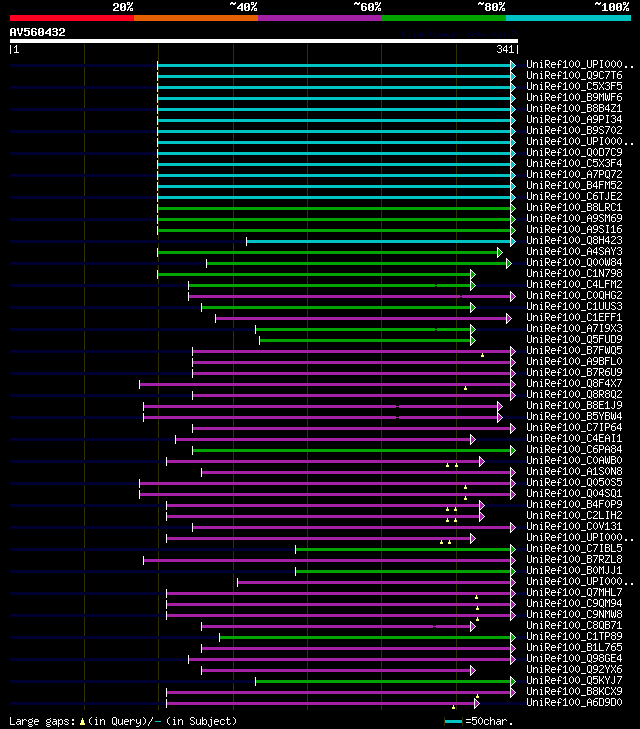
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV560432 SQ134e11F
(341 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_UPI000019718A oxidoreductase family protein n=1 Tax=Ar... 166 6e-40
UniRef100_Q9C7T6 Phosphoglycerate dehydrogenase, putative; 33424... 166 6e-40
UniRef100_C5X3F5 Putative uncharacterized protein Sb02g008670 n=... 137 3e-31
UniRef100_B9MWF6 Predicted protein n=1 Tax=Populus trichocarpa R... 135 2e-30
UniRef100_B8B4Z1 Putative uncharacterized protein n=1 Tax=Oryza ... 134 2e-30
UniRef100_A9PI34 Putative uncharacterized protein n=1 Tax=Populu... 134 2e-30
UniRef100_B9S702 Phosphoglycerate dehydrogenase, putative n=1 Ta... 133 5e-30
UniRef100_UPI0001985516 PREDICTED: hypothetical protein n=1 Tax=... 132 2e-29
UniRef100_Q0D7C9 Os07g0264100 protein n=2 Tax=Oryza sativa Japon... 132 2e-29
UniRef100_C5X3F4 Putative uncharacterized protein Sb02g008660 n=... 132 2e-29
UniRef100_A7PQ72 Chromosome chr18 scaffold_24, whole genome shot... 132 2e-29
UniRef100_B4FM52 Putative uncharacterized protein n=1 Tax=Zea ma... 126 7e-28
UniRef100_C6TJE2 Putative uncharacterized protein n=1 Tax=Glycin... 124 3e-27
UniRef100_B8LRC1 Putative uncharacterized protein n=1 Tax=Picea ... 116 9e-25
UniRef100_A9SM69 Predicted protein n=1 Tax=Physcomitrella patens... 108 1e-22
UniRef100_A9SI16 Predicted protein n=1 Tax=Physcomitrella patens... 105 1e-21
UniRef100_Q8H423 Putative phosphoglycerate dehydrogenase n=1 Tax... 102 2e-20
UniRef100_A4SAY3 Predicted protein n=1 Tax=Ostreococcus lucimari... 81 4e-14
UniRef100_Q00W84 Oxidoreductase family protein (ISS) n=1 Tax=Ost... 70 9e-11
UniRef100_C1N798 Predicted protein n=1 Tax=Micromonas pusilla CC... 65 2e-09
UniRef100_C4LFM2 D-isomer specific 2-hydroxyacid dehydrogenase N... 64 7e-09
UniRef100_C0QHG2 SerA2 n=1 Tax=Desulfobacterium autotrophicum HR... 63 1e-08
UniRef100_C1UUS3 Lactate dehydrogenase-like oxidoreductase n=1 T... 63 1e-08
UniRef100_C1EFF1 Predicted protein n=1 Tax=Micromonas sp. RCC299... 62 2e-08
UniRef100_A7I9X3 D-isomer specific 2-hydroxyacid dehydrogenase, ... 60 6e-08
UniRef100_Q5FUD9 D-3-phosphoglycerate dehydrogenase n=1 Tax=Gluc... 59 2e-07
UniRef100_B7FWQ5 Predicted protein n=1 Tax=Phaeodactylum tricorn... 58 4e-07
UniRef100_A9BFL0 D-isomer specific 2-hydroxyacid dehydrogenase N... 57 6e-07
UniRef100_B7R6U9 D-isomer specific 2-hydroxyacid dehydrogenase, ... 57 6e-07
UniRef100_Q8F4X7 D-3-phosphoglycerate dehydrogenase n=2 Tax=Lept... 57 8e-07
UniRef100_Q8R8Q2 Lactate dehydrogenase and related dehydrogenase... 56 1e-06
UniRef100_B8E1J9 D-isomer specific 2-hydroxyacid dehydrogenase N... 56 1e-06
UniRef100_B5YBW4 Glyoxylate reductase n=1 Tax=Dictyoglomus therm... 56 1e-06
UniRef100_C7IP64 D-isomer specific 2-hydroxyacid dehydrogenase N... 56 1e-06
UniRef100_C4EAI1 Phosphoglycerate dehydrogenase-like oxidoreduct... 56 1e-06
UniRef100_C6PA84 D-isomer specific 2-hydroxyacid dehydrogenase N... 56 1e-06
UniRef100_C0AWB0 Putative uncharacterized protein n=1 Tax=Proteu... 56 1e-06
UniRef100_A1S0N8 D-isomer specific 2-hydroxyacid dehydrogenase, ... 55 2e-06
UniRef100_Q050S5 Dehydrogenase n=1 Tax=Leptospira borgpetersenii... 55 2e-06
UniRef100_Q04SQ1 Dehydrogenase n=1 Tax=Leptospira borgpetersenii... 55 2e-06
UniRef100_B4F0P9 D-3-phosphoglycerate dehydrogenase n=1 Tax=Prot... 55 3e-06
UniRef100_C2LIH2 D-3-phosphoglycerate dehydrogenase n=1 Tax=Prot... 55 3e-06
UniRef100_C0V131 Lactate dehydrogenase-like oxidoreductase n=1 T... 54 4e-06
UniRef100_UPI000197BF4B hypothetical protein PROVRETT_00383 n=1 ... 54 5e-06
UniRef100_C7IBL5 D-isomer specific 2-hydroxyacid dehydrogenase N... 54 5e-06
UniRef100_B7RZL8 D-isomer specific 2-hydroxyacid dehydrogenase, ... 54 5e-06
UniRef100_B0MJJ1 Putative uncharacterized protein n=1 Tax=Anaero... 54 5e-06
UniRef100_UPI00016ACE02 D-isomer specific 2-hydroxyacid dehydrog... 54 7e-06
UniRef100_Q7MHL7 Phosphoglycerate dehydrogenase n=1 Tax=Vibrio v... 54 7e-06
UniRef100_C9QM94 D-3-phosphoglycerate dehydrogenase n=1 Tax=Vibr... 54 7e-06
UniRef100_C9NMW8 D-3-phosphoglycerate dehydrogenase n=1 Tax=Vibr... 54 7e-06
UniRef100_C8QB71 D-isomer specific 2-hydroxyacid dehydrogenase N... 54 7e-06
UniRef100_C1TP89 Lactate dehydrogenase-like oxidoreductase n=1 T... 54 7e-06
UniRef100_B1L765 Glyoxylate reductase n=1 Tax=Candidatus Korarch... 54 7e-06
UniRef100_Q98GE4 Phosphoglycerate dehydrogenase n=1 Tax=Mesorhiz... 53 9e-06
UniRef100_Q92YX6 Dehydrogenase n=1 Tax=Sinorhizobium meliloti Re... 53 9e-06
UniRef100_Q5KYJ7 Dehydrogenase n=1 Tax=Geobacillus kaustophilus ... 53 9e-06
UniRef100_B8KCX9 Chain A, D-3-Phosphoglycerate Dehydrogenase n=1... 53 9e-06
UniRef100_A6D9D0 D-3-phosphoglycerate dehydrogenase n=1 Tax=Vibr... 53 9e-06
UniRef100_A6C853 D-3-phosphoglycerate dehydrogenase n=1 Tax=Plan... 53 9e-06