[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV442141 APZ11f05_r
(568 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
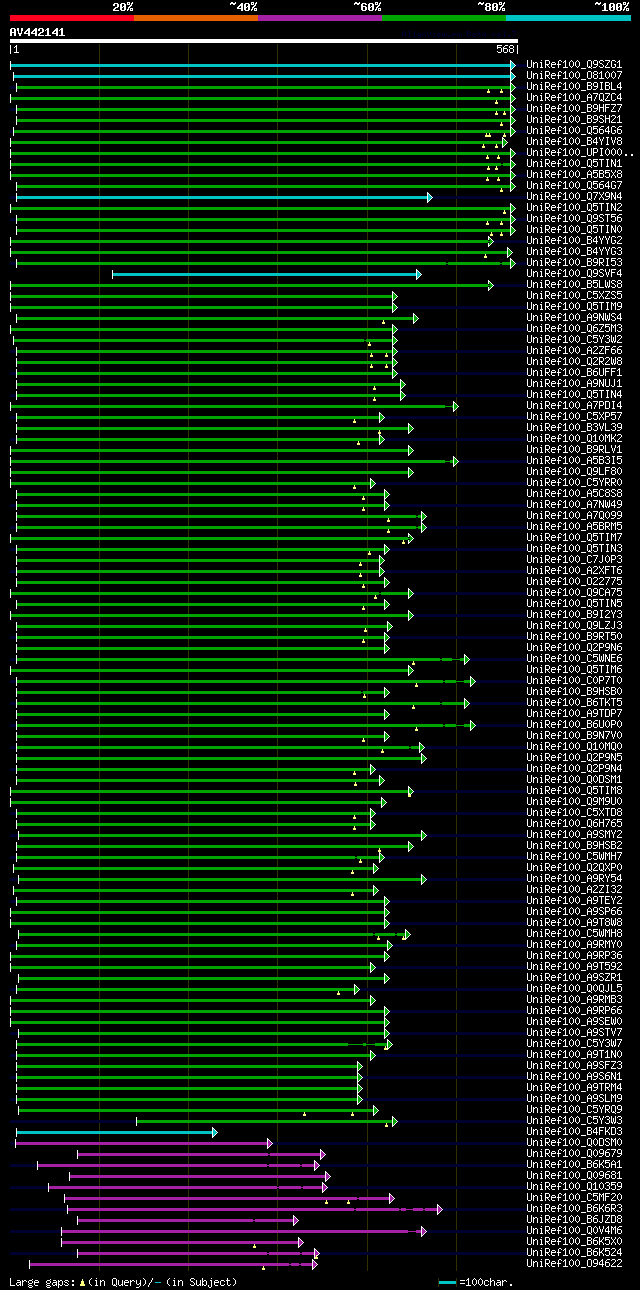
significant alignments:[graphical|details]
UniRef100_Q9SZG1 Glycosyltransferase 6 n=2 Tax=Arabidopsis thali... 413 e-114
UniRef100_O81007 Putative glycosyltransferase 7 n=1 Tax=Arabidop... 307 3e-82
UniRef100_B9IBL4 Predicted protein n=1 Tax=Populus trichocarpa R... 270 4e-71
UniRef100_A7QZC4 Chromosome chr7 scaffold_270, whole genome shot... 265 1e-69
UniRef100_B9HFZ7 Predicted protein n=1 Tax=Populus trichocarpa R... 263 5e-69
UniRef100_B9SH21 Transferase, putative n=1 Tax=Ricinus communis ... 263 9e-69
UniRef100_Q564G6 Galactomannan galactosyltransferase n=1 Tax=Sen... 261 3e-68
UniRef100_B4YIV8 Putative galactomannan galactosyl transferase n... 260 6e-68
UniRef100_UPI0001983A6E PREDICTED: hypothetical protein n=1 Tax=... 256 1e-66
UniRef100_Q5TIN1 Alpha-6-galactosyltransferase n=1 Tax=Nicotiana... 254 2e-66
UniRef100_A5B5X8 Putative uncharacterized protein n=1 Tax=Vitis ... 254 2e-66
UniRef100_Q564G7 Galactomannan galactosyltransferase 1 n=1 Tax=C... 248 3e-64
UniRef100_Q7X9N4 Galactomannan galactosyltransferase n=1 Tax=Lot... 245 2e-63
UniRef100_Q5TIN2 Alpha-6-galactosyltransferase n=1 Tax=Solanum t... 243 7e-63
UniRef100_Q9ST56 Alpha galactosyltransferase (Fragment) n=1 Tax=... 240 5e-62
UniRef100_Q5TIN0 Alpha-6-galactosyltransferase n=1 Tax=Medicago ... 239 8e-62
UniRef100_B4YYG2 Putative galactosyl transferase (Fragment) n=1 ... 239 1e-61
UniRef100_B4YYG3 Putative galactomannan galactosyl transferase n... 237 4e-61
UniRef100_B9RI53 Transferase, putative n=1 Tax=Ricinus communis ... 237 5e-61
UniRef100_Q9SVF4 Putative uncharacterized protein AT4g38310 n=1 ... 233 8e-60
UniRef100_B5LWS8 Galactosyl transferase n=1 Tax=Coffea canephora... 233 8e-60
UniRef100_C5XZS5 Putative uncharacterized protein Sb04g029500 n=... 216 1e-54
UniRef100_Q5TIM9 Putative uncharacterized protein n=1 Tax=Zea ma... 214 3e-54
UniRef100_A9NWS4 Putative uncharacterized protein n=1 Tax=Picea ... 210 5e-53
UniRef100_Q6Z5M3 Os02g0723200 protein n=3 Tax=Oryza sativa RepID... 206 1e-51
UniRef100_C5Y3W2 Putative uncharacterized protein Sb05g020910 n=... 189 2e-46
UniRef100_A2ZF66 Putative uncharacterized protein n=1 Tax=Oryza ... 184 4e-45
UniRef100_Q2R2W8 Glycosyltransferase 6, putative, expressed n=1 ... 182 1e-44
UniRef100_B6UFF1 Glycosyltransferase 6 n=1 Tax=Zea mays RepID=B6... 181 3e-44
UniRef100_A9NUJ1 Putative uncharacterized protein n=1 Tax=Picea ... 180 6e-44
UniRef100_Q5TIN4 Alpha-1,6-xylosyltransferase n=1 Tax=Pinus taed... 179 2e-43
UniRef100_A7PDI4 Chromosome chr17 scaffold_12, whole genome shot... 175 2e-42
UniRef100_C5XP57 Putative uncharacterized protein Sb03g005110 n=... 174 3e-42
UniRef100_B3VL39 Putative xylosyl transferase n=1 Tax=Coffea can... 174 4e-42
UniRef100_Q10MK2 Os03g0305800 protein n=2 Tax=Oryza sativa RepID... 174 4e-42
UniRef100_B9RLV1 Xyloglucan 6-xylosyltransferase, putative n=1 T... 174 5e-42
UniRef100_A5B3I5 Putative uncharacterized protein n=1 Tax=Vitis ... 173 7e-42
UniRef100_Q9LF80 Putative glycosyltransferase 3 n=2 Tax=Arabidop... 173 9e-42
UniRef100_C5YRR0 Putative uncharacterized protein Sb08g003090 n=... 172 1e-41
UniRef100_A5C8S8 Putative uncharacterized protein n=1 Tax=Vitis ... 172 2e-41
UniRef100_A7NW49 Chromosome chr5 scaffold_2, whole genome shotgu... 171 3e-41
UniRef100_A7Q099 Chromosome chr7 scaffold_42, whole genome shotg... 171 5e-41
UniRef100_A5BRM5 Putative uncharacterized protein n=1 Tax=Vitis ... 171 5e-41
UniRef100_Q5TIM7 Putative glycosyltransferase n=1 Tax=Solanum tu... 170 6e-41
UniRef100_Q5TIN3 Alpha-1,6-xylosyltransferase (Fragment) n=1 Tax... 169 1e-40
UniRef100_C7J0P3 Os03g0306100 protein n=2 Tax=Oryza sativa Japon... 169 1e-40
UniRef100_A2XFT6 Putative uncharacterized protein n=1 Tax=Oryza ... 169 1e-40
UniRef100_O22775 Putative glycosyltransferase 2 n=1 Tax=Arabidop... 169 1e-40
UniRef100_Q9CA75 Putative glycosyltransferase 5 n=1 Tax=Arabidop... 169 1e-40
UniRef100_Q5TIN5 Alpha-1,6-xylosyltransferase n=1 Tax=Vitis vini... 169 2e-40
UniRef100_B9I2Y3 Predicted protein n=1 Tax=Populus trichocarpa R... 168 2e-40
UniRef100_Q9LZJ3 Xyloglucan 6-xylosyltransferase n=2 Tax=Arabido... 168 3e-40
UniRef100_B9RT50 Xyloglucan 6-xylosyltransferase, putative n=1 T... 167 4e-40
UniRef100_Q2P9N6 Alpha-1,6-xylosyltransferase n=1 Tax=Physcomitr... 167 5e-40
UniRef100_C5WNE6 Putative uncharacterized protein Sb01g038000 n=... 167 5e-40
UniRef100_Q5TIM6 Putative glycosyltransferase n=1 Tax=Nicotiana ... 167 7e-40
UniRef100_C0P7T0 Putative uncharacterized protein n=1 Tax=Zea ma... 167 7e-40
UniRef100_B9HSB0 Predicted protein n=1 Tax=Populus trichocarpa R... 167 7e-40
UniRef100_B6TKT5 Glycosyltransferase 5 n=1 Tax=Zea mays RepID=B6... 167 7e-40
UniRef100_A9TDP7 Predicted protein n=1 Tax=Physcomitrella patens... 167 7e-40
UniRef100_B6U0P0 Glycosyltransferase 5 n=1 Tax=Zea mays RepID=B6... 166 9e-40
UniRef100_B9N7V0 Predicted protein n=1 Tax=Populus trichocarpa R... 165 2e-39
UniRef100_Q10MQ0 Putative uncharacterized protein n=2 Tax=Oryza ... 164 4e-39
UniRef100_Q2P9N5 Alpha-1,6-xylosyltransferase n=1 Tax=Physcomitr... 163 7e-39
UniRef100_Q2P9N4 Putative glycosyltransferase n=1 Tax=Zea mays R... 163 7e-39
UniRef100_Q0DSM1 Os03g0300000 protein (Fragment) n=1 Tax=Oryza s... 163 7e-39
UniRef100_Q5TIM8 Putative glycosyltransferase n=1 Tax=Lotus japo... 162 2e-38
UniRef100_Q9M9U0 Putative glycosyltransferase 4 n=1 Tax=Arabidop... 162 2e-38
UniRef100_C5XTD8 Putative uncharacterized protein Sb04g021570 n=... 162 2e-38
UniRef100_Q6H765 Os02g0529600 protein n=2 Tax=Oryza sativa RepID... 161 3e-38
UniRef100_A9SMY2 Predicted protein n=1 Tax=Physcomitrella patens... 161 4e-38
UniRef100_B9HSB2 Predicted protein n=1 Tax=Populus trichocarpa R... 160 5e-38
UniRef100_C5WMH7 Putative uncharacterized protein Sb01g037530 n=... 160 8e-38
UniRef100_Q2QXP0 Os12g0149300 protein n=1 Tax=Oryza sativa Japon... 159 1e-37
UniRef100_A9RY54 Predicted protein n=1 Tax=Physcomitrella patens... 159 1e-37
UniRef100_A2ZI32 Putative uncharacterized protein n=1 Tax=Oryza ... 159 1e-37
UniRef100_A9TEY2 Predicted protein n=1 Tax=Physcomitrella patens... 157 7e-37
UniRef100_A9SP66 Predicted protein n=1 Tax=Physcomitrella patens... 157 7e-37
UniRef100_A9T8W8 Predicted protein n=1 Tax=Physcomitrella patens... 155 3e-36
UniRef100_C5WMH8 Putative uncharacterized protein Sb01g037540 n=... 152 2e-35
UniRef100_A9RMY0 Predicted protein n=1 Tax=Physcomitrella patens... 152 2e-35
UniRef100_A9RP36 Predicted protein n=1 Tax=Physcomitrella patens... 152 2e-35
UniRef100_A9T592 Predicted protein n=1 Tax=Physcomitrella patens... 150 5e-35
UniRef100_A9SZR1 Predicted protein n=1 Tax=Physcomitrella patens... 150 5e-35
UniRef100_Q0QJL5 Putative galactosyl transferase n=1 Tax=Ceratop... 150 8e-35
UniRef100_A9RMB3 Predicted protein n=1 Tax=Physcomitrella patens... 150 8e-35
UniRef100_A9RP66 Predicted protein n=1 Tax=Physcomitrella patens... 149 2e-34
UniRef100_A9SEW0 Predicted protein n=1 Tax=Physcomitrella patens... 147 5e-34
UniRef100_A9STV7 Predicted protein n=1 Tax=Physcomitrella patens... 145 2e-33
UniRef100_C5Y3W7 Putative uncharacterized protein Sb05g020930 n=... 145 3e-33
UniRef100_A9T1N0 Predicted protein n=1 Tax=Physcomitrella patens... 145 3e-33
UniRef100_A9SFZ3 Predicted protein (Fragment) n=1 Tax=Physcomitr... 132 2e-29
UniRef100_A9S6N1 Predicted protein n=1 Tax=Physcomitrella patens... 130 5e-29
UniRef100_A9TRM4 Predicted protein (Fragment) n=1 Tax=Physcomitr... 127 5e-28
UniRef100_A9SLM9 Predicted protein n=1 Tax=Physcomitrella patens... 127 5e-28
UniRef100_C5YRQ9 Putative uncharacterized protein Sb08g003080 n=... 127 6e-28
UniRef100_C5Y3W3 Putative uncharacterized protein Sb05g020911 (F... 114 7e-24
UniRef100_B4FKD3 Putative uncharacterized protein n=1 Tax=Zea ma... 106 1e-21
UniRef100_Q0DSM0 Os03g0300100 protein n=1 Tax=Oryza sativa Japon... 65 3e-09
UniRef100_Q09679 Probable alpha-1,2-galactosyltransferase gmh1 n... 61 5e-08
UniRef100_B6K5A1 Alpha-1,2-galactosyltransferase gmh3 n=1 Tax=Sc... 60 9e-08
UniRef100_Q09681 Probable alpha-1,2-galactosyltransferase gmh2 n... 60 9e-08
UniRef100_Q10359 Alpha-1,2-galactosyltransferase gmh3 n=1 Tax=Sc... 60 2e-07
UniRef100_C5MF20 Putative uncharacterized protein n=1 Tax=Candid... 59 3e-07
UniRef100_B6K6R3 Alpha-1,2-galactosyltransferase gmh3 n=1 Tax=Sc... 58 4e-07
UniRef100_B6JZD8 Alpha-1,2-galactosyltransferase gmh3 n=1 Tax=Sc... 58 4e-07
UniRef100_Q0V4M6 Putative uncharacterized protein n=1 Tax=Phaeos... 56 2e-06
UniRef100_B6K5X0 Alpha-1,2-galactosyltransferase n=1 Tax=Schizos... 56 2e-06
UniRef100_B6K524 Alpha-1,2-galactosyltransferase gmh3 n=1 Tax=Sc... 56 2e-06
UniRef100_O94622 Uncharacterized alpha-1,2-galactosyltransferase... 56 2e-06
UniRef100_Q6C1C8 YALI0F17402p n=1 Tax=Yarrowia lipolytica RepID=... 54 8e-06