[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV555198 SQ008a01F
(541 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
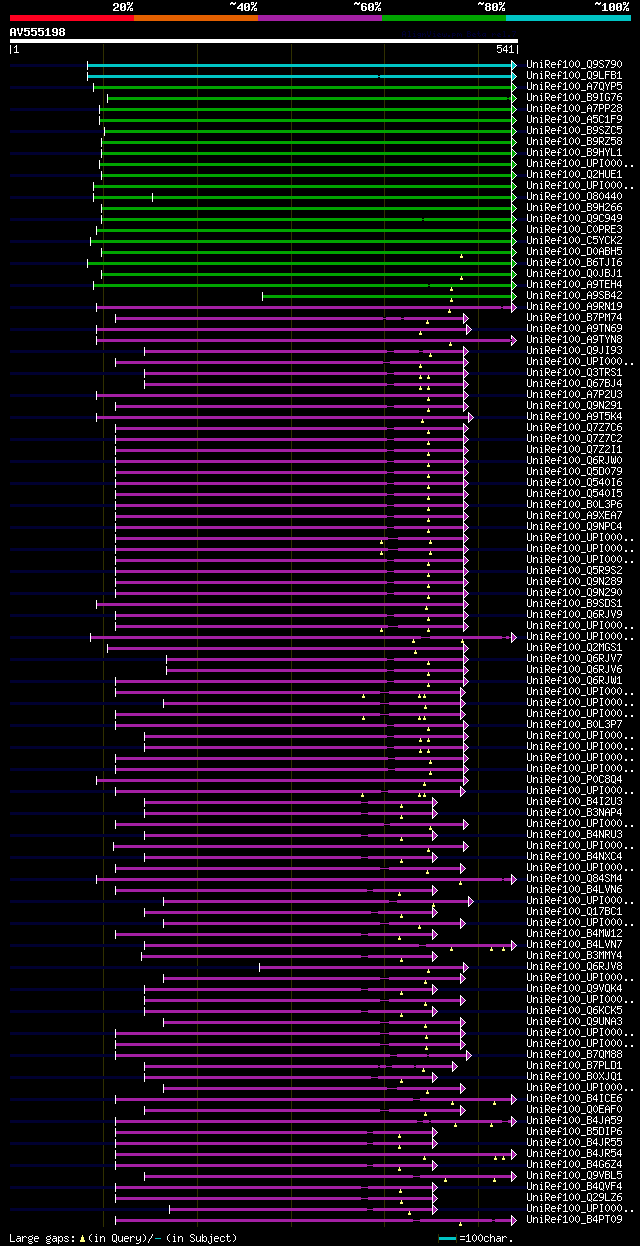
significant alignments:[graphical|details]
UniRef100_Q9S790 MZB10.5 protein n=1 Tax=Arabidopsis thaliana Re... 315 2e-84
UniRef100_Q9LFB1 Putative uncharacterized protein F7J8_230 n=1 T... 246 1e-63
UniRef100_A7QYP5 Chromosome undetermined scaffold_252, whole gen... 193 6e-48
UniRef100_B9IG76 Predicted protein (Fragment) n=1 Tax=Populus tr... 189 1e-46
UniRef100_A7PP28 Chromosome chr8 scaffold_23, whole genome shotg... 187 3e-46
UniRef100_A5C1F9 Putative uncharacterized protein n=1 Tax=Vitis ... 187 3e-46
UniRef100_B9SZC5 Lactosylceramide 4-alpha-galactosyltransferase,... 184 3e-45
UniRef100_B9RZ58 Lactosylceramide 4-alpha-galactosyltransferase,... 183 8e-45
UniRef100_B9HYL1 Predicted protein n=1 Tax=Populus trichocarpa R... 182 2e-44
UniRef100_UPI0001A7B374 galactosyltransferase n=1 Tax=Arabidopsi... 181 2e-44
UniRef100_Q2HUE1 Glycosyltransferase sugar-binding region contai... 181 3e-44
UniRef100_UPI000150588F transferase/ transferase, transferring g... 175 2e-42
UniRef100_O80440 Putative uncharacterized protein At2g38150 n=1 ... 175 2e-42
UniRef100_B9H266 Predicted protein n=1 Tax=Populus trichocarpa R... 172 1e-41
UniRef100_Q9C949 Putative uncharacterized protein T7P1.18 n=1 Ta... 167 3e-40
UniRef100_C0PRE3 Putative uncharacterized protein n=1 Tax=Picea ... 167 3e-40
UniRef100_C5YCK2 Putative uncharacterized protein Sb06g023350 n=... 157 5e-37
UniRef100_D0ABH5 OO_Ba0013J05-OO_Ba0033A15.32 protein n=1 Tax=Or... 156 8e-37
UniRef100_B6TJI6 Transferase/ transferase, transferring glycosyl... 150 6e-35
UniRef100_Q0JBJ1 Os04g0529700 protein n=4 Tax=Oryza sativa RepID... 150 7e-35
UniRef100_A9TEH4 Predicted protein n=1 Tax=Physcomitrella patens... 132 2e-29
UniRef100_A9SB42 Predicted protein (Fragment) n=1 Tax=Physcomitr... 95 4e-18
UniRef100_A9RN19 Predicted protein n=1 Tax=Physcomitrella patens... 84 6e-15
UniRef100_B7PM74 Secreted protein, putative (Fragment) n=1 Tax=I... 81 4e-14
UniRef100_A9TN69 Predicted protein (Fragment) n=1 Tax=Physcomitr... 76 1e-12
UniRef100_A9TYN8 Predicted protein n=1 Tax=Physcomitrella patens... 74 5e-12
UniRef100_Q9JI93 Lactosylceramide 4-alpha-galactosyltransferase ... 71 4e-11
UniRef100_UPI0001797B7B PREDICTED: similar to Lactosylceramide 4... 71 6e-11
UniRef100_Q3TRS1 Putative uncharacterized protein n=1 Tax=Mus mu... 70 1e-10
UniRef100_Q67BJ4 Lactosylceramide 4-alpha-galactosyltransferase ... 70 1e-10
UniRef100_A7P2U3 Chromosome chr1 scaffold_5, whole genome shotgu... 70 1e-10
UniRef100_Q9N291 Lactosylceramide 4-alpha-galactosyltransferase ... 70 1e-10
UniRef100_A9T5K4 Predicted protein (Fragment) n=1 Tax=Physcomitr... 69 2e-10
UniRef100_Q7Z7C6 Alpha-1,4-galactosyltransferase n=1 Tax=Homo sa... 69 2e-10
UniRef100_Q7Z7C2 Alpha-1,4-galactosyltransferase (Fragment) n=1 ... 69 2e-10
UniRef100_Q7Z2I1 Alpha-1,4-galactosyltransferase n=1 Tax=Homo sa... 69 2e-10
UniRef100_Q6RJW0 Alpha-1,4-galactosyltransferase n=1 Tax=Homo sa... 69 2e-10
UniRef100_Q5D079 A4GALT protein n=1 Tax=Homo sapiens RepID=Q5D07... 69 2e-10
UniRef100_Q540I6 Alpha-1,4-galactosyltransferase n=1 Tax=Homo sa... 69 2e-10
UniRef100_Q540I5 Alpha-1,4-galactosyltransferase n=1 Tax=Homo sa... 69 2e-10
UniRef100_B0L3P6 Alpha-1,4-galactosyltransferase n=1 Tax=Homo sa... 69 2e-10
UniRef100_A9XEA7 Alpha-1,4-galactosyltransferase n=1 Tax=Homo sa... 69 2e-10
UniRef100_Q9NPC4 Lactosylceramide 4-alpha-galactosyltransferase ... 69 2e-10
UniRef100_UPI000194E2E6 PREDICTED: similar to alpha 1,4-galactos... 69 3e-10
UniRef100_UPI000194E2E7 PREDICTED: similar to alpha 1,4-galactos... 68 4e-10
UniRef100_UPI00006D17A5 PREDICTED: alpha 1,4-galactosyltransfera... 68 5e-10
UniRef100_Q5R9S2 Putative uncharacterized protein DKFZp469C0812 ... 68 5e-10
UniRef100_Q9N289 Lactosylceramide 4-alpha-galactosyltransferase ... 68 5e-10
UniRef100_Q9N290 Lactosylceramide 4-alpha-galactosyltransferase ... 68 5e-10
UniRef100_B9SDS1 Lactosylceramide 4-alpha-galactosyltransferase,... 67 6e-10
UniRef100_Q6RJV9 Alpha-1,4-galactosyltransferase n=1 Tax=Homo sa... 67 6e-10
UniRef100_UPI000194E2E8 PREDICTED: similar to alpha 1,4-galactos... 67 8e-10
UniRef100_UPI0001555E5A PREDICTED: similar to alpha-1,4-N-acetyl... 67 8e-10
UniRef100_Q2MGS1 Glycosyltransferase sugar-binding region contai... 67 8e-10
UniRef100_Q6RJV7 Alpha-1,4-galactosyltransferase n=1 Tax=Homo sa... 66 1e-09
UniRef100_Q6RJV6 Alpha-1,4-galactosyltransferase n=1 Tax=Homo sa... 66 1e-09
UniRef100_Q6RJW1 Alpha-1,4-galactosyltransferase n=1 Tax=Homo sa... 66 2e-09
UniRef100_UPI0000E80B37 PREDICTED: similar to Alpha-1,4-N-acetyl... 65 2e-09
UniRef100_UPI00004F3D7F PREDICTED: similar to Alpha-1,4-N-acetyl... 65 2e-09
UniRef100_UPI0000ECB0AC Alpha-1,4-N-acetylglucosaminyltransferas... 65 2e-09
UniRef100_B0L3P7 Alpha-1,4-galactosyltransferase n=1 Tax=Homo sa... 65 3e-09
UniRef100_UPI00005A2159 PREDICTED: similar to Lactosylceramide 4... 65 4e-09
UniRef100_UPI0000EB2B1F Lactosylceramide 4-alpha-galactosyltrans... 65 4e-09
UniRef100_UPI0000E7F958 PREDICTED: similar to alpha-1,4-N-acetyl... 64 5e-09
UniRef100_UPI0000ECD2AB Lactosylceramide 4-alpha-galactosyltrans... 64 5e-09
UniRef100_P0C8Q4 Uncharacterized protein At4g19900 n=1 Tax=Arabi... 64 5e-09
UniRef100_UPI000194CC8B PREDICTED: similar to rCG25233 n=1 Tax=T... 63 1e-08
UniRef100_B4I2U3 GM18175 n=1 Tax=Drosophila sechellia RepID=B4I2... 63 1e-08
UniRef100_B3NAP4 GG24467 n=1 Tax=Drosophila erecta RepID=B3NAP4_... 62 2e-08
UniRef100_UPI00006A19A9 Lactosylceramide 4-alpha-galactosyltrans... 62 3e-08
UniRef100_B4NRU3 GD11999 n=1 Tax=Drosophila simulans RepID=B4NRU... 62 3e-08
UniRef100_UPI0001982966 PREDICTED: similar to glycosyl transfera... 62 3e-08
UniRef100_B4NXC4 GE14957 n=1 Tax=Drosophila yakuba RepID=B4NXC4_... 61 5e-08
UniRef100_UPI00015553C9 PREDICTED: similar to Alpha-1,4-N-acetyl... 61 6e-08
UniRef100_Q84SM4 Putative uncharacterized protein OJ1092_A07.115... 61 6e-08
UniRef100_B4LVN6 GJ17463 n=1 Tax=Drosophila virilis RepID=B4LVN6... 60 8e-08
UniRef100_UPI00006A0533 Alpha-1,4-N-acetylglucosaminyltransferas... 60 1e-07
UniRef100_Q17BC1 Lactosylceramide 4-alpha-galactosyltransferase ... 59 2e-07
UniRef100_UPI00006D23EE PREDICTED: alpha-1,4-N-acetylglucosaminy... 59 2e-07
UniRef100_B4MW12 GK15176 n=1 Tax=Drosophila willistoni RepID=B4M... 59 2e-07
UniRef100_B4LVN7 GJ17464 n=1 Tax=Drosophila virilis RepID=B4LVN7... 59 2e-07
UniRef100_B3MMY4 GF14753 n=1 Tax=Drosophila ananassae RepID=B3MM... 59 2e-07
UniRef100_Q6RJV8 Alpha-1,4-galactosyltransferase n=1 Tax=Homo sa... 59 2e-07
UniRef100_UPI000036B79D PREDICTED: alpha-1,4-N-acetylglucosaminy... 59 3e-07
UniRef100_Q9VQK4 Alpha4GT1 n=1 Tax=Drosophila melanogaster RepID... 59 3e-07
UniRef100_UPI00017973AB alpha 1,4-N-Acetylglucosaminyltransferas... 58 4e-07
UniRef100_Q6KCK5 Alpha1,4-N-acetylgalactosaminyltransferase n=1 ... 58 4e-07
UniRef100_Q9UNA3 Alpha-1,4-N-acetylglucosaminyltransferase n=1 T... 58 4e-07
UniRef100_UPI00005A4369 PREDICTED: similar to Alpha-1,4-N-acetyl... 58 5e-07
UniRef100_UPI0000EB139F Alpha-1,4-N-acetylglucosaminyltransferas... 58 5e-07
UniRef100_B7QM88 Lactosylceramide 4-alpha-galactosyltransferase,... 57 7e-07
UniRef100_B7PLD1 Lactosylceramide 4-alpha-galactosyltransferase,... 57 7e-07
UniRef100_B0XJQ1 Lactosylceramide 4-alpha-galactosyltransferase ... 57 8e-07
UniRef100_UPI00006A0531 Alpha-1,4-N-acetylglucosaminyltransferas... 57 1e-06
UniRef100_B4ICE6 GM10267 n=1 Tax=Drosophila sechellia RepID=B4IC... 57 1e-06
UniRef100_Q0EAF0 Alpha 1,4-N-Acetylglucosaminyltransferase (Frag... 56 1e-06
UniRef100_B4JA59 GH10340 n=1 Tax=Drosophila grimshawi RepID=B4JA... 56 2e-06
UniRef100_B5DIP6 GA26016 n=1 Tax=Drosophila pseudoobscura pseudo... 55 2e-06
UniRef100_B4JR55 GH13059 n=1 Tax=Drosophila grimshawi RepID=B4JR... 55 2e-06
UniRef100_B4JR54 GH13060 n=1 Tax=Drosophila grimshawi RepID=B4JR... 55 2e-06
UniRef100_B4G6Z4 GL19576 n=1 Tax=Drosophila persimilis RepID=B4G... 55 2e-06
UniRef100_Q9VBL5 Alpha4GT2 n=1 Tax=Drosophila melanogaster RepID... 55 3e-06
UniRef100_B4QVF4 GD21237 n=1 Tax=Drosophila simulans RepID=B4QVF... 55 3e-06
UniRef100_Q29LZ6 GA14400 n=2 Tax=pseudoobscura subgroup RepID=Q2... 55 3e-06
UniRef100_UPI0000D56C5B PREDICTED: similar to GA14400-PA n=1 Tax... 54 6e-06
UniRef100_B4PT09 GE23622 n=1 Tax=Drosophila yakuba RepID=B4PT09_... 54 7e-06
UniRef100_Q4KLB8 A4gnt protein n=1 Tax=Xenopus laevis RepID=Q4KL... 54 9e-06