[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV640052 HCL009d08_r
(320 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
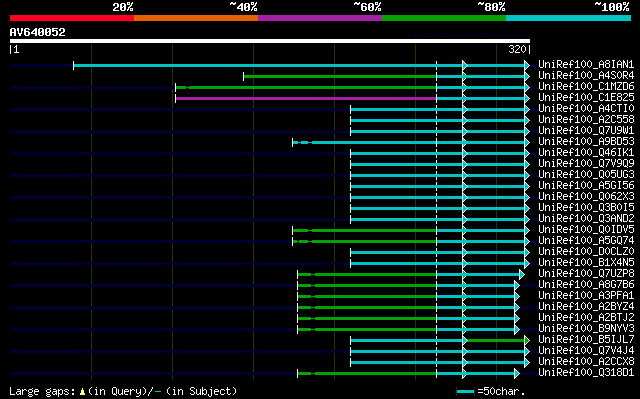
significant alignments:[graphical|details]
UniRef100_A8IAN1 Transketolase n=1 Tax=Chlamydomonas reinhardtii... 157 1e-43
UniRef100_A4S0R4 Predicted protein n=1 Tax=Ostreococcus lucimari... 58 1e-11
UniRef100_C1MZD6 Predicted protein n=1 Tax=Micromonas pusilla CC... 57 1e-11
UniRef100_C1E825 Transketolase chloroplast n=1 Tax=Micromonas sp... 55 6e-11
UniRef100_A4CTI0 Transketolase n=1 Tax=Synechococcus sp. WH 7805... 49 1e-09
UniRef100_A2C558 Transketolase n=1 Tax=Prochlorococcus marinus s... 47 4e-09
UniRef100_Q7U9W1 Transketolase n=1 Tax=Synechococcus sp. WH 8102... 47 4e-09
UniRef100_A9BD53 Transketolase n=1 Tax=Prochlorococcus marinus s... 47 4e-09
UniRef100_Q46IK1 Transketolase n=1 Tax=Prochlorococcus marinus s... 47 5e-09
UniRef100_Q7V9Q9 Transketolase n=1 Tax=Prochlorococcus marinus R... 47 5e-09
UniRef100_Q05UG3 Transketolase n=1 Tax=Synechococcus sp. RS9916 ... 47 5e-09
UniRef100_A5GI56 Transketolase n=1 Tax=Synechococcus sp. WH 7803... 47 8e-09
UniRef100_Q062X3 Transketolase n=1 Tax=Synechococcus sp. BL107 R... 45 1e-08
UniRef100_Q3B0I5 Transketolase n=1 Tax=Synechococcus sp. CC9902 ... 45 1e-08
UniRef100_Q3AND2 Transketolase n=1 Tax=Synechococcus sp. CC9605 ... 45 1e-08
UniRef100_Q0IDV5 Transketolase n=1 Tax=Synechococcus sp. CC9311 ... 45 1e-08
UniRef100_A5GQ74 Transketolase n=1 Tax=Synechococcus sp. RCC307 ... 45 1e-08
UniRef100_D0CLZ0 Transketolase n=1 Tax=Synechococcus sp. WH 8109... 45 1e-08
UniRef100_B1X4N5 Transketolase n=1 Tax=Paulinella chromatophora ... 45 1e-08
UniRef100_Q7UZP8 Transketolase n=1 Tax=Prochlorococcus marinus s... 45 2e-08
UniRef100_A8G7B6 Transketolase n=1 Tax=Prochlorococcus marinus s... 45 2e-08
UniRef100_A3PFA1 Transketolase n=1 Tax=Prochlorococcus marinus s... 45 2e-08
UniRef100_A2BYZ4 Transketolase n=1 Tax=Prochlorococcus marinus s... 45 2e-08
UniRef100_A2BTJ2 Transketolase n=1 Tax=Prochlorococcus marinus s... 45 2e-08
UniRef100_B9NYV3 Transketolase n=1 Tax=Prochlorococcus marinus s... 45 2e-08
UniRef100_B5IJL7 Transketolase n=1 Tax=Cyanobium sp. PCC 7001 Re... 48 3e-08
UniRef100_Q7V4J4 Transketolase n=1 Tax=Prochlorococcus marinus s... 44 3e-08
UniRef100_A2CCX8 Transketolase n=1 Tax=Prochlorococcus marinus s... 44 3e-08
UniRef100_Q318D1 Transketolase n=1 Tax=Prochlorococcus marinus s... 45 3e-08
UniRef100_Q5ENN6 Chloroplast transketolase n=1 Tax=Heterocapsa t... 56 7e-08