
BLAST2 result
BLASTP 2.1.2 [Oct-19-2000]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BAB33192.1 34 aa
(34 letters)
Database: nr
640,428 sequences; 201,912,487 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
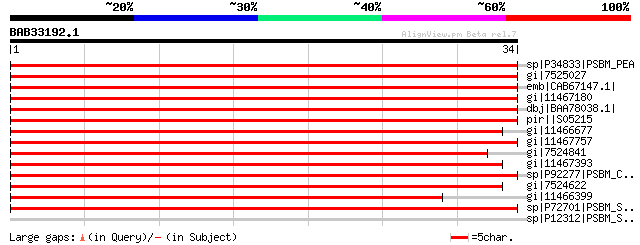
sp|P34833|PSBM_PEA PHOTOSYSTEM II REACTION CENTER M PROTEIN >gi|... 66 4e-11
gi|7525027 PSII low MW protein [Arabidopsis thaliana] >gi|114659... 65 1e-10
emb|CAB67147.1| (AJ271079) PSII M-protein [Oenothera elata subsp... 65 1e-10
gi|11467180 PSII low MW protein [Zea mays] >gi|1346867|sp|P48189... 64 3e-10
dbj|BAA78038.1| (AB027572) PSII M-protein [Triticum aestivum] 62 8e-10
pir||S05215 photosystem II protein psbM - Synechococcus sp. (str... 59 7e-09
gi|11466677 ORF34 [Marchantia polymorpha] >gi|131371|sp|P12168|P... 59 1e-08
gi|11467757 M protein of photosystem II [Nephroselmis olivacea] ... 54 2e-07
gi|7524841 photosystem II PsbM protein [Chlorella vulgaris] >gi|... 53 3e-07
gi|11467393 PsbM subunit of the photosystem II reaction center c... 50 3e-06
sp|P92277|PSBM_CHLRE PHOTOSYSTEM II REACTION CENTER M PROTEIN >g... 50 4e-06
gi|7524622 PSII M protein [Pinus thunbergii] >gi|1172689|sp|P416... 49 8e-06
gi|11466399 M protein of photosystem II [Mesostigma viride] >gi|... 46 4e-05
sp|P72701|PSBM_SYNY3 PHOTOSYSTEM II REACTION CENTER M PROTEIN >g... 45 9e-05
sp|P12312|PSBM_SYNVU PHOTOSYSTEM II REACTION CENTER M PROTEIN 33 0.53
>sp|P34833|PSBM_PEA PHOTOSYSTEM II REACTION CENTER M PROTEIN
dbj|BAA02098.1| (D12535) photosystem II polypeptide M [Pisum sativum]
Length = 34
Score = 66.3 bits (159), Expect = 4e-11
Identities = 34/34 (100%), Positives = 34/34 (100%)
Query: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQSD 34
MEVNILAFIATALFILVPTAFLLIIYVKTVSQSD
Sbjct: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQSD 34
>gi|7525027 PSII low MW protein [Arabidopsis thaliana]
ref|NP_054490.1| PSII M-protein [Nicotiana tabacum]
ref|NP_039371.1| PSII low MW protein [Oryza sativa]
ref|NP_054926.1| PSII M-protein [Spinacia oleracea]
sp|P12169|PSBM_ORYSA PHOTOSYSTEM II REACTION CENTER M PROTEIN
pir||F2RZM photosystem II protein psbM - rice chloroplast
emb|CAA33984.1| (X15901) PSII low MW protein [Oryza sativa]
emb|CAA77413.1| (Z00044) PSII M-protein [Nicotiana tabacum]
dbj|BAA84379.1| (AP000423) PSII low MW protein [Arabidopsis thaliana]
emb|CAB88719.1| (AJ400848) PSII M-protein [Spinacia oleracea]
prf||1603356M photosystem II low MW protein [Oryza sativa]
Length = 34
Score = 65.2 bits (156), Expect = 1e-10
Identities = 33/34 (97%), Positives = 34/34 (99%)
Query: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQSD 34
MEVNILAFIATALFILVPTAFLLIIYVKTVSQ+D
Sbjct: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQND 34
>emb|CAB67147.1| (AJ271079) PSII M-protein [Oenothera elata subsp. hookeri]
Length = 34
Score = 64.8 bits (155), Expect = 1e-10
Identities = 33/34 (97%), Positives = 34/34 (99%)
Query: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQSD 34
MEVNILAFIATALFILVPTAFLLIIYVKTVSQS+
Sbjct: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQSE 34
>gi|11467180 PSII low MW protein [Zea mays]
sp|P48189|PSBM_MAIZE PHOTOSYSTEM II REACTION CENTER M PROTEIN
pir||S58540 photosystem II protein psbM - maize chloroplast
emb|CAA60274.1| (X86563) PSII low MW protein [Zea mays]
Length = 34
Score = 63.6 bits (152), Expect = 3e-10
Identities = 32/34 (94%), Positives = 33/34 (96%)
Query: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQSD 34
MEVNILAFIATALFILVPTAFLLIIYVKT SQ+D
Sbjct: 1 MEVNILAFIATALFILVPTAFLLIIYVKTASQND 34
>dbj|BAA78038.1| (AB027572) PSII M-protein [Triticum aestivum]
Length = 34
Score = 62.1 bits (148), Expect = 8e-10
Identities = 31/34 (91%), Positives = 34/34 (99%)
Query: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQSD 34
MEVNILAFIATALFILVPT+FLLIIYVKTVSQ++
Sbjct: 1 MEVNILAFIATALFILVPTSFLLIIYVKTVSQNN 34
>pir||S05215 photosystem II protein psbM - Synechococcus sp. (strain Copeland)
Length = 34
Score = 58.9 bits (140), Expect = 7e-09
Identities = 29/34 (85%), Positives = 32/34 (93%)
Query: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQSD 34
MEVN L FIATALF+LVP+AFLLIIYVKTVSQ+D
Sbjct: 1 MEVNQLGFIATALFVLVPSAFLLIIYVKTVSQND 34
>gi|11466677 ORF34 [Marchantia polymorpha]
sp|P12168|PSBM_MARPO PHOTOSYSTEM II REACTION CENTER M PROTEIN
pir||A05007 photosystem II protein psbM - liverwort (Marchantia polymorpha)
chloroplast
emb|CAA28059.1| (X04465) ORF34 [Marchantia polymorpha]
Length = 34
Score = 58.6 bits (139), Expect = 1e-08
Identities = 28/33 (84%), Positives = 32/33 (96%)
Query: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQS 33
MEVNILAFIATALFIL+PTAFLLI+YV+T SQ+
Sbjct: 1 MEVNILAFIATALFILIPTAFLLILYVQTASQN 33
>gi|11467757 M protein of photosystem II [Nephroselmis olivacea]
gb|AAD54779.1|AF137379_2 (AF137379) M protein of photosystem II [Nephroselmis olivacea]
Length = 34
Score = 54.3 bits (128), Expect = 2e-07
Identities = 25/34 (73%), Positives = 30/34 (87%)
Query: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQSD 34
MEVNIL IATALFI++PT+FLLI+YVKT SQ +
Sbjct: 1 MEVNILGLIATALFIIIPTSFLLILYVKTASQQN 34
>gi|7524841 photosystem II PsbM protein [Chlorella vulgaris]
sp|P56325|PSBM_CHLVU PHOTOSYSTEM II REACTION CENTER M PROTEIN
pir||T07271 photosystem II protein psbM - Chlorella vulgaris chloroplast
dbj|BAA57919.1| (AB001684) photosystem II PsbM protein [Chlorella vulgaris]
Length = 36
Score = 53.5 bits (126), Expect = 3e-07
Identities = 25/32 (78%), Positives = 29/32 (90%)
Query: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQ 32
MEVNIL IATALFI++PT+FLLI+YVKT SQ
Sbjct: 1 MEVNILGVIATALFIIIPTSFLLILYVKTASQ 32
>gi|11467393 PsbM subunit of the photosystem II reaction center complex
[Cyanophora paradoxa]
sp|P48107|PSBM_CYAPA PHOTOSYSTEM II REACTION CENTER M PROTEIN
pir||T06938 photosystem II protein psbM - Cyanophora paradoxa cyanelle
gb|AAA81281.1| (U30821) PsbM subunit of the photosystem II reaction center
complex [Cyanophora paradoxa]
Length = 38
Score = 50.4 bits (118), Expect = 3e-06
Identities = 22/33 (66%), Positives = 30/33 (90%)
Query: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQS 33
MEVNIL IATALFI++PT+FLLI+Y++TV+ +
Sbjct: 1 MEVNILGLIATALFIVIPTSFLLILYIQTVASA 33
>sp|P92277|PSBM_CHLRE PHOTOSYSTEM II REACTION CENTER M PROTEIN
pir||T08029 photosystem II protein psbM - Chlamydomonas reinhardtii
chloroplast
gb|AAB39941.1| (U81552) photosystem II subunit [Chlamydomonas reinhardtii]
Length = 34
Score = 50.0 bits (117), Expect = 4e-06
Identities = 23/34 (67%), Positives = 27/34 (78%)
Query: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQSD 34
MEVNI ATALFI++PT+FLLI+YVKT S D
Sbjct: 1 MEVNIYGLTATALFIIIPTSFLLILYVKTASTQD 34
>gi|7524622 PSII M protein [Pinus thunbergii]
sp|P41608|PSBM_PINTH PHOTOSYSTEM II REACTION CENTER M PROTEIN
pir||T07455 photosystem II protein psbM - Japanese black pine chloroplast
dbj|BAA04333.1| (D17510) PSII M protein [Pinus thunbergii]
Length = 37
Score = 48.8 bits (114), Expect = 8e-06
Identities = 25/33 (75%), Positives = 26/33 (78%)
Query: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQS 33
MEVN LAFIA LF+ VPTAFLLI YVKT S S
Sbjct: 1 MEVNTLAFIAVLLFLAVPTAFLLIPYVKTASAS 33
>gi|11466399 M protein of photosystem II [Mesostigma viride]
gb|AAF43843.1|AF166114_55 (AF166114) M protein of photosystem II [Mesostigma viride]
Length = 31
Score = 46.5 bits (108), Expect = 4e-05
Identities = 20/29 (68%), Positives = 26/29 (88%)
Query: 1 MEVNILAFIATALFILVPTAFLLIIYVKT 29
ME NILA +ATALFI++PTAFL+I+Y +T
Sbjct: 1 METNILALMATALFIIIPTAFLIILYAQT 29
>sp|P72701|PSBM_SYNY3 PHOTOSYSTEM II REACTION CENTER M PROTEIN
pir||S74556 photosystem II psbM protein - Synechocystis sp. (strain PCC 6803)
dbj|BAA16708.1| (D90900) photosystem II PsbM protein [Synechocystis sp.]
Length = 35
Score = 45.3 bits (105), Expect = 9e-05
Identities = 20/34 (58%), Positives = 28/34 (81%)
Query: 1 MEVNILAFIATALFILVPTAFLLIIYVKTVSQSD 34
M+VN L FIA+ LF+LVPT FLLI++++T QS+
Sbjct: 1 MQVNNLGFIASILFVLVPTVFLLILFIQTGKQSE 34
>sp|P12312|PSBM_SYNVU PHOTOSYSTEM II REACTION CENTER M PROTEIN
Length = 19
Score = 32.8 bits (73), Expect = 0.53
Identities = 15/19 (78%), Positives = 17/19 (88%)
Query: 1 MEVNILAFIATALFILVPT 19
MEVN L FIATALF+LVP+
Sbjct: 1 MEVNQLGFIATALFVLVPS 19
Database: nr
Posted date: Mar 14, 2001 1:25 PM
Number of letters in database: 201,912,487
Number of sequences in database: 640,428
Lambda K H
0.334 0.144 0.379
Gapped
Lambda K H
0.270 0.0470 0.230
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8410219
Number of Sequences: 640428
Number of extensions: 117154
Number of successful extensions: 829
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 814
Number of HSP's gapped (non-prelim): 15
length of query: 34
length of database: 201,912,487
effective HSP length: 13
effective length of query: 21
effective length of database: 193,586,923
effective search space: 4065325383
effective search space used: 4065325383
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.8 bits)
X3: 64 (24.9 bits)
S1: 39 (21.6 bits)
S2: 63 (29.0 bits)
Lotus japonicus(Cp)