RCC299... 67 6e-10
UniRef100_B3H4H9 Uncharacterized protein At2g28671.1 n=1 Tax=Ara... 67 6e-10
UniRef100_A1CMR3 Actin associated protein Wsp1, putative n=1 Tax... 67 6e-10
UniRef100_UPI0001983015 PREDICTED: hypothetical protein n=1 Tax=... 67 7e-10
UniRef100_Q6ZD62 Os08g0108300 protein n=1 Tax=Oryza sativa Japon... 67 7e-10
UniRef100_C0P2F3 Putative uncharacterized protein n=1 Tax=Zea ma... 67 7e-10
UniRef100_A8J1N3 Cell wall protein pherophorin-C10 (Fragment) n=... 67 7e-10
UniRef100_B3M929 GF25073 n=1 Tax=Drosophila ananassae RepID=B3M9... 67 7e-10
UniRef100_C9JMG9 Putative uncharacterized protein IQSEC1 n=1 Tax... 67 7e-10
UniRef100_Q6FTP1 Similar to uniprot|P37370 Saccharomyces cerevis... 67 7e-10
UniRef100_UPI00006A151D UPI00006A151D related cluster n=1 Tax=Xe... 67 1e-09
UniRef100_UPI0000EB007F UPI0000EB007F related cluster n=1 Tax=Ca... 67 1e-09
UniRef100_Q3HTK5 Pherophorin-C2 protein n=1 Tax=Chlamydomonas re... 67 1e-09
UniRef100_C1EA37 Predicted protein n=1 Tax=Micromonas sp. RCC299... 67 1e-09
UniRef100_B5DYM0 GA27216 n=1 Tax=Drosophila pseudoobscura pseudo... 67 1e-09
UniRef100_Q7T5D9 Very large tegument protein n=1 Tax=Macacine he... 66 1e-09
UniRef100_Q4A2Z7 Putative membrane protein n=2 Tax=Emiliania hux... 66 1e-09
UniRef100_Q6ND96 Possible OmpA family member n=1 Tax=Rhodopseudo... 66 1e-09
UniRef100_Q07UA8 OmpA/MotB domain protein n=1 Tax=Rhodopseudomon... 66 1e-09
UniRef100_C5YB77 Putative uncharacterized protein Sb06g000250 n=... 66 1e-09
UniRef100_C5XJ70 Putative uncharacterized protein Sb03g034040 n=... 66 1e-09
UniRef100_B9RKQ3 ATP binding protein, putative n=1 Tax=Ricinus c... 66 1e-09
UniRef100_A9P3B8 Anther-specific proline rich protein n=1 Tax=Br... 66 1e-09
UniRef100_A9P3B6 Anther-specific proline rich protein n=1 Tax=Br... 66 1e-09
UniRef100_Q9BIU0 Major ampullate spidroin 1 (Fragment) n=1 Tax=L... 66 1e-09
UniRef100_Q5UNI4 Dragline silk spidroin 1 (Fragment) n=1 Tax=Nep... 66 1e-09
UniRef100_Q206M2 Major ampullate spidroin 1 (Fragment) n=1 Tax=L... 66 1e-09
UniRef100_B6KAY2 Putative uncharacterized protein n=2 Tax=Toxopl... 66 1e-09
UniRef100_Q7SCZ7 WASP-interacting protein-like protein vrp1p n=1... 66 1e-09
UniRef100_UPI0000DF06C6 Os02g0456000 n=1 Tax=Oryza sativa Japoni... 66 2e-09
UniRef100_Q4A2U1 Putative membrane protein n=2 Tax=Emiliania hux... 66 2e-09
UniRef100_Q89KP2 Bll4862 protein n=1 Tax=Bradyrhizobium japonicu... 66 2e-09
UniRef100_Q01AC1 Meltrins, fertilins and related Zn-dependent me... 66 2e-09
UniRef100_B9TKS2 Putative uncharacterized protein (Fragment) n=1... 66 2e-09
UniRef100_B9HIU4 Predicted protein (Fragment) n=1 Tax=Populus tr... 66 2e-09
UniRef100_B8ATX9 Putative uncharacterized protein n=1 Tax=Oryza ... 66 2e-09
UniRef100_Q6CEK4 YALI0B14971p n=1 Tax=Yarrowia lipolytica RepID=... 66 2e-09
UniRef100_Q2HG22 Putative uncharacterized protein n=1 Tax=Chaeto... 66 2e-09
UniRef100_C6HS16 Actin associated protein Wsp1 n=1 Tax=Ajellomyc... 66 2e-09
UniRef100_A1DL75 Actin associated protein Wsp1, putative n=1 Tax... 66 2e-09
UniRef100_P03211 Epstein-Barr nuclear antigen 1 n=1 Tax=Human he... 66 2e-09
UniRef100_UPI0000D9EA10 PREDICTED: hypothetical protein n=1 Tax=... 65 2e-09
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 65 2e-09
UniRef100_Q2VLH3 Major ampullate spidroin 1-like (Fragment) n=1 ... 65 2e-09
UniRef100_Q0UMH1 Putative uncharacterized protein n=1 Tax=Phaeos... 65 2e-09
UniRef100_C0SFQ9 Putative uncharacterized protein n=1 Tax=Paraco... 65 2e-09
UniRef100_B0CYD2 WASP-interacting protein VRP1/WIP n=1 Tax=Lacca... 65 2e-09
UniRef100_A6R0R5 Predicted protein n=1 Tax=Ajellomyces capsulatu... 65 2e-09
UniRef100_UPI0001A7B3C7 unknown protein n=1 Tax=Arabidopsis thal... 65 3e-09
UniRef100_UPI00015B501E PREDICTED: hypothetical protein n=1 Tax=... 65 3e-09
UniRef100_UPI00015538CB PREDICTED: hypothetical protein n=1 Tax=... 65 3e-09
UniRef100_Q4A2S6 Putative membrane protein n=2 Tax=Emiliania hux... 65 3e-09
UniRef100_C4E3E6 RNA polymerase sigma factor, sigma-70 family n=... 65 3e-09
UniRef100_Q9LV48 Protein kinase-like protein n=1 Tax=Arabidopsis... 65 3e-09
UniRef100_Q94JZ6 At3g24600 n=1 Tax=Arabidopsis thaliana RepID=Q9... 65 3e-09
UniRef100_Q6QJ28 Putative uncharacterized protein (Fragment) n=1... 65 3e-09
UniRef100_B9Q1D7 Chloride channel, putative n=1 Tax=Toxoplasma g... 65 3e-09
UniRef100_Q6ZCX3 Formin-like protein 6 n=1 Tax=Oryza sativa Japo... 65 3e-09
UniRef100_A6YIY1 Major ampullate spidroin 1 n=1 Tax=Latrodectus ... 64 3e-09
UniRef100_UPI0000DD9486 Os08g0280200 n=1 Tax=Oryza sativa Japoni... 65 4e-09
UniRef100_B8JAJ3 General secretory system II protein E domain pr... 65 4e-09
UniRef100_Q8W158 Anther-specific proline-rich protein n=1 Tax=Br... 65 4e-09
UniRef100_C5XK46 Putative uncharacterized protein Sb03g034710 n=... 65 4e-09
UniRef100_A9P3B5 Anther-specific proline rich protein n=1 Tax=Br... 65 4e-09
UniRef100_A8JKC9 Predicted protein (Fragment) n=1 Tax=Chlamydomo... 65 4e-09
UniRef100_B0F642 Major ampullate spidroin 1 locus 1 (Fragment) n... 65 4e-09
UniRef100_Q6MYW6 Basic proline-rich protein n=1 Tax=Aspergillus ... 65 4e-09
UniRef100_C8VTC6 Proline-rich, actin-associated protein Vrp1, pu... 65 4e-09
UniRef100_C5P5P0 WH2 motif family protein n=1 Tax=Coccidioides p... 65 4e-09
UniRef100_B0XQT8 Conserved proline-rich protein n=2 Tax=Aspergil... 65 4e-09
UniRef100_UPI000198414B PREDICTED: hypothetical protein isoform ... 64 5e-09
UniRef100_UPI000198414A PREDICTED: hypothetical protein isoform ... 64 5e-09
UniRef100_Q7TYG8 PE-PGRS FAMILY PROTEIN [SECOND PART] n=1 Tax=My... 64 5e-09
UniRef100_C1AEV5 PE-PGRS family protein n=1 Tax=Mycobacterium bo... 64 5e-09
UniRef100_A1KLI4 PE-PGRS family protein [second part] n=1 Tax=My... 64 5e-09
UniRef100_C5XEJ9 Putative uncharacterized protein Sb03g029150 n=... 64 5e-09
UniRef100_C1MMN0 DNAJ family chaperone protein n=1 Tax=Micromona... 64 5e-09
UniRef100_B9RLU7 Putative uncharacterized protein n=1 Tax=Ricinu... 64 5e-09
UniRef100_A1KR25 Cell wall glycoprotein GP2 (Fragment) n=2 Tax=C... 64 5e-09
UniRef100_Q15G91 MaSp2b (Fragment) n=1 Tax=Deinopis spinosa RepI... 64 5e-09
UniRef100_B5DCV3 Major ampullate spidroin-like protein (Fragment... 64 5e-09
UniRef100_Q6CDQ5 YALI0B22110p n=1 Tax=Yarrowia lipolytica RepID=... 64 5e-09
UniRef100_Q1DRR0 Putative uncharacterized protein n=1 Tax=Coccid... 64 5e-09
UniRef100_A1D2Q3 Conserved proline-rich protein n=1 Tax=Neosarto... 64 5e-09
UniRef100_UPI0000E59252 mucin 2 precursor n=1 Tax=Homo sapiens R... 64 6e-09
UniRef100_UPI0000E12BCF Os07g0596300 n=1 Tax=Oryza sativa Japoni... 64 6e-09
UniRef100_UPI0000252E11 AER176Wp n=1 Tax=Ashbya gossypii ATCC 10... 64 6e-09
UniRef100_UPI0001AE6B68 Mucin-2 precursor (Intestinal mucin-2). ... 64 6e-09
UniRef100_UPI00006C10F2 UPI00006C10F2 related cluster n=1 Tax=Ho... 64 6e-09
UniRef100_UPI000179EE74 UPI000179EE74 related cluster n=1 Tax=Bo... 64 6e-09
UniRef100_Q8FM53 Putative uncharacterized protein n=1 Tax=Coryne... 64 6e-09
UniRef100_C2CRU3 Possible Fe-S dehydrogenase n=1 Tax=Corynebacte... 64 6e-09
UniRef100_Q41805 Extensin-like protein n=1 Tax=Zea mays RepID=Q4... 64 6e-09
UniRef100_Q010M7 Predicted membrane protein (Patched superfamily... 64 6e-09
UniRef100_C5Y7E8 Putative uncharacterized protein Sb05g025945 (F... 64 6e-09
UniRef100_B9H3S0 Predicted protein (Fragment) n=1 Tax=Populus tr... 64 6e-09
UniRef100_A9PGI2 Putative uncharacterized protein n=1 Tax=Populu... 64 6e-09
UniRef100_A9P8J4 Predicted protein n=1 Tax=Populus trichocarpa R... 64 6e-09
UniRef100_B9QEP3 Chloride channel protein k, putative n=1 Tax=To... 64 6e-09
UniRef100_A9VAA9 Predicted protein n=1 Tax=Monosiga brevicollis ... 64 6e-09
UniRef100_B6H3W8 Pc13g11020 protein n=1 Tax=Penicillium chrysoge... 64 6e-09
UniRef100_Q6SPF0 Atherin n=1 Tax=Homo sapiens RepID=SAMD1_HUMAN 64 6e-09
UniRef100_Q7SAT8 Actin cytoskeleton-regulatory complex protein p... 64 6e-09
UniRef100_Q84ZL0-2 Isoform 2 of Formin-like protein 5 n=1 Tax=Or... 64 6e-09
UniRef100_Q84ZL0 Formin-like protein 5 n=1 Tax=Oryza sativa Japo... 64 6e-09
UniRef100_A3AB67 Formin-like protein 16 n=1 Tax=Oryza sativa Jap... 64 6e-09
UniRef100_UPI0001982E0B PREDICTED: hypothetical protein n=1 Tax=... 64 8e-09
UniRef100_UPI0000F1FA8B PREDICTED: hypothetical protein n=1 Tax=... 64 8e-09
UniRef100_UPI0000E4A721 PREDICTED: similar to LOC397922 protein ... 64 8e-09
UniRef100_UPI0000E48217 PREDICTED: similar to LOC397922 protein,... 64 8e-09
UniRef100_UPI0000DB6FAC PREDICTED: similar to Protein cappuccino... 64 8e-09
UniRef100_UPI0000EB3C03 UPI0000EB3C03 related cluster n=1 Tax=Ca... 64 8e-09
UniRef100_UPI0000EB072B UPI0000EB072B related cluster n=1 Tax=Ca... 64 8e-09
UniRef100_UPI000179CE55 Integrator complex subunit 1 (Int1). n=1... 64 8e-09
UniRef100_Q6NTV3 MGC81512 protein n=1 Tax=Xenopus laevis RepID=Q... 64 8e-09
UniRef100_A8U535 Envelope glycoprotein G n=1 Tax=Human herpesvir... 64 8e-09
UniRef100_Q2J3C8 OmpA/MotB n=1 Tax=Rhodopseudomonas palustris Ha... 64 8e-09
UniRef100_Q2IGU1 General secretory system II, protein E-like n=1... 64 8e-09
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 64 8e-09
UniRef100_O22514 Proline rich protein n=1 Tax=Santalum album Rep... 64 8e-09
UniRef100_B8B912 Putative uncharacterized protein n=1 Tax=Oryza ... 64 8e-09
UniRef100_A7QW77 Chromosome chr3 scaffold_199, whole genome shot... 64 8e-09
UniRef100_B0F645 Major ampullate spidroin 1 locus 2 (Fragment) n... 64 8e-09
UniRef100_B0F639 Major ampullate spidroin 1 locus 3 (Fragment) n... 64 8e-09
UniRef100_A8P059 Pherophorin-dz1 protein, putative n=1 Tax=Brugi... 64 8e-09
UniRef100_Q96VI4 Protease 1 n=1 Tax=Pneumocystis carinii RepID=Q... 64 8e-09
UniRef100_UPI00019829A2 PREDICTED: hypothetical protein n=1 Tax=... 63 1e-08
UniRef100_UPI000180C8F8 PREDICTED: similar to CG16995 CG16995-PA... 63 1e-08
UniRef100_UPI0000E48C31 PREDICTED: hypothetical protein n=1 Tax=... 63 1e-08
UniRef100_UPI0000D9FB77 PREDICTED: hypothetical protein, partial... 63 1e-08
UniRef100_UPI00005DC1BB PERK10 (PROLINE-RICH EXTENSIN-LIKE RECEP... 63 1e-08
UniRef100_UPI00016E1896 UPI00016E1896 related cluster n=1 Tax=Ta... 63 1e-08
UniRef100_Q6PF41 MGC68961 protein n=1 Tax=Xenopus laevis RepID=Q... 63 1e-08
UniRef100_Q9Q0B5 Glycoprotein G-2 (Fragment) n=1 Tax=Human herpe... 63 1e-08
UniRef100_A8U5F7 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_C1WLN2 Putative uncharacterized protein n=1 Tax=Kribbe... 63 1e-08
UniRef100_Q9FPQ5 Gamete-specific hydroxyproline-rich glycoprotei... 63 1e-08
UniRef100_Q9C660 Pto kinase interactor, putative n=1 Tax=Arabido... 63 1e-08
UniRef100_Q7XNA5 OSJNBa0011E07.13 protein n=1 Tax=Oryza sativa J... 63 1e-08
UniRef100_Q5ZDR5 Os01g0329300 protein n=1 Tax=Oryza sativa Japon... 63 1e-08
UniRef100_Q41814 Hydroxyproline-rich glycoprotein n=1 Tax=Zea ma... 63 1e-08
UniRef100_B8B832 Putative uncharacterized protein n=1 Tax=Oryza ... 63 1e-08
UniRef100_B4JDR2 GH10507 n=1 Tax=Drosophila grimshawi RepID=B4JD... 63 1e-08
UniRef100_B4J8I1 GH21940 n=1 Tax=Drosophila grimshawi RepID=B4J8... 63 1e-08
UniRef100_B3MVK1 GF23569 n=1 Tax=Drosophila ananassae RepID=B3MV... 63 1e-08
UniRef100_A2QQW4 Contig An08c0110, complete genome n=1 Tax=Asper... 63 1e-08
UniRef100_A6YP79 Minor ampullate spidroin-like protein (Fragment... 60 1e-08
UniRef100_UPI0001B44C4E hypothetical protein MtubK_14054 n=1 Tax... 63 1e-08
UniRef100_UPI0001926FBD PREDICTED: hypothetical protein n=1 Tax=... 63 1e-08
UniRef100_UPI000060456A TAF4A RNA polymerase II, TATA box bindin... 63 1e-08
UniRef100_Q9Q0C7 Glycoprotein G-2 (Fragment) n=2 Tax=Human herpe... 63 1e-08
UniRef100_Q9Q0C2 Glycoprotein G-2 (Fragment) n=2 Tax=Human herpe... 63 1e-08
UniRef100_Q9Q0C0 Glycoprotein G-2 (Fragment) n=1 Tax=Human herpe... 63 1e-08
UniRef100_Q9Q0B9 Glycoprotein G-2 (Fragment) n=1 Tax=Human herpe... 63 1e-08
UniRef100_Q9Q0B6 Glycoprotein G-2 (Fragment) n=1 Tax=Human herpe... 63 1e-08
UniRef100_Q9Q0B2 Glycoprotein G-2 (Fragment) n=3 Tax=Human herpe... 63 1e-08
UniRef100_A8U5H2 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U5G9 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U5G6 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U5G0 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U5F5 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U5F3 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U5F1 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U5D7 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U5D0 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U5C4 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U5B9 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U5A7 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U5A2 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U596 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U592 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U561 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U556 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U552 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U548 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U517 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U514 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U509 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8U504 Envelope glycoprotein G n=1 Tax=Human herpesvir... 63 1e-08
UniRef100_A8L089 Putative uncharacterized protein n=1 Tax=Franki... 63 1e-08
UniRef100_A8HYN7 Putative uncharacterized protein n=1 Tax=Azorhi... 63 1e-08
UniRef100_A5U5I6 PE-PGRS family protein n=3 Tax=Mycobacterium tu... 63 1e-08
UniRef100_C6DMZ3 Putative uncharacterized protein n=1 Tax=Mycoba... 63 1e-08
UniRef100_A5WQA0 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 63 1e-08
UniRef100_C5X8F7 Putative uncharacterized protein Sb02g032910 n=... 63 1e-08
UniRef100_C0PJF1 Putative uncharacterized protein n=1 Tax=Zea ma... 63 1e-08
UniRef100_B9GEU2 Predicted protein n=1 Tax=Populus trichocarpa R... 63 1e-08
UniRef100_B4FYJ4 Putative uncharacterized protein n=1 Tax=Zea ma... 63 1e-08
UniRef100_A2F502 Formin Homology 2 Domain containing protein n=1... 63 1e-08
UniRef100_Q2KFY0 Putative uncharacterized protein n=1 Tax=Magnap... 63 1e-08
UniRef100_A4RBS7 Putative uncharacterized protein n=1 Tax=Magnap... 63 1e-08
UniRef100_A1DF14 Cell wall protein, putative n=1 Tax=Neosartorya... 63 1e-08
UniRef100_Q6ZQP7 Uncharacterized protein LOC284861 n=1 Tax=Homo ... 63 1e-08
UniRef100_C1B313 Translation initiation factor IF-2 n=1 Tax=Rhod... 63 1e-08
UniRef100_P13290 Envelope glycoprotein G n=2 Tax=Human herpesvir... 63 1e-08
UniRef100_P14918 Extensin n=1 Tax=Zea mays RepID=EXTN_MAIZE 63 1e-08
UniRef100_Q3KSS4 Epstein-Barr nuclear antigen 1 n=1 Tax=Human he... 63 1e-08
UniRef100_UPI0001A7B0C1 protein binding / structural constituent... 62 2e-08
UniRef100_UPI0000F2EB19 PREDICTED: hypothetical protein n=1 Tax=... 62 2e-08
UniRef100_UPI0000E240E0 PREDICTED: hypothetical protein n=1 Tax=... 62 2e-08
UniRef100_UPI00015DF937 CDNA: FLJ22184 fis, clone HRC00983. n=1 ... 62 2e-08
UniRef100_UPI0000EB41C9 UPI0000EB41C9 related cluster n=1 Tax=Ca... 62 2e-08
UniRef100_Q9Q0C4 Glycoprotein G-2 (Fragment) n=3 Tax=Human herpe... 62 2e-08
UniRef100_A8U5G3 Envelope glycoprotein G n=1 Tax=Human herpesvir... 62 2e-08
UniRef100_A7U885 Envelope glycoprotein G n=1 Tax=Human herpesvir... 62 2e-08
UniRef100_Q21D92 OmpA/MotB n=1 Tax=Rhodopseudomonas palustris Bi... 62 2e-08
UniRef100_B1VHX6 Putative Fe-S oxidoreductase n=1 Tax=Corynebact... 62 2e-08
UniRef100_C1YUC1 Putative uncharacterized protein n=1 Tax=Nocard... 62 2e-08
UniRef100_B5GHX2 Predicted protein n=1 Tax=Streptomyces sp. SPB7... 62 2e-08
UniRef100_B4D0M3 Putative uncharacterized protein n=1 Tax=Chthon... 62 2e-08
UniRef100_Q9SPM0 Extensin-like protein n=1 Tax=Zea mays RepID=Q9... 62 2e-08
UniRef100_Q9SN46 Extensin-like protein n=1 Tax=Arabidopsis thali... 62 2e-08
UniRef100_C0PKV4 Putative uncharacterized protein n=1 Tax=Zea ma... 62 2e-08
UniRef100_B9FY87 Putative uncharacterized protein n=1 Tax=Oryza ... 62 2e-08
UniRef100_B9FLI1 Putative uncharacterized protein n=1 Tax=Oryza ... 62 2e-08
UniRef100_A9PG83 Putative uncharacterized protein n=1 Tax=Populu... 62 2e-08
UniRef100_A2Y786 Putative uncharacterized protein n=1 Tax=Oryza ... 62 2e-08
UniRef100_Q9VZC2 CG15021 n=1 Tax=Drosophila melanogaster RepID=Q... 62 2e-08
UniRef100_Q5CQC1 Uncharacterized secreted protein with thr rich ... 62 2e-08
UniRef100_Q4QE97 Formin A n=1 Tax=Leishmania major RepID=Q4QE97_... 62 2e-08
UniRef100_Q206M1 Major ampullate spidroin 2 (Fragment) n=1 Tax=L... 62 2e-08
UniRef100_O17434 Minor ampullate silk protein MiSp1 (Fragment) n... 62 2e-08
UniRef100_C3YWB8 Putative uncharacterized protein n=1 Tax=Branch... 62 2e-08
UniRef100_C3YC75 Putative uncharacterized protein n=1 Tax=Branch... 62 2e-08
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 62 2e-08
UniRef100_A8NN84 Putative uncharacterized protein n=1 Tax=Coprin... 62 2e-08
UniRef100_Q6ZQP7-2 Isoform 2 of Uncharacterized protein LOC28486... 62 2e-08
UniRef100_Q9H6K5 Putative uncharacterized protein FLJ22184 n=1 T... 62 2e-08
UniRef100_Q02817 Mucin-2 n=1 Tax=Homo sapiens RepID=MUC2_HUMAN 62 2e-08
UniRef100_Q1QHE7 OmpA/MotB n=1 Tax=Nitrobacter hamburgensis X14 ... 62 2e-08
UniRef100_UPI0001A5EB4A PREDICTED: hypothetical protein FLJ22184... 62 2e-08
UniRef100_UPI0000E82048 PREDICTED: similar to WASP-family protei... 62 2e-08
UniRef100_UPI0001A2BBDD hypothetical protein LOC100009637 n=1 Ta... 62 2e-08
UniRef100_UPI0000ECC14B UPI0000ECC14B related cluster n=1 Tax=Ga... 62 2e-08
UniRef100_Q4SEV7 Chromosome undetermined SCAF14611, whole genome... 62 2e-08
UniRef100_A2RUY4 Zgc:158395 protein n=1 Tax=Danio rerio RepID=A2... 62 2e-08
UniRef100_Q9Q0C1 Glycoprotein G-2 (Fragment) n=1 Tax=Human herpe... 62 2e-08
UniRef100_A8U5F8 Envelope glycoprotein G n=1 Tax=Human herpesvir... 62 2e-08
UniRef100_A8U531 Envelope glycoprotein G n=1 Tax=Human herpesvir... 62 2e-08
UniRef100_A9GHQ0 Cellulose synthase 1 operon protein C n=1 Tax=S... 62 2e-08
UniRef100_A3Q0W3 Putative uncharacterized protein n=1 Tax=Mycoba... 62 2e-08
UniRef100_Q9C946 Putative uncharacterized protein T7P1.21 n=1 Ta... 62 2e-08
UniRef100_Q9ARH1 Receptor protein kinase PERK1 n=1 Tax=Brassica ... 62 2e-08
UniRef100_B9RZI2 Protein binding protein, putative n=1 Tax=Ricin... 62 2e-08
UniRef100_B8A6X3 Putative uncharacterized protein n=1 Tax=Oryza ... 62 2e-08
UniRef100_B6SUZ1 Putative uncharacterized protein n=1 Tax=Zea ma... 62 2e-08
UniRef100_Q5CLM1 Putative uncharacterized protein n=1 Tax=Crypto... 62 2e-08
UniRef100_B9QFG7 Subtilisin-like protein TgSUB1, putative n=1 Ta... 62 2e-08
UniRef100_B0F654 Major ampullate spidroin 1 locus 1 (Fragment) n... 62 2e-08
UniRef100_B0F652 Major ampullate spidroin 1 locus 1 (Fragment) n... 62 2e-08
UniRef100_B0F640 Major ampullate spidroin 1 locus 3 (Fragment) n... 62 2e-08
UniRef100_Q2GVT8 Protein transport protein SEC31 n=1 Tax=Chaetom... 62 2e-08
UniRef100_A4FM34 Translation initiation factor IF-2 n=1 Tax=Sacc... 62 2e-08
UniRef100_UPI0000E7FCDE PREDICTED: frizzled homolog 8 (Drosophil... 62 3e-08
UniRef100_UPI0000E1257F Os05g0516400 n=1 Tax=Oryza sativa Japoni... 62 3e-08
UniRef100_UPI000023F096 hypothetical protein FG08656.1 n=1 Tax=G... 62 3e-08
UniRef100_UPI00016E65E1 UPI00016E65E1 related cluster n=1 Tax=Ta... 62 3e-08
UniRef100_UPI00016E65E0 UPI00016E65E0 related cluster n=1 Tax=Ta... 62 3e-08
UniRef100_UPI00016E65DF UPI00016E65DF related cluster n=1 Tax=Ta... 62 3e-08
UniRef100_Q9DVW0 PxORF73 peptide n=1 Tax=Plutella xylostella gra... 62 3e-08
UniRef100_Q070J3 Virion core protein n=1 Tax=Crocodilepox virus ... 62 3e-08
UniRef100_Q0RQ47 Modular polyketide synthase n=1 Tax=Frankia aln... 62 3e-08
UniRef100_Q0BUS1 TonB family proteins n=1 Tax=Granulibacter beth... 62 3e-08
UniRef100_Q41719 Hydroxyproline-rich glycoprotein n=1 Tax=Zea di... 62 3e-08
UniRef100_C7J265 Os05g0516400 protein (Fragment) n=1 Tax=Oryza s... 62 3e-08
UniRef100_A7PAK3 Chromosome chr14 scaffold_9, whole genome shotg... 62 3e-08
UniRef100_Q8MQG9 Prion-like-(Q/n-rich)-domain-bearing protein pr... 62 3e-08
UniRef100_Q4G1Y1 Major ampullate spidroin 2 (Fragment) n=1 Tax=L... 62 3e-08
UniRef100_Q16987 Fibroin-3 (Fragment) n=1 Tax=Araneus diadematus... 62 3e-08
UniRef100_O46171 Spidroin 1 (Fragment) n=1 Tax=Nephila clavipes ... 62 3e-08
UniRef100_O02123 Prion-like-(Q/n-rich)-domain-bearing protein pr... 62 3e-08
UniRef100_B4MMR0 GK17563 n=1 Tax=Drosophila willistoni RepID=B4M... 62 3e-08
UniRef100_B0F653 Major ampullate spidroin 1 locus 1 (Fragment) n... 62 3e-08
UniRef100_B0F651 Major ampullate spidroin 1 locus 1 (Fragment) n... 62 3e-08
UniRef100_A9URA4 Predicted protein n=1 Tax=Monosiga brevicollis ... 62 3e-08
UniRef100_A6YIY0 Major ampullate spidroin 2 n=1 Tax=Latrodectus ... 62 3e-08
UniRef100_Q6C7Q8 YALI0D26191p n=1 Tax=Yarrowia lipolytica RepID=... 62 3e-08
UniRef100_Q2U304 Predicted protein n=1 Tax=Aspergillus oryzae Re... 62 3e-08
UniRef100_Q0D114 Predicted protein n=1 Tax=Aspergillus terreus N... 62 3e-08
UniRef100_B2AYQ6 Predicted CDS Pa_1_11890 n=1 Tax=Podospora anse... 62 3e-08
UniRef100_A7TP17 Putative uncharacterized protein n=1 Tax=Vander... 62 3e-08
UniRef100_A4R5L4 Putative uncharacterized protein n=1 Tax=Magnap... 62 3e-08
UniRef100_UPI0000E7F7FF PREDICTED: similar to N-WASP n=1 Tax=Gal... 61 4e-08
UniRef100_Q2IIV6 Zinc finger/thioredoxin putative n=1 Tax=Anaero... 61 4e-08
UniRef100_Q0RRK1 Putative serine/threonine protein kinase n=1 Ta... 61 4e-08
UniRef100_C4DMD4 Translation initiation factor IF-2 n=1 Tax=Stac... 61 4e-08
UniRef100_Q2QVX3 Transposon protein, putative, CACTA, En/Spm sub... 61 4e-08
UniRef100_Q10I10 Os03g0568800 protein n=1 Tax=Oryza sativa Japon... 61 4e-08
UniRef100_C7BGM8 Formin 2A n=1 Tax=Physcomitrella patens RepID=C... 61 4e-08
UniRef100_A9P9Z1 Predicted protein n=1 Tax=Populus trichocarpa R... 61 4e-08
UniRef100_A5AVB2 Putative uncharacterized protein n=1 Tax=Vitis ... 61 4e-08
UniRef100_B4KRP9 GI19648 n=1 Tax=Drosophila mojavensis RepID=B4K... 61 4e-08
UniRef100_A7M8K7 Major ampullate spidroin-like (Fragment) n=1 Ta... 61 4e-08
UniRef100_UPI000198522B PREDICTED: hypothetical protein n=1 Tax=... 61 5e-08
UniRef100_UPI0001982EE5 PREDICTED: hypothetical protein n=1 Tax=... 61 5e-08
UniRef100_UPI000194E282 PREDICTED: similar to Wiskott-Aldrich sy... 61 5e-08
UniRef100_UPI00017B435B UPI00017B435B related cluster n=1 Tax=Te... 61 5e-08
UniRef100_UPI00016E2CCB UPI00016E2CCB related cluster n=1 Tax=Ta... 61 5e-08
UniRef100_Q4S986 Chromosome 3 SCAF14700, whole genome shotgun se... 61 5e-08
UniRef100_B8IP63 Putative uncharacterized protein n=1 Tax=Methyl... 61 5e-08
UniRef100_Q8VKN7 Putative uncharacterized protein n=1 Tax=Mycoba... 61 5e-08
UniRef100_B5HES0 Translation initiation factor IF-2 n=1 Tax=Stre... 61 5e-08
UniRef100_Q9FFW5 Similarity to protein kinase n=1 Tax=Arabidopsi... 61 5e-08
UniRef100_C0PJH9 Putative uncharacterized protein n=1 Tax=Zea ma... 61 5e-08
UniRef100_B9RL99 Putative uncharacterized protein n=1 Tax=Ricinu... 61 5e-08
UniRef100_A4S1Y9 Predicted protein n=1 Tax=Ostreococcus lucimari... 61 5e-08
UniRef100_Q4G1Y2 Major ampullate spidroin 1 (Fragment) n=1 Tax=L... 61 5e-08
UniRef100_Q4FX85 Putative uncharacterized protein n=1 Tax=Leishm... 61 5e-08
UniRef100_B4PM42 GE26088 n=1 Tax=Drosophila yakuba RepID=B4PM42_... 61 5e-08
UniRef100_B4PI74 GE21449 n=1 Tax=Drosophila yakuba RepID=B4PI74_... 61 5e-08
UniRef100_B0F643 Major ampullate spidroin 1 locus 3 (Fragment) n... 61 5e-08
UniRef100_Q9P944 Kexin-like protease KEX1 n=1 Tax=Pneumocystis m... 61 5e-08
UniRef100_C5P6Q6 WH1 domain containing protein n=1 Tax=Coccidioi... 61 5e-08
UniRef100_C1GKW7 Predicted protein n=1 Tax=Paracoccidioides bras... 61 5e-08
UniRef100_B8NIL1 Proline-rich, actin-associated protein Vrp1, pu... 61 5e-08
UniRef100_B1B5Z7 Kexin-like protease KEX1 (Fragment) n=1 Tax=Pne... 61 5e-08
UniRef100_A8NWX4 Putative uncharacterized protein n=1 Tax=Coprin... 61 5e-08
UniRef100_A4QSC9 Predicted protein n=1 Tax=Magnaporthe grisea Re... 61 5e-08
UniRef100_Q6MWG9 Formin-like protein 18 n=1 Tax=Oryza sativa Jap... 61 5e-08
UniRef100_P13983 Extensin n=1 Tax=Nicotiana tabacum RepID=EXTN_T... 61 5e-08
UniRef100_P40603 Anther-specific proline-rich protein APG (Fragm... 61 5e-08
UniRef100_UPI0001900D5F hypothetical protein Mtub9_12983 n=1 Tax... 60 7e-08
UniRef100_UPI0001797E8F PREDICTED: similar to piccolo n=1 Tax=Eq... 60 7e-08
UniRef100_UPI0000E125D3 Os05g0552600 n=1 Tax=Oryza sativa Japoni... 60 7e-08
UniRef100_UPI0001951704 UPI0001951704 related cluster n=1 Tax=Ho... 60 7e-08
UniRef100_Q9Q0B7 Glycoprotein G-2 (Fragment) n=1 Tax=Human herpe... 60 7e-08
UniRef100_Q8B9G6 Putative uncharacterized protein n=1 Tax=Rachip... 60 7e-08
UniRef100_A8U5F4 Envelope glycoprotein G n=1 Tax=Human herpesvir... 60 7e-08
UniRef100_A8U526 Envelope glycoprotein G n=1 Tax=Human herpesvir... 60 7e-08
UniRef100_A1XYV9 Latency-associated nuclear antigen n=1 Tax=Retr... 60 7e-08
UniRef100_Q1B7N1 Putative uncharacterized protein n=1 Tax=Mycoba... 60 7e-08
UniRef100_A8IEX2 Putative uncharacterized protein n=1 Tax=Azorhi... 60 7e-08
UniRef100_Q8GFF2 Putative uncharacterized protein n=1 Tax=Strept... 60 7e-08
UniRef100_Q40552 Pistil extensin like protein (Fragment) n=1 Tax... 60 7e-08
UniRef100_Q01N06 OSIGBa0126J24.3 protein n=1 Tax=Oryza sativa Re... 60 7e-08
UniRef100_C7BGM9 Formin 2B n=1 Tax=Physcomitrella patens RepID=C... 60 7e-08
UniRef100_B9T5W9 ATP binding protein, putative n=1 Tax=Ricinus c... 60 7e-08
UniRef100_B9S1L1 DNA binding protein, putative n=1 Tax=Ricinus c... 60 7e-08
UniRef100_B4LXM4 GJ23422 n=1 Tax=Drosophila virilis RepID=B4LXM4... 60 7e-08
UniRef100_B0F649 Major ampullate spidroin 1 locus 2 (Fragment) n... 60 7e-08
UniRef100_A4HZX9 Putative uncharacterized protein n=1 Tax=Leishm... 60 7e-08
UniRef100_A0FK61 Major ampullate gland dragline silk protein (Fr... 60 7e-08
UniRef100_A0DA74 Chromosome undetermined scaffold_43, whole geno... 60 7e-08
UniRef100_Q55RM6 Putative uncharacterized protein n=1 Tax=Filoba... 60 7e-08
UniRef100_Q2H4B1 Putative uncharacterized protein n=1 Tax=Chaeto... 60 7e-08
UniRef100_Q0CQD0 Predicted protein n=1 Tax=Aspergillus terreus N... 60 7e-08
UniRef100_C7YGW0 Putative uncharacterized protein n=1 Tax=Nectri... 60 7e-08
UniRef100_C5DNX4 ZYRO0A12386p n=1 Tax=Zygosaccharomyces rouxii C... 60 7e-08
UniRef100_B6QCB9 Protein transport protein (SEC31), putative n=1... 60 7e-08
UniRef100_B2W1J3 Putative uncharacterized protein n=1 Tax=Pyreno... 60 7e-08
UniRef100_A1CPW0 Conserved proline-rich protein n=1 Tax=Aspergil... 60 7e-08
UniRef100_Q9UF83 Uncharacterized protein DKFZP434B061 n=1 Tax=Ho... 60 7e-08
UniRef100_Q6SPE9 Atherin n=1 Tax=Oryctolagus cuniculus RepID=SAM... 60 7e-08
UniRef100_P81780 Envelope glycoprotein G n=2 Tax=Human herpesvir... 60 7e-08
UniRef100_UPI000151B778 hypothetical protein PGUG_03728 n=1 Tax=... 60 9e-08
UniRef100_UPI00006A1DCC UPI00006A1DCC related cluster n=1 Tax=Xe... 60 9e-08
UniRef100_UPI00006A1DCA UPI00006A1DCA related cluster n=1 Tax=Xe... 60 9e-08
UniRef100_UPI0000566A27 formin 1 n=1 Tax=Mus musculus RepID=UPI0... 60 9e-08
UniRef100_UPI0000566A26 formin 1 n=1 Tax=Mus musculus RepID=UPI0... 60 9e-08
UniRef100_B7ZW99 Fmn1 protein n=1 Tax=Mus musculus RepID=B7ZW99_... 60 9e-08
UniRef100_Q2RWE5 Putative uncharacterized protein n=1 Tax=Rhodos... 60 9e-08
UniRef100_B4UHA9 General secretory system II protein E domain pr... 60 9e-08
UniRef100_B0C060 Putative uncharacterized protein n=1 Tax=Acaryo... 60 9e-08
UniRef100_A4YUJ3 Putative uncharacterized protein n=1 Tax=Bradyr... 60 9e-08
UniRef100_O68872 Putative uncharacterized protein n=1 Tax=Myxoco... 60 9e-08
UniRef100_C0WGM5 Fe-S oxidoreductase n=1 Tax=Corynebacterium acc... 60 9e-08
UniRef100_Q9SPM1 Extensin-like protein n=1 Tax=Solanum lycopersi... 60 9e-08
UniRef100_Q9CAL8 Putative uncharacterized protein F24J13.3 n=1 T... 60 9e-08
UniRef100_Q5JN60 Os01g0750600 protein n=1 Tax=Oryza sativa Japon... 60 9e-08
UniRef100_Q40385 Pistil extensin-like protein n=1 Tax=Nicotiana ... 60 9e-08
UniRef100_B9MST9 GASA-like protein n=1 Tax=Gossypium hirsutum Re... 60 9e-08
UniRef100_B9HAA2 Predicted protein n=1 Tax=Populus trichocarpa R... 60 9e-08
UniRef100_A9P3B9 Anther-specific proline rich protein n=1 Tax=Br... 60 9e-08
UniRef100_A9P3B7 Anther-specific proline rich protein n=1 Tax=Br... 60 9e-08
UniRef100_A9P3B4 Anther-specific proline rich protein n=1 Tax=Br... 60 9e-08
UniRef100_A9P3B0 Anther-specific proline rich protein n=1 Tax=Br... 60 9e-08
UniRef100_A9P3A9 Anther-specific proline rich protein n=4 Tax=Br... 60 9e-08
UniRef100_A9P3A8 Anther-specific proline rich protein n=1 Tax=Br... 60 9e-08
UniRef100_C3ZT83 Putative uncharacterized protein n=1 Tax=Branch... 60 9e-08
UniRef100_C3XQ94 Putative uncharacterized protein n=1 Tax=Branch... 60 9e-08
UniRef100_B4G4U4 GL23304 n=1 Tax=Drosophila persimilis RepID=B4G... 60 9e-08
UniRef100_A8Y2E4 Putative uncharacterized protein (Fragment) n=1... 60 9e-08
UniRef100_A7M8L5 Major ampullate spidroin 1 (Fragment) n=1 Tax=E... 60 9e-08
UniRef100_A7M8L3 Major ampullate spidroin 1 (Fragment) n=1 Tax=E... 60 9e-08
UniRef100_A7M8L1 Major ampullate spidroin 1 (Fragment) n=1 Tax=E... 60 9e-08
UniRef100_Q4P876 Putative uncharacterized protein n=1 Tax=Ustila... 60 9e-08
UniRef100_C5P8S7 Pherophorin-dz1 protein, putative n=2 Tax=Cocci... 60 9e-08
UniRef100_C5FD36 Proline-rich protein LAS17 n=1 Tax=Microsporum ... 60 9e-08
UniRef100_B8LZJ9 Protein transport protein (SEC31), putative n=1... 60 9e-08
UniRef100_A5DKC7 Putative uncharacterized protein n=1 Tax=Pichia... 60 9e-08
UniRef100_Q05860-5 Isoform 5 of Formin-1 n=1 Tax=Mus musculus Re... 60 9e-08
UniRef100_Q05860-2 Isoform 2 of Formin-1 n=1 Tax=Mus musculus Re... 60 9e-08
UniRef100_Q05860-3 Isoform 3 of Formin-1 n=1 Tax=Mus musculus Re... 60 9e-08
UniRef100_Q05860-6 Isoform 6 of Formin-1 n=1 Tax=Mus musculus Re... 60 9e-08
UniRef100_Q05860 Formin-1 n=1 Tax=Mus musculus RepID=FMN1_MOUSE 60 9e-08
UniRef100_A0PVK0 PE-PGRS family protein n=1 Tax=Mycobacterium ul... 56 1e-07
UniRef100_UPI0001BB056E translation initiation factor IF-2 n=1 T... 60 1e-07
UniRef100_UPI0001924514 PREDICTED: hypothetical protein, partial... 60 1e-07
UniRef100_UPI0000F2B8DB PREDICTED: similar to GABARAPL2 protein ... 60 1e-07
UniRef100_UPI0000E24F27 PREDICTED: hypothetical protein n=1 Tax=... 60 1e-07
UniRef100_Q9IMX9 EBNA-3B n=1 Tax=Macacine herpesvirus 4 RepID=Q9... 60 1e-07
UniRef100_Q0BT49 Hypothetical secreted protein n=1 Tax=Granuliba... 60 1e-07
UniRef100_Q3EC03 Uncharacterized protein At2g14890.2 n=1 Tax=Ara... 60 1e-07
UniRef100_Q339R1 cDNA clone:001-107-D08, full insert sequence n=... 60 1e-07
UniRef100_C7IYE4 Os02g0557000 protein (Fragment) n=1 Tax=Oryza s... 60 1e-07
UniRef100_C1FH15 Predicted protein n=1 Tax=Micromonas sp. RCC299... 60 1e-07
UniRef100_C1DZV1 Predicted protein n=1 Tax=Micromonas sp. RCC299... 60 1e-07
UniRef100_B3XYC5 Chitin-binding lectin n=1 Tax=Solanum lycopersi... 60 1e-07
UniRef100_A8J9H7 Mastigoneme-like flagellar protein n=1 Tax=Chla... 60 1e-07
UniRef100_A7R0E9 Chromosome undetermined scaffold_306, whole gen... 60 1e-07
UniRef100_A3KD20 Leucine-rich repeat/extensin n=1 Tax=Nicotiana ... 60 1e-07
UniRef100_Q95UQ2 Subtilisin-like protein n=1 Tax=Toxoplasma gond... 60 1e-07
UniRef100_B9PU53 Subtilase family protein n=1 Tax=Toxoplasma gon... 60 1e-07
UniRef100_A9UNP0 SH3, pleckstrin-like and PDZ/DHR/GLGF domain-co... 60 1e-07
UniRef100_C5E0R1 ZYRO0G14894p n=1 Tax=Zygosaccharomyces rouxii C... 60 1e-07
UniRef100_A6SQQ1 Putative uncharacterized protein n=1 Tax=Botryo... 60 1e-07
UniRef100_Q9C5S0 Classical arabinogalactan protein 9 n=1 Tax=Ara... 60 1e-07
UniRef100_UPI0001795F41 PREDICTED: similar to WASL protein n=1 T... 59 2e-07