
BLAST2 result
BLASTP 2.1.2 [Oct-19-2000]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BAB33186.1 750 aa
(750 letters)
Database: nr
640,428 sequences; 201,912,487 total letters
Searching..................................................done
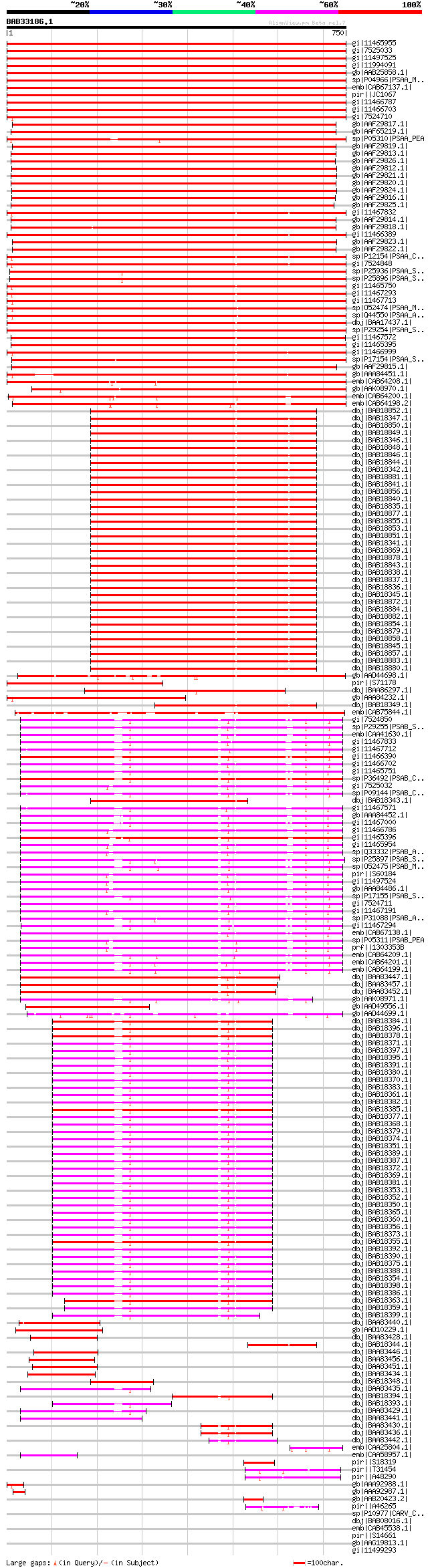
Score E
Sequences producing significant alignments: (bits) Value
gi|11465955 PSI P700 apoprotein A1 [Nicotiana tabacum] >gi|13114... 1552 0.0
gi|7525033 PSI P700 apoprotein A1 [Arabidopsis thaliana] >gi|668... 1551 0.0
gi|11497525 PSI P700 apoprotein A1 [Spinacia oleracea] >gi|13113... 1541 0.0
gi|11994091 PSI P700 apoprotein A1 [Zea mays] >gi|11990233|emb|C... 1532 0.0
gb|AAB25858.1| photosystem I reaction center protein psaA produc... 1532 0.0
sp|P04966|PSAA_MAIZE PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 1527 0.0
emb|CAB67137.1| (AJ271079) PSI P700 apoprotein A1 [Oenothera ela... 1525 0.0
pir||JC1067 psaA protein - Sorghum chloroplast 1521 0.0
gi|11466787 PSI P700 apoprotein A1 [Oryza sativa] >gi|131136|sp|... 1519 0.0
gi|11466703 psaA [Marchantia polymorpha] >gi|131135|sp|P06406|PS... 1494 0.0
gi|7524710 PSI P700 apoprotein A1 [Pinus thunbergii] >gi|1172656... 1492 0.0
gb|AAF29817.1| (AF180016) photosystem I P700 apoprotein A1 [Drim... 1484 0.0
gb|AAF65219.1| (AF223227) photosystem protein [Pisum sativum] 1480 0.0
sp|P05310|PSAA_PEA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A... 1480 0.0
gb|AAF29819.1| (AF180018) photosystem I P700 apoprotein A1 [Arau... 1439 0.0
gb|AAF29813.1| (AF180012) photosystem I P700 apoprotein A1 [Sequ... 1439 0.0
gb|AAF29826.1| (AF180025) photosystem I P700 apoprotein A1 [Torr... 1436 0.0
gb|AAF29812.1| (AF180011) photosystem I P700 apoprotein A1 [Ence... 1433 0.0
gb|AAF29821.1| (AF180020) photosystem I P700 apoprotein A1 [Angi... 1432 0.0
gb|AAF29820.1| (AF180019) photosystem I P700 apoprotein A1 [Equi... 1421 0.0
gb|AAF29824.1| (AF180023) photosystem I P700 apoprotein A1 [Psil... 1417 0.0
gb|AAF29816.1| (AF180015) photosystem I P700 apoprotein A1 [Cyca... 1414 0.0
gb|AAF29825.1| (AF180024) photosystem I P700 apoprotein A1 [Hupe... 1412 0.0
gi|11467832 P700 apoprotein A1 of photosystem I [Nephroselmis ol... 1411 0.0
gb|AAF29814.1| (AF180013) photosystem I P700 apoprotein A1 [Welw... 1406 0.0
gb|AAF29818.1| (AF180017) photosystem I P700 apoprotein A1 [Ephe... 1403 0.0
gi|11466389 P700 apoprotein A1 of photosystem I [Mesostigma viri... 1398 0.0
gb|AAF29823.1| (AF180022) photosystem I P700 apoprotein A1 [Adia... 1398 0.0
gb|AAF29822.1| (AF180021) photosystem I P700 apoprotein A1 [Aspl... 1393 0.0
sp|P12154|PSAA_CHLRE PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 1390 0.0
gi|7524848 P700 apoprotein subunit Ia [Chlorella vulgaris] >gi|3... 1388 0.0
sp|P25936|PSAA_SYNVU PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 1354 0.0
sp|P25896|PSAA_SYNEN PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 1352 0.0
gi|11465750 Photosystem I p700 chlorophyll A apoprotein A1 [Porp... 1352 0.0
gi|11467293 PsaA subunit of photosystem I reaction center [Cyano... 1349 0.0
gi|11467713 PSI P700 apoprotein A1 [Guillardia theta] >gi|609379... 1335 0.0
sp|O52474|PSAA_MASLA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 1322 0.0
sp|Q44550|PSAA_ANAVA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 1322 0.0
dbj|BAA17437.1| (D90906) P700 apoprotein subunit Ia [Synechocyst... 1321 0.0
sp|P29254|PSAA_SYNY3 PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 1321 0.0
gi|11467572 PSI, P700 apoprotein A1 [Odontella sinensis] >gi|134... 1307 0.0
gi|11465395 unknown; Photosystem I p700 chlorophyll A apoprotein... 1306 0.0
gi|11466999 PSI P700 apoprotein, subunit 1a [Euglena gracilis] >... 1303 0.0
sp|P17154|PSAA_SYNP2 PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 1291 0.0
gb|AAF29815.1| (AF180014) photosystem I P700 apoprotein A1 [Mars... 1291 0.0
gb|AAA84451.1| (M37526) alpha-apoprotein [Euglena gracilis] 1209 0.0
emb|CAB64208.1| (AJ133190) photosystem I subunit PsaA [Synechoco... 1185 0.0
gb|AAK08970.1| (AY026898) photosystem I P700 protein subunit A [... 1140 0.0
emb|CAB64200.1| (AJ133191) photosystem I subunit PsaA [Prochloro... 1080 0.0
emb|CAB64198.2| (AJ133192) photosystem I subunit PsaA [Prochloro... 1044 0.0
dbj|BAB18852.1| (AB044231) P700 chlorophyll a-protein A1 [Pandor... 936 0.0
dbj|BAB18347.1| (AB044421) photosystem I P700 chlorophyll a apop... 936 0.0
dbj|BAB18850.1| (AB044227) P700 chlorophyll a-protein A1 [Pandor... 933 0.0
dbj|BAB18849.1| (AB044226) P700 chlorophyll a-protein A1 [Pandor... 932 0.0
dbj|BAB18346.1| (AB044420) photosystem I P700 chlorophyll a apop... 930 0.0
dbj|BAB18848.1| (AB044225) P700 chlorophyll a-protein A1 [Volvul... 929 0.0
dbj|BAB18846.1| (AB044223) P700 chlorophyll a-protein A1 [Volvul... 929 0.0
dbj|BAB18844.1| (AB044216) P700 chlorophyll a-protein A1 [Yamagi... 929 0.0
dbj|BAB18342.1| (AB044416) photosystem I P700 chlorophyll a apop... 929 0.0
dbj|BAB18881.1| (AB044230) P700 chlorophyll a-protein A1 [Pandor... 929 0.0
dbj|BAB18841.1| (AB044199) P700 chlorophyll a-protein A1 [Eudori... 929 0.0
dbj|BAB18856.1| (AB044242) P700 chlorophyll a-protein A1 [Gonium... 928 0.0
dbj|BAB18840.1| (AB044198) P700 chlorophyll a-protein A1 [Eudori... 928 0.0
dbj|BAB18835.1| (AB044182) P700 chlorophyll a-protein A1 [Volvox... 928 0.0
dbj|BAB18877.1| (AB044212) P700 chlorophyll a-protein A1 [Platyd... 927 0.0
dbj|BAB18855.1| (AB044241) P700 chlorophyll a-protein A1 [Gonium... 927 0.0
dbj|BAB18853.1| (AB044232) P700 chlorophyll a-protein A1 [Pandor... 927 0.0
dbj|BAB18851.1| (AB044228) P700 chlorophyll a-protein A1 [Pandor... 927 0.0
dbj|BAB18341.1| (AB044415) photosystem I P700 chlorophyll a apop... 927 0.0
dbj|BAB18869.1| (AB044185) P700 chlorophyll a-protein A1 [Volvox... 927 0.0
dbj|BAB18878.1| (AB044215) unnamed protein product [Yamagishiell... 927 0.0
dbj|BAB18843.1| (AB044213) P700 chlorophyll a-protein A1 [Yamagi... 927 0.0
dbj|BAB18838.1| (AB044187) P700 chlorophyll a-protein A1 [Volvox... 927 0.0
dbj|BAB18837.1| (AB044186) P700 chlorophyll a-protein A1 [Volvox... 927 0.0
dbj|BAB18836.1| (AB044183) P700 chlorophyll a-protein A1 [Volvox... 927 0.0
dbj|BAB18345.1| (AB044419) photosystem I P700 chlorophyll a apop... 926 0.0
dbj|BAB18872.1| (AB044197) P700 chlorophyll a-protein A1 [Pleodo... 925 0.0
dbj|BAB18884.1| (AB044240) P700 chlorophyll a-protein A1 [Gonium... 924 0.0
dbj|BAB18882.1| (AB044234) P700 chlorophyll a-protein A1 [Astrep... 923 0.0
dbj|BAB18854.1| (AB044235) P700 chlorophyll a-protein A1 [Astrep... 922 0.0
dbj|BAB18879.1| (AB044219) P700 chlorophyll a-protein A1 [Volvul... 922 0.0
dbj|BAB18858.1| (AB044244) P700 chlorophyll a-protein A1 [Gonium... 922 0.0
dbj|BAB18845.1| (AB044220) P700 chlorophyll a-protein A1 [Volvul... 922 0.0
dbj|BAB18857.1| (AB044243) P700 chlorophyll a-protein A1 [Gonium... 922 0.0
dbj|BAB18883.1| (AB044238) P700 chlorophyll a-protein A1 [Astrep... 917 0.0
dbj|BAB18880.1| (AB044222) P700 chlorophyll a-protein A1 [Volvul... 917 0.0
gb|AAD44698.1| (AF130031) PSI P700 apoprotein A1 [Heterocapsa tr... 809 0.0
pir||S71178 photosystem I protein A1 - Arabidopsis thaliana chlo... 714 0.0
dbj|BAA86297.1| (AB027235) P700 chlorophyll a apoprotein [Gymnod... 684 0.0
gb|AAA84232.1| (M17309) P700 chlorophyll a-protein [Euglena grac... 680 0.0
dbj|BAB18349.1| (AB044423) photosystem I P700 chlorophyll a apop... 673 0.0
emb|CAB75844.1| (AJ250264) photosystem I P700 chlorophyll a apop... 661 0.0
gi|7524850 P700 apoprotein subunit Ib [Chlorella vulgaris] >gi|3... 654 0.0
sp|P29255|PSAB_SYNY3 PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 651 0.0
emb|CAA41630.1| (X58825) P700 apoprotein subunit Ib (PSA-B) [Sy... 649 0.0
gi|11467833 P700 apoprotein A2 of photosystem I [Nephroselmis ol... 647 0.0
gi|11467712 PSI P700 apoprotein A2 [Guillardia theta] >gi|609379... 644 0.0
gi|11466390 P700 apoprotein A2 of photosystem I [Mesostigma viri... 642 0.0
gi|11466702 psaB [Marchantia polymorpha] >gi|131147|sp|P06408|PS... 641 0.0
gi|11465751 Photosystem I p700 chlorophyll A apoprotein A2 [Porp... 638 0.0
sp|P36492|PSAB_CHLMO PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 637 0.0
gi|7525032 PSI P700 apoprotein A2 [Arabidopsis thaliana] >gi|668... 636 0.0
sp|P09144|PSAB_CHLRE PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 634 e-180
dbj|BAB18343.1| (AB044417) photosystem I P700 chlorophyll a apop... 633 e-180
gi|11467571 PSI, P700 apoprotein A2 [Odontella sinensis] >gi|134... 632 e-180
gb|AAA84452.1| (M37526) alpha-apoprotein [Euglena gracilis] 631 e-180
gi|11467000 PSI P700 apoprotein, subunit 1b [Euglena gracilis] >... 631 e-180
gi|11466786 PSI P700 apoprotein A2 [Oryza sativa] >gi|131148|sp|... 631 e-180
gi|11465396 unknown; Photosystem I p700 chlorophyll A apoprotein... 631 e-180
gi|11465954 PSI P700 apoprotein A2 [Nicotiana tabacum] >gi|13115... 631 e-179
sp|Q33332|PSAB_ANTMA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 629 e-179
sp|P25897|PSAB_SYNEN PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 628 e-179
sp|O52475|PSAB_MASLA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 627 e-178
pir||S60184 photosystem I protein A2 - garden snapdragon chlorop... 627 e-178
gi|11497524 PSI P700 apoprotein A2 [Spinacia oleracea] >gi|13115... 627 e-178
gb|AAA84486.1| (M11203) P700 chlorophyll a-protein PSI-A2 [Zea m... 626 e-178
sp|P17155|PSAB_SYNP2 PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 626 e-178
gi|7524711 PSI P700 apoprotein A2 [Pinus thunbergii] >gi|1172658... 625 e-178
gi|11467191 PSI P700 apoprotein A2 [Zea mays] >gi|1172657|sp|P04... 625 e-178
sp|P31088|PSAB_ANAVA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN... 624 e-177
gi|11467294 PsaB subunit of photosystem I reaction center [Cyano... 623 e-177
emb|CAB67138.1| (AJ271079) PSI P700 apoprotein A2 [Oenothera ela... 621 e-176
sp|P05311|PSAB_PEA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A... 615 e-175
prf||1303353B gene psaA2 [Pisum sativum] 614 e-174
emb|CAB64209.1| (AJ133190) photosystem I subunit PsaB [Synechoco... 613 e-174
emb|CAB64201.1| (AJ133191) photosystem I subunit PsaB [Prochloro... 593 e-168
emb|CAB64199.1| (AJ133192) photosystem I subunit PsaB [Prochloro... 585 e-166
dbj|BAA83447.1| (AB013671) photosystem I P700 apoprotein A2 [Sph... 540 e-152
dbj|BAA83457.1| (AB013681) photosystem I P700 apoprotein A2 [Adi... 527 e-148
dbj|BAA83452.1| (AB013676) photosystem I P700 apoprotein A2 [Hap... 524 e-147
gb|AAK08971.1| (AY026898) photosystem I P700 protein subunit B [... 516 e-145
gb|AAD49556.1| (AF107783) PsaA [Trichodesmium sp. IMS101] 510 e-143
gb|AAD44699.1| (AF130032) PSI P700 apoprotein A2 [Heterocapsa tr... 469 e-131
dbj|BAB18384.1| (AB044458) photosystem I P700 chlorophyll a apop... 462 e-129
dbj|BAB18396.1| (AB044470) photosystem I P700 chlorophyll a apop... 462 e-129
dbj|BAB18378.1| (AB044452) photosystem I P700 chlorophyll a apop... 461 e-128
dbj|BAB18371.1| (AB044445) photosystem I P700 chlorophyll a apop... 460 e-128
dbj|BAB18397.1| (AB044471) photosystem I P700 chlorophyll a apop... 459 e-128
dbj|BAB18395.1| (AB044469) photosystem I P700 chlorophyll a apop... 459 e-128
dbj|BAB18391.1| (AB044465) photosystem I P700 chlorophyll a apop... 459 e-128
dbj|BAB18380.1| (AB044454) photosystem I P700 chlorophyll a apop... 459 e-128
dbj|BAB18370.1| (AB044444) photosystem I P700 chlorophyll a apop... 459 e-128
dbj|BAB18383.1| (AB044457) photosystem I P700 chlorophyll a apop... 459 e-128
dbj|BAB18361.1| (AB044435) photosystem I P700 chlorophyll a apop... 459 e-128
dbj|BAB18382.1| (AB044456) photosystem I P700 chlorophyll a apop... 458 e-128
dbj|BAB18385.1| (AB044459) photosystem I P700 chlorophyll a apop... 458 e-127
dbj|BAB18377.1| (AB044451) photosystem I P700 chlorophyll a apop... 458 e-127
dbj|BAB18368.1| (AB044442) photosystem I P700 chlorophyll a apop... 458 e-127
dbj|BAB18379.1| (AB044453) photosystem I P700 chlorophyll a apop... 457 e-127
dbj|BAB18374.1| (AB044448) photosystem I P700 chlorophyll a apop... 457 e-127
dbj|BAB18351.1| (AB044425) photosystem I P700 chlorophyll a apop... 457 e-127
dbj|BAB18389.1| (AB044463) photosystem I P700 chlorophyll a apop... 457 e-127
dbj|BAB18387.1| (AB044461) photosystem I P700 chlorophyll a apop... 457 e-127
dbj|BAB18372.1| (AB044446) photosystem I P700 chlorophyll a apop... 457 e-127
dbj|BAB18369.1| (AB044443) photosystem I P700 chlorophyll a apop... 457 e-127
dbj|BAB18381.1| (AB044455) photosystem I P700 chlorophyll a apop... 456 e-127
dbj|BAB18353.1| (AB044427) photosystem I P700 chlorophyll a apop... 456 e-127
dbj|BAB18352.1| (AB044426) photosystem I P700 chlorophyll a apop... 456 e-127
dbj|BAB18350.1| (AB044424) photosystem I P700 chlorophyll a apop... 456 e-127
dbj|BAB18365.1| (AB044439) photosystem I P700 chlorophyll a apop... 456 e-127
dbj|BAB18360.1| (AB044434) photosystem I P700 chlorophyll a apop... 456 e-127
dbj|BAB18356.1| (AB044430) photosystem I P700 chlorophyll a apop... 456 e-127
dbj|BAB18373.1| (AB044447) photosystem I P700 chlorophyll a apop... 455 e-127
dbj|BAB18355.1| (AB044429) photosystem I P700 chlorophyll a apop... 455 e-127
dbj|BAB18392.1| (AB044466) photosystem I P700 chlorophyll a apop... 455 e-127
dbj|BAB18390.1| (AB044464) photosystem I P700 chlorophyll a apop... 455 e-127
dbj|BAB18375.1| (AB044449) photosystem I P700 chlorophyll a apop... 455 e-127
dbj|BAB18388.1| (AB044462) photosystem I P700 chlorophyll a apop... 455 e-126
dbj|BAB18354.1| (AB044428) photosystem I P700 chlorophyll a apop... 453 e-126
dbj|BAB18398.1| (AB044472) photosystem I P700 chlorophyll a apop... 451 e-125
dbj|BAB18386.1| (AB044460) photosystem I P700 chlorophyll a apop... 450 e-125
dbj|BAB18363.1| (AB044437) photosystem I P700 chlorophyll a apop... 441 e-122
dbj|BAB18359.1| (AB044433) photosystem I P700 chlorophyll a apop... 436 e-121
dbj|BAB18399.1| (AB044473) photosystem I P700 chlorophyll a apop... 413 e-114
dbj|BAA83440.1| (AB013664) photosystem I P700 apoprotein A1 [Ant... 346 6e-94
gb|AAD10229.1| (U72056) photosystem I protein A1 [Anabaena sp. CA] 328 1e-88
dbj|BAA83428.1| (AB013652) photosystem I P700 apoprotein A1 [Phy... 298 2e-79
dbj|BAB18344.1| (AB044418) photosystem I P700 chlorophyll a apop... 294 2e-78
dbj|BAA83446.1| (AB013670) photosystem I P700 apoprotein A1 [Sph... 287 4e-76
dbj|BAA83456.1| (AB013680) photosystem I P700 apoprotein A1 [Adi... 285 1e-75
dbj|BAA83451.1| (AB013675) photosystem I P700 apoprotein A1 [Hap... 279 9e-74
dbj|BAA83434.1| (AB013658) photosystem I P700 apoprotein A1 [Col... 278 2e-73
dbj|BAB18348.1| (AB044422) photosystem I P700 chlorophyll a apop... 249 1e-64
dbj|BAA83435.1| (AB013659) photosystem I P700 apoprotein A2 [Col... 243 4e-63
dbj|BAB18394.1| (AB044468) photosystem I P700 chlorophyll a apop... 236 6e-61
dbj|BAB18393.1| (AB044467) photosystem I P700 chlorophyll a apop... 221 3e-56
dbj|BAA83429.1| (AB013653) photosystem I P700 apoprotein A2 [Phy... 215 2e-54
dbj|BAA83441.1| (AB013665) photosystem I P700 apoprotein A2 [Ant... 198 2e-49
dbj|BAA83430.1| (AB013654) photosystem I P700 apoprotein A2 [Phy... 168 2e-40
dbj|BAA83436.1| (AB013660) photosystem I P700 apoprotein A2 [Col... 166 9e-40
dbj|BAA83442.1| (AB013666) photosystem I P700 apoprotein A2 [Ant... 140 5e-32
emb|CAA25804.1| (X01647) open reading frame (ORF 704) [Marchanti... 109 9e-23
emb|CAA58957.1| (X84152) photosystem I subunit [Antirrhinum majus] 98 4e-19
pir||S18319 photosystem I protein A2 - Chlamydomonas reinhardtii... 93 1e-17
pir||T31454 reaction center core polypeptide pshA - Heliobacillu... 84 5e-15
pir||A48290 reaction center core polypeptide - Heliobacillus mob... 71 4e-11
gb|AAA92988.1| (L27262) P700 chlorophyll a-apoprotein A1 [Chlamy... 57 1e-06
gb|AAA92987.1| (L27259) P700 chlorophyll a-apoprotein A1 [Chlamy... 55 4e-06
gb|AAB20423.2| (S67792) polypeptide of the photosystem I reactio... 52 3e-05
pir||A46265 P840 (photosystem 1-like) reaction center large chai... 49 2e-04
sp|P10977|CARV_CANAL VACUOLAR ASPARTIC PROTEASE PRECURSOR (ASPAR... 35 3.1
dbj|BAB08016.1| (AB026513) cytochrome c oxidase subunit I [Campa... 35 4.0
emb|CAB45538.1| (AJ242660) photosystem I P700 chlorophyll [Sinap... 35 4.0
pir||S14661 photosystem I protein psaA - maize (fragment) >gi|12... 35 4.0
gb|AAG19813.1| (AE005065) Vng1524c [Halobacterium sp. NRC-1] 34 6.9
gi|11499293 hypothetical protein [Archaeoglobus fulgidus] >gi|74... 34 6.9
>gi|11465955 PSI P700 apoprotein A1 [Nicotiana tabacum]
sp|P06405|PSAA_TOBAC PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||A1NTP7 photosystem I P700 apoprotein A1 - common tobacco chloroplast
emb|CAA77352.1| (Z00044) PSI P700 apoprotein A1 [Nicotiana tabacum]
prf||1211235AC photosystem I P700 apoprotein A1 [Nicotiana tabacum]
Length = 750
Score = 1552 bits (3973), Expect = 0.0
Identities = 730/750 (97%), Positives = 741/750 (98%)
Query: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
MIIRSPEPEVKILVDRDP+KTSFEEWA+PGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT
Sbjct: 1 MIIRSPEPEVKILVDRDPVKTSFEEWARPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
Query: 61 SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVG 120
SDLE+ISRK+FSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHI PSAQVVWPIVG
Sbjct: 61 SDLEEISRKVFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIGPSAQVVWPIVG 120
Query: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHK 180
QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIG LVFAALMLFAGWFHYHK
Sbjct: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGALVFAALMLFAGWFHYHK 180
Query: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 240
AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF
Sbjct: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 240
Query: 241 ILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
ILNRDLLAQLYPSFAEGATPFFTLNW+KYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI
Sbjct: 241 ILNRDLLAQLYPSFAEGATPFFTLNWSKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
Query: 301 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSL 360
LFLIAGHMYRTNWGIGHG+KDILEAHKGPFTGQGHKGLYEILTTSWHAQLS+NLAMLGSL
Sbjct: 301 LFLIAGHMYRTNWGIGHGLKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSLNLAMLGSL 360
Query: 361 TIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
TI+VAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY
Sbjct: 361 TIVVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
Query: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
NDLLDRVLRHRDAIISHLNW CIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+F
Sbjct: 421 NDLLDRVLRHRDAIISHLNWACIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVF 480
Query: 481 AQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 540
AQWIQNTHALAPG TAPGAT TSLTWGGGDLVAVG KVALLPIPLGTADFLVHHIHAFT
Sbjct: 481 AQWIQNTHALAPGATAPGATASTSLTWGGGDLVAVGGKVALLPIPLGTADFLVHHIHAFT 540
Query: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
IHVT LILLKGVLFARSSRL PDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA
Sbjct: 541 IHVTALILLKGVLFARSSRLTPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
Query: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
ISVVIFHFSWKMQSDVWGS+SDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY
Sbjct: 601 ISVVIFHFSWKMQSDVWGSVSDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
Query: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIV 720
GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSI+
Sbjct: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSII 720
Query: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
QGRAVGVTHYLLGGIATTWAFFLARIIAVG
Sbjct: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
>gi|7525033 PSI P700 apoprotein A1 [Arabidopsis thaliana]
sp|P56766|PSAA_ARATH PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
dbj|BAA84385.1| (AP000423) PSI P700 apoprotein A1 [Arabidopsis thaliana]
Length = 750
Score = 1551 bits (3971), Expect = 0.0
Identities = 732/750 (97%), Positives = 742/750 (98%)
Query: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT
Sbjct: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
Query: 61 SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVG 120
SDLE+ISRK+FSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHI PSAQVVWPIVG
Sbjct: 61 SDLEEISRKVFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIGPSAQVVWPIVG 120
Query: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHK 180
QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIG LVFAALMLFAGWFHYHK
Sbjct: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGALVFAALMLFAGWFHYHK 180
Query: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 240
AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF
Sbjct: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 240
Query: 241 ILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
ILNRDLLAQLYPSFAEGATPFFTLNW+KY++FLTFRGGLDPVTGGLWLTDIAHHHLAIAI
Sbjct: 241 ILNRDLLAQLYPSFAEGATPFFTLNWSKYSEFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
Query: 301 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSL 360
LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLS+NLAMLGSL
Sbjct: 301 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSLNLAMLGSL 360
Query: 361 TIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
TIIVAHHMYSMPPYPYLATDY TQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPT RY
Sbjct: 361 TIIVAHHMYSMPPYPYLATDYATQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTNRY 420
Query: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+F
Sbjct: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVF 480
Query: 481 AQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 540
AQWIQNTHALAPG+TAPG T TSLTWGGG+LVAVG KVALLPIPLGTADFLVHHIHAFT
Sbjct: 481 AQWIQNTHALAPGVTAPGETASTSLTWGGGELVAVGGKVALLPIPLGTADFLVHHIHAFT 540
Query: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA
Sbjct: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
Query: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY
Sbjct: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
Query: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIV 720
GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSI+
Sbjct: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSII 720
Query: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
QGRAVGVTHYLLGGIATTWAFFLARIIAVG
Sbjct: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
>gi|11497525 PSI P700 apoprotein A1 [Spinacia oleracea]
sp|P06511|PSAA_SPIOL PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||S00444 photosystem I protein A1 - spinach chloroplast
emb|CAB88726.1| (AJ400848) PSI P700 apoprotein A1 [Spinacia oleracea]
prf||1303218A gene psaA [Spinacia oleracea]
Length = 750
Score = 1541 bits (3945), Expect = 0.0
Identities = 726/750 (96%), Positives = 738/750 (97%)
Query: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
MIIRSPEPEVKILVDRDP+KTSFE WAKPGHFSRTIAKGP+TTTWIWNLHADAHDFDSHT
Sbjct: 1 MIIRSPEPEVKILVDRDPVKTSFEAWAKPGHFSRTIAKGPETTTWIWNLHADAHDFDSHT 60
Query: 61 SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVG 120
SDLE+ISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHI PSAQVVWPIVG
Sbjct: 61 SDLEEISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIGPSAQVVWPIVG 120
Query: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHK 180
QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIG LVFAALMLFAGWFHYHK
Sbjct: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGALVFAALMLFAGWFHYHK 180
Query: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 240
AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQ+HVSLPINQFLNAGVDPKEIPLPHE
Sbjct: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQIHVSLPINQFLNAGVDPKEIPLPHEL 240
Query: 241 ILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
ILNRDLLAQLYPSFAEGATPFFTLNW+KYADFLTFRGGLDPVTGGLWLTD AHHHLAIAI
Sbjct: 241 ILNRDLLAQLYPSFAEGATPFFTLNWSKYADFLTFRGGLDPVTGGLWLTDTAHHHLAIAI 300
Query: 301 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSL 360
LFLIAGHMYRTNWGIGHG+KDILEAHKGPFTGQGHKGLYEILTTSWHAQL++NLAMLGSL
Sbjct: 301 LFLIAGHMYRTNWGIGHGLKDILEAHKGPFTGQGHKGLYEILTTSWHAQLALNLAMLGSL 360
Query: 361 TIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
TI+VAHHMY+MPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY
Sbjct: 361 TIVVAHHMYAMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
Query: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
NDLLDRVLRHRDAIISHLNW CIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+F
Sbjct: 421 NDLLDRVLRHRDAIISHLNWACIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVF 480
Query: 481 AQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 540
AQWIQNTHALAP TAPGAT TSLTWGG DLVAVG KVALLPIPLGTADFLVHHIHAFT
Sbjct: 481 AQWIQNTHALAPSATAPGATASTSLTWGGSDLVAVGGKVALLPIPLGTADFLVHHIHAFT 540
Query: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+
Sbjct: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNS 600
Query: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY
Sbjct: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
Query: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIV 720
GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIV
Sbjct: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIV 720
Query: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
QGRAVGVTHYLLGGIATTWAFFLARIIAVG
Sbjct: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
>gi|11994091 PSI P700 apoprotein A1 [Zea mays]
emb|CAA60286.2| (X86563) PSI P700 apoprotein A1 [Zea mays]
Length = 750
Score = 1532 bits (3922), Expect = 0.0
Identities = 717/750 (95%), Positives = 739/750 (97%)
Query: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
MIIRS EPEVKI VDRDPIKTSFEEWA+PGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT
Sbjct: 1 MIIRSSEPEVKIAVDRDPIKTSFEEWARPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
Query: 61 SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVG 120
DLE+ISRK+FSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHI PSAQVVWPIVG
Sbjct: 61 GDLEEISRKVFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIGPSAQVVWPIVG 120
Query: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHK 180
QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIG L+FA+LMLFAGWFHYHK
Sbjct: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGALIFASLMLFAGWFHYHK 180
Query: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 240
AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQ+HVSLPINQFL+AGVDPKEIPLPHEF
Sbjct: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQIHVSLPINQFLDAGVDPKEIPLPHEF 240
Query: 241 ILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
ILNRDLLAQLYPSFAEGATPFFTLNW+KYA+FL+FRGGLDP+TGGLWL+DIAHHHLAIAI
Sbjct: 241 ILNRDLLAQLYPSFAEGATPFFTLNWSKYAEFLSFRGGLDPITGGLWLSDIAHHHLAIAI 300
Query: 301 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSL 360
LFLIAGHMYRTNWGIGHG+KDILEAHKGPFTGQGHKGLYEILTTSWHAQLS+NLAMLGS
Sbjct: 301 LFLIAGHMYRTNWGIGHGLKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSLNLAMLGST 360
Query: 361 TIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
TI+VAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY
Sbjct: 361 TIVVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
Query: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSD AIQLQPIF
Sbjct: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDAAIQLQPIF 480
Query: 481 AQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 540
AQWIQN HA APG+TAPGATT TSLTWGGG+LVA+G KVALLPIPLGTADFLVHHIHAFT
Sbjct: 481 AQWIQNIHAGAPGVTAPGATTSTSLTWGGGELVAIGGKVALLPIPLGTADFLVHHIHAFT 540
Query: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+
Sbjct: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNS 600
Query: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
ISVVIFHFSWKMQSDVWG+ISDQG+VTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY
Sbjct: 601 ISVVIFHFSWKMQSDVWGTISDQGIVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
Query: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIV 720
GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSI+
Sbjct: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSII 720
Query: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
QGRAVGVTHYLLGGIATTWAFFLARIIAVG
Sbjct: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
>gb|AAB25858.1| photosystem I reaction center protein psaA product [Sorghum
bicolor, Qiuji No. 5, Peptide Chloroplast, 750 aa]
Length = 750
Score = 1532 bits (3922), Expect = 0.0
Identities = 717/750 (95%), Positives = 739/750 (97%)
Query: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
MIIR EPEVKI VDRDP+KTSFEEWA+PGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT
Sbjct: 1 MIIRPSEPEVKIAVDRDPVKTSFEEWARPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
Query: 61 SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVG 120
DLE+ISRK+FSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHI PSAQVVWPIVG
Sbjct: 61 GDLEEISRKVFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIGPSAQVVWPIVG 120
Query: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHK 180
QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIG L+FA+LMLFAGWFHYHK
Sbjct: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGALIFASLMLFAGWFHYHK 180
Query: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 240
AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQ+HVSLPINQFL+AGVDPKEIPLPHEF
Sbjct: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQIHVSLPINQFLDAGVDPKEIPLPHEF 240
Query: 241 ILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
ILNRDLLAQLYPSFAEGATPFFTLNW+KYA+FL+FRGGLDP+TGGLWL+DIAHHHLAIAI
Sbjct: 241 ILNRDLLAQLYPSFAEGATPFFTLNWSKYAEFLSFRGGLDPITGGLWLSDIAHHHLAIAI 300
Query: 301 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSL 360
LFLIAGHMYRTNWGIGHG+KDILEAHKGPFTGQGHKGLYEILTTSWHAQLS+NLAMLGS
Sbjct: 301 LFLIAGHMYRTNWGIGHGLKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSLNLAMLGST 360
Query: 361 TIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
TI+VAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY
Sbjct: 361 TIVVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
Query: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF
Sbjct: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
Query: 481 AQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 540
AQWIQN HA APG+TAPGATT TSLTWGGG+LVAVG KVALLPIPLGTADFLVHHIHAFT
Sbjct: 481 AQWIQNIHAGAPGVTAPGATTSTSLTWGGGELVAVGGKVALLPIPLGTADFLVHHIHAFT 540
Query: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+
Sbjct: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNS 600
Query: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
ISVVIFHFSWKMQSDVWG+ISDQG+VTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY
Sbjct: 601 ISVVIFHFSWKMQSDVWGTISDQGIVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
Query: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIV 720
GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSI+
Sbjct: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSII 720
Query: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
QGRAVGVTHYLLGGIATTWAFFLARIIAVG
Sbjct: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
>sp|P04966|PSAA_MAIZE PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||S58552 photosystem I protein A1 - maize chloroplast
gb|AAA84485.1| (M11203) P700 chlorophyll a-protein PSI-A1 [Zea mays]
Length = 751
Score = 1527 bits (3910), Expect = 0.0
Identities = 717/751 (95%), Positives = 739/751 (97%), Gaps = 1/751 (0%)
Query: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKG-PDTTTWIWNLHADAHDFDSH 59
MIIRS EPEVKI VDRDPIKTSFEEWA+PGHFSRTIAKG PDTTTWIWNLHADAHDFDSH
Sbjct: 1 MIIRSSEPEVKIAVDRDPIKTSFEEWARPGHFSRTIAKGNPDTTTWIWNLHADAHDFDSH 60
Query: 60 TSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIV 119
T DLE+ISRK+FSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHI PSAQVVWPIV
Sbjct: 61 TGDLEEISRKVFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIGPSAQVVWPIV 120
Query: 120 GQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYH 179
GQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIG L+FA+LMLFAGWFHYH
Sbjct: 121 GQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGALIFASLMLFAGWFHYH 180
Query: 180 KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHE 239
KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQ+HVSLPINQFL+AGVDPKEIPLPHE
Sbjct: 181 KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQIHVSLPINQFLDAGVDPKEIPLPHE 240
Query: 240 FILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIA 299
FILNRDLLAQLYPSFAEGATPFFTLNW+KYA+FL+FRGGLDP+TGGLWL+DIAHHHLAIA
Sbjct: 241 FILNRDLLAQLYPSFAEGATPFFTLNWSKYAEFLSFRGGLDPITGGLWLSDIAHHHLAIA 300
Query: 300 ILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGS 359
ILFLIAGHMYRTNWGIGHG+KDILEAHKGPFTGQGHKGLYEILTTSWHAQLS+NLAMLGS
Sbjct: 301 ILFLIAGHMYRTNWGIGHGLKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSLNLAMLGS 360
Query: 360 LTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTR 419
TI+VAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTR
Sbjct: 361 TTIVVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTR 420
Query: 420 YNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPI 479
YNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSD AIQLQPI
Sbjct: 421 YNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDAAIQLQPI 480
Query: 480 FAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAF 539
FAQWIQN HA APG+TAPGATT TSLTWGGG+LVA+G KVALLPIPLGTADFLVHHIHAF
Sbjct: 481 FAQWIQNIHAGAPGVTAPGATTSTSLTWGGGELVAIGGKVALLPIPLGTADFLVHHIHAF 540
Query: 540 TIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN 599
TIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN
Sbjct: 541 TIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN 600
Query: 600 AISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQS 659
+ISVVIFHFSWKMQSDVWG+ISDQG+VTHITGGNFAQSSITINGWLRDFLWAQASQVIQS
Sbjct: 601 SISVVIFHFSWKMQSDVWGTISDQGIVTHITGGNFAQSSITINGWLRDFLWAQASQVIQS 660
Query: 660 YGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSI 719
YGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSI
Sbjct: 661 YGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSI 720
Query: 720 VQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
+QGRAVGVTHYLLGGIATTWAFFLARIIAVG
Sbjct: 721 IQGRAVGVTHYLLGGIATTWAFFLARIIAVG 751
>emb|CAB67137.1| (AJ271079) PSI P700 apoprotein A1 [Oenothera elata subsp. hookeri]
Length = 751
Score = 1525 bits (3904), Expect = 0.0
Identities = 723/751 (96%), Positives = 737/751 (97%), Gaps = 1/751 (0%)
Query: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGP+TTTWIWNLHADAHDFDSHT
Sbjct: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPETTTWIWNLHADAHDFDSHT 60
Query: 61 SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVG 120
SD+E ISRK+ SAHFGQLS+IFLWLSGMY+ A FSNYEAWLSDPTHI PSAQVVWPIVG
Sbjct: 61 SDVEAISRKVSSAHFGQLSLIFLWLSGMYWGFALFSNYEAWLSDPTHIGPSAQVVWPIVG 120
Query: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHK 180
QEILNGDVGGGFRGIQITSGFFQ+WRASGITSELQLYCTAIG L+FAALMLFAGWFHYHK
Sbjct: 121 QEILNGDVGGGFRGIQITSGFFQLWRASGITSELQLYCTAIGALIFAALMLFAGWFHYHK 180
Query: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 240
AAPKLAWFQDV+SMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPL HEF
Sbjct: 181 AAPKLAWFQDVQSMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLLHEF 240
Query: 241 ILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
ILNRDLLAQLYPSFAEGATPFFTLNW+KYA+FLTFRGGLDPVTGGLWLTDIAHHHLAIAI
Sbjct: 241 ILNRDLLAQLYPSFAEGATPFFTLNWSKYAEFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
Query: 301 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSL 360
LFLIAGHMYRTNWGIGHG+KDILEAHKGPFTGQGHKGLYEILTTSWHAQLS+NLAMLGSL
Sbjct: 301 LFLIAGHMYRTNWGIGHGLKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSLNLAMLGSL 360
Query: 361 TIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
TI+VAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY
Sbjct: 361 TIVVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
Query: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+F
Sbjct: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVF 480
Query: 481 AQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 540
AQWIQNTHALAPG TAPGATT TSL WGGGDLVAVG KVALLPIPLGTADFLVHHIHAFT
Sbjct: 481 AQWIQNTHALAPGATAPGATTSTSLAWGGGDLVAVGGKVALLPIPLGTADFLVHHIHAFT 540
Query: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA
Sbjct: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
Query: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY
Sbjct: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
Query: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFS-GRGYWQELIESIVWAHNKLKVAPATQPRALSI 719
GSSLSAYGLFFLGAHFVWAFSLMFLFS GRGYWQELIESIVWAHNKLKVAPATQPRALSI
Sbjct: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSCGRGYWQELIESIVWAHNKLKVAPATQPRALSI 720
Query: 720 VQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
VQGR VGVTHYLLGGIATTWAFFLARIIAVG
Sbjct: 721 VQGRTVGVTHYLLGGIATTWAFFLARIIAVG 751
>pir||JC1067 psaA protein - Sorghum chloroplast
Length = 750
Score = 1521 bits (3894), Expect = 0.0
Identities = 713/750 (95%), Positives = 735/750 (97%)
Query: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
MIIR EPEVKI VDRDP+KTSFEEWA+PGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT
Sbjct: 1 MIIRPSEPEVKIAVDRDPVKTSFEEWARPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
Query: 61 SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVG 120
DLE+ISRK+FSAHFGQLSIIFLWLSG YFHGARFSNYEAWLSDPTHI PSAQVVWPIVG
Sbjct: 61 GDLEEISRKVFSAHFGQLSIIFLWLSGNYFHGARFSNYEAWLSDPTHIGPSAQVVWPIVG 120
Query: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHK 180
QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIG L+FA+L LFAGWFHYHK
Sbjct: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGALIFASLNLFAGWFHYHK 180
Query: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 240
AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQ+HVSLPINQFL+AGVDPKEIPLPHEF
Sbjct: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQIHVSLPINQFLDAGVDPKEIPLPHEF 240
Query: 241 ILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
ILNRDLLAQLYPSFAEGATPFFTLNW+KYA+FL+FRGGLDP+TGGLWL+DIAHHHLAIAI
Sbjct: 241 ILNRDLLAQLYPSFAEGATPFFTLNWSKYAEFLSFRGGLDPITGGLWLSDIAHHHLAIAI 300
Query: 301 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSL 360
LFLIAGHMYRTNWGIGHG+KDILEAHKGPFTGQGHKGLYEILTTSWHAQLS+NLAMLGS
Sbjct: 301 LFLIAGHMYRTNWGIGHGLKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSLNLAMLGST 360
Query: 361 TIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
TI+VAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY
Sbjct: 361 TIVVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
Query: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF
Sbjct: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
Query: 481 AQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 540
AQWIQN HA APG+TAPGATT TSLTWGGG+LVAVG KVALLPIPLGTADFLVHHIHAFT
Sbjct: 481 AQWIQNIHAGAPGVTAPGATTSTSLTWGGGELVAVGGKVALLPIPLGTADFLVHHIHAFT 540
Query: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFW YN+
Sbjct: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWNYNS 600
Query: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
ISVVIFHFSWKMQSDVWG+ISDQG+VTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY
Sbjct: 601 ISVVIFHFSWKMQSDVWGTISDQGIVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
Query: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIV 720
GSSLSAYGLFFLGAHFVWAFSL FLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSI+
Sbjct: 661 GSSLSAYGLFFLGAHFVWAFSLNFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSII 720
Query: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
QGRAVGVTHYLLGGIATTWAFFLARIIAVG
Sbjct: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
>gi|11466787 PSI P700 apoprotein A1 [Oryza sativa]
sp|P12155|PSAA_ORYSA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||A1RZP7 photosystem I P700 apoprotein A1 - rice chloroplast
emb|CAA33996.1| (X15901) PSI P700 apoprotein A1 [Oryza sativa]
prf||1603356AB photosystem I P700 apoprotein A1 [Oryza sativa]
Length = 750
Score = 1519 bits (3890), Expect = 0.0
Identities = 711/750 (94%), Positives = 737/750 (97%)
Query: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
M+IRSPEPEVKI+VDRDP+KTSFEEWA+PGHFSRT+AKGPDTTTWIWNLHADAHDFDSHT
Sbjct: 1 MMIRSPEPEVKIVVDRDPVKTSFEEWARPGHFSRTLAKGPDTTTWIWNLHADAHDFDSHT 60
Query: 61 SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVG 120
DLE+ISRK+FSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHI PSAQVVWPIVG
Sbjct: 61 GDLEEISRKVFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIGPSAQVVWPIVG 120
Query: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHK 180
QEILNGDVGGGFRGIQI+SGFFQIWRASGITSELQLYCTAIG L+FA+LMLFAGWFHYHK
Sbjct: 121 QEILNGDVGGGFRGIQISSGFFQIWRASGITSELQLYCTAIGALIFASLMLFAGWFHYHK 180
Query: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 240
AAPKLAW DVESMLNHHLAGLLGLGSLSWAGHQ+HVSLPINQFL+AGVDPKEIPLPHEF
Sbjct: 181 AAPKLAWSHDVESMLNHHLAGLLGLGSLSWAGHQIHVSLPINQFLDAGVDPKEIPLPHEF 240
Query: 241 ILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
ILNRDLLAQLYPSFAE ATPFFTLNW+KYA+FL+FRGGLDP+TGGLWL+DIAHHHLAIAI
Sbjct: 241 ILNRDLLAQLYPSFAERATPFFTLNWSKYAEFLSFRGGLDPITGGLWLSDIAHHHLAIAI 300
Query: 301 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSL 360
LFLIAGHMYRTNWGIGHG+KDILEAHKGPFTGQ HKGLYEILTTSWHAQLS+NLAMLGS
Sbjct: 301 LFLIAGHMYRTNWGIGHGLKDILEAHKGPFTGQRHKGLYEILTTSWHAQLSLNLAMLGST 360
Query: 361 TIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
TI+VAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY
Sbjct: 361 TIVVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
Query: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF
Sbjct: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
Query: 481 AQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 540
AQW+QN HA AP +TAPGATT TSLTWGGG+LVAVG KVALLPIPLGTADFLVHHIHAFT
Sbjct: 481 AQWVQNLHAGAPRLTAPGATTSTSLTWGGGELVAVGGKVALLPIPLGTADFLVHHIHAFT 540
Query: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+
Sbjct: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNS 600
Query: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
ISVVIFHFSWKMQSDVWG+ISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY
Sbjct: 601 ISVVIFHFSWKMQSDVWGTISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
Query: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIV 720
GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSI+
Sbjct: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSII 720
Query: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
QGRAVGVTHYLLGGIATTWAFFLARIIAVG
Sbjct: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
>gi|11466703 psaA [Marchantia polymorpha]
sp|P06406|PSAA_MARPO PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||A1LVP7 photosystem I P700 apoprotein A1 - liverwort (Marchantia
polymorpha) chloroplast
emb|CAA28085.1| (X04465) psaA [Marchantia polymorpha]
Length = 750
Score = 1494 bits (3825), Expect = 0.0
Identities = 694/750 (92%), Positives = 732/750 (97%)
Query: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
M IRSPEPEVKI+V++DP+KTSFE+WAKPGHFSRT+AKGP TTTWIWNLHADAHDFDSHT
Sbjct: 1 MTIRSPEPEVKIVVEKDPVKTSFEKWAKPGHFSRTLAKGPSTTTWIWNLHADAHDFDSHT 60
Query: 61 SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVG 120
+DLE+ISRK+FSAHFGQL+IIF+WLSGMYFHGARFSNYEAWLSDPTHI+PSAQVVWPIVG
Sbjct: 61 NDLEEISRKVFSAHFGQLAIIFIWLSGMYFHGARFSNYEAWLSDPTHIKPSAQVVWPIVG 120
Query: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHK 180
QEILNGDVGGGF+GIQITSGFFQ+WRASGITSELQLY TAIGGLVFAALMLFAGWFHYHK
Sbjct: 121 QEILNGDVGGGFQGIQITSGFFQLWRASGITSELQLYSTAIGGLVFAALMLFAGWFHYHK 180
Query: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 240
AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQ L+AGVDPKEIPLPHEF
Sbjct: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQLLDAGVDPKEIPLPHEF 240
Query: 241 ILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
ILNRDLLA+LYPSFA+G TPFFTLNW++Y+DFLTFRGGL+PVTGGLWLTD AHHHLAIA+
Sbjct: 241 ILNRDLLAELYPSFAKGLTPFFTLNWSEYSDFLTFRGGLNPVTGGLWLTDTAHHHLAIAV 300
Query: 301 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSL 360
LFL+AGHMYRTNWGIGH K+ILEAHKGPFTG+GHKGLYEILTTSWHAQL++NLAMLGSL
Sbjct: 301 LFLVAGHMYRTNWGIGHSFKEILEAHKGPFTGEGHKGLYEILTTSWHAQLALNLAMLGSL 360
Query: 361 TIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
TIIVAHHMY+MPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTT+Y
Sbjct: 361 TIIVAHHMYAMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTQY 420
Query: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
N+LLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+F
Sbjct: 421 NNLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVF 480
Query: 481 AQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 540
AQWIQNTHALAP TAP A TSLTWGGGD++AVG+KVALLPIPLGTADFLVHHIHAFT
Sbjct: 481 AQWIQNTHALAPNFTAPNALASTSLTWGGGDVIAVGSKVALLPIPLGTADFLVHHIHAFT 540
Query: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+
Sbjct: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNS 600
Query: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
ISVVIFHFSWKMQSDVWG+IS+QGVVTHITGGNFAQS+ITINGWLRDFLWAQASQVIQSY
Sbjct: 601 ISVVIFHFSWKMQSDVWGTISEQGVVTHITGGNFAQSAITINGWLRDFLWAQASQVIQSY 660
Query: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIV 720
GSSLSAYGL FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRALSI
Sbjct: 661 GSSLSAYGLLFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRALSIT 720
Query: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
QGRAVGV HYLLGGIATTWAFFLARIIAVG
Sbjct: 721 QGRAVGVAHYLLGGIATTWAFFLARIIAVG 750
>gi|7524710 PSI P700 apoprotein A1 [Pinus thunbergii]
sp|P41639|PSAA_PINTH PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||T07543 photosystem I protein A1 - Japanese black pine chloroplast
dbj|BAA04419.1| (D17510) PSI P700 apoprotein A1 [Pinus thunbergii]
Length = 753
Score = 1492 bits (3820), Expect = 0.0
Identities = 694/753 (92%), Positives = 731/753 (96%), Gaps = 3/753 (0%)
Query: 1 MIIRSPEPEVK---ILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFD 57
M IRSPEPEVK ++VDRD +KTSFE+WAKPGHFSRT+AKGPDTTTWIWNLHADAHDFD
Sbjct: 1 MTIRSPEPEVKKVKVVVDRDTVKTSFEKWAKPGHFSRTLAKGPDTTTWIWNLHADAHDFD 60
Query: 58 SHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWP 117
SHT++LEDISRKIFSAHFGQL+IIF+WLSGMY+HGARFSNYEAWL+DPTHI+PSAQ+VWP
Sbjct: 61 SHTNNLEDISRKIFSAHFGQLAIIFIWLSGMYYHGARFSNYEAWLADPTHIKPSAQIVWP 120
Query: 118 IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFH 177
IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIG L+FAALMLFAGWFH
Sbjct: 121 IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGALIFAALMLFAGWFH 180
Query: 178 YHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLP 237
YHKAAPKLAWFQ+VESMLNHHLAGLLGLGSLSWAGHQ+HVSLPINQ L+AGVDPKEIPLP
Sbjct: 181 YHKAAPKLAWFQEVESMLNHHLAGLLGLGSLSWAGHQIHVSLPINQLLDAGVDPKEIPLP 240
Query: 238 HEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLA 297
HEFI NRDLLAQLYPSFA+G TPF TLNW++Y+DFLTFRGGL+PVTGGLWLTD AHHHLA
Sbjct: 241 HEFIFNRDLLAQLYPSFAKGVTPFLTLNWSEYSDFLTFRGGLNPVTGGLWLTDTAHHHLA 300
Query: 298 IAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAML 357
IA+LFLIAGHMY+TNW IGH +KD+LEAHKGPFTG+GHKGLYEILTTSWHAQL++NLAML
Sbjct: 301 IAVLFLIAGHMYKTNWRIGHNLKDLLEAHKGPFTGEGHKGLYEILTTSWHAQLAVNLAML 360
Query: 358 GSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPT 417
GSLTI+VAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGF+IVGAAAHAAIFMVRDYDPT
Sbjct: 361 GSLTIVVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFIIVGAAAHAAIFMVRDYDPT 420
Query: 418 TRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ 477
T+YN+LLDRVLRHRDAI+SHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ
Sbjct: 421 TQYNNLLDRVLRHRDAIVSHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ 480
Query: 478 PIFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIH 537
PIFAQWIQNTHA APG TAPGAT TSLTWGGGDLV VG+KVALLPIPLGTADFLVHHIH
Sbjct: 481 PIFAQWIQNTHASAPGSTAPGATASTSLTWGGGDLVTVGSKVALLPIPLGTADFLVHHIH 540
Query: 538 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 597
AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM
Sbjct: 541 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 600
Query: 598 YNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVI 657
YNAISVVIFHFSWKMQSDVWG+ISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVI
Sbjct: 601 YNAISVVIFHFSWKMQSDVWGNISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVI 660
Query: 658 QSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRAL 717
QSYGSSLSAYGL FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRAL
Sbjct: 661 QSYGSSLSAYGLLFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRAL 720
Query: 718 SIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
SIVQGRAVGV HYLLGGI TTWAFFLARIIA+G
Sbjct: 721 SIVQGRAVGVAHYLLGGIVTTWAFFLARIIAIG 753
>gb|AAF29817.1| (AF180016) photosystem I P700 apoprotein A1 [Drimys winteri]
Length = 717
Score = 1484 bits (3799), Expect = 0.0
Identities = 696/717 (97%), Positives = 709/717 (98%)
Query: 13 LVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKIFS 72
LVDRDP+KTSFEEWA+PGHFSRTIAKGP+TTTWIWNLHADAHDFDSHTSDLE+ISRK+FS
Sbjct: 1 LVDRDPVKTSFEEWARPGHFSRTIAKGPETTTWIWNLHADAHDFDSHTSDLEEISRKVFS 60
Query: 73 AHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGGF 132
AHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHI PSAQVVWPIVGQEILNGDVGGGF
Sbjct: 61 AHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIGPSAQVVWPIVGQEILNGDVGGGF 120
Query: 133 RGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDVE 192
RGIQITSGFFQIWRASGITSELQLYCTAIG LVFA LMLFAGWFHYHKAAPKLAWFQDVE
Sbjct: 121 RGIQITSGFFQIWRASGITSELQLYCTAIGALVFAGLMLFAGWFHYHKAAPKLAWFQDVE 180
Query: 193 SMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYP 252
SMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFL+AGVDPKEIPLPHEFILNRDLLAQLYP
Sbjct: 181 SMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLDAGVDPKEIPLPHEFILNRDLLAQLYP 240
Query: 253 SFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTN 312
SFAEGATPFFTLNW+KYA+FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTN
Sbjct: 241 SFAEGATPFFTLNWSKYAEFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTN 300
Query: 313 WGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMP 372
WGIGHG+KDILEAHKGPFTGQGHKGLYEILTTSWHAQLS+NLAMLGSLTI+VAHHMYSMP
Sbjct: 301 WGIGHGLKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSLNLAMLGSLTIVVAHHMYSMP 360
Query: 373 PYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRD 432
PYPYLA DYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRD
Sbjct: 361 PYPYLAIDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRD 420
Query: 433 AIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALAP 492
AIISHLNW CIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQW+QNTHALAP
Sbjct: 421 AIISHLNWACIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWVQNTHALAP 480
Query: 493 GITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGV 552
G TAPGATT TSLTWGGGDL+AVG KVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGV
Sbjct: 481 GATAPGATTSTSLTWGGGDLIAVGGKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGV 540
Query: 553 LFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKM 612
LFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKM
Sbjct: 541 LFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKM 600
Query: 613 QSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFFL 672
QSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFFL
Sbjct: 601 QSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFFL 660
Query: 673 GAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTH 729
GAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTH
Sbjct: 661 GAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTH 717
>gb|AAF65219.1| (AF223227) photosystem protein [Pisum sativum]
Length = 719
Score = 1480 bits (3790), Expect = 0.0
Identities = 697/719 (96%), Positives = 708/719 (97%)
Query: 11 KILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKI 70
KI VDRDPIKTSFE+WAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLE+ISRK+
Sbjct: 1 KIXVDRDPIKTSFEQWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEEISRKV 60
Query: 71 FSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGG 130
FSAHFGQLSIIFLWLSGMYFHGARFSNYEAWL+DPTHI PSAQVVWPIVGQEILNGDVGG
Sbjct: 61 FSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLNDPTHIGPSAQVVWPIVGQEILNGDVGG 120
Query: 131 GFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQD 190
GFRGIQITSGFFQIWRASGITSELQLYCTAIG LVFA LMLFAGWFHYHKAAPKLAWFQD
Sbjct: 121 GFRGIQITSGFFQIWRASGITSELQLYCTAIGALVFAGLMLFAGWFHYHKAAPKLAWFQD 180
Query: 191 VESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQL 250
VESMLNHHLAGLLGLGSLSWA HQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQL
Sbjct: 181 VESMLNHHLAGLLGLGSLSWARHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQL 240
Query: 251 YPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYR 310
YPSFAEGATPFFTLNW+KYADFLTFRGGLDP+TGGLWLTDIAHHHLAIAILFLIAGHMYR
Sbjct: 241 YPSFAEGATPFFTLNWSKYADFLTFRGGLDPLTGGLWLTDIAHHHLAIAILFLIAGHMYR 300
Query: 311 TNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYS 370
TNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTI+VA HMYS
Sbjct: 301 TNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIVVAQHMYS 360
Query: 371 MPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRH 430
MPPYPYLATDY TQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRH
Sbjct: 361 MPPYPYLATDYATQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRH 420
Query: 431 RDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHAL 490
RDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQNTHAL
Sbjct: 421 RDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQNTHAL 480
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
APG TAPGAT TSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK
Sbjct: 481 APGTTAPGATASTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 540
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSW 610
GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+ISVVIFHFSW
Sbjct: 541 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSISVVIFHFSW 600
Query: 611 KMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLF 670
KMQSDVWG+I+DQGVVTHIT GNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLF
Sbjct: 601 KMQSDVWGTINDQGVVTHITAGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLF 660
Query: 671 FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTH 729
FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTH
Sbjct: 661 FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTH 719
>sp|P05310|PSAA_PEA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||S00703 photosystem I protein A1 - garden pea chloroplast
emb|CAA29003.1| (X05423) P700 chlorophyll a-apoproteins 84 KD protein [Pisum
sativum]
prf||1303353A gene psaA1 [Pisum sativum]
Length = 761
Score = 1480 bits (3789), Expect = 0.0
Identities = 714/776 (92%), Positives = 724/776 (93%), Gaps = 41/776 (5%)
Query: 1 MIIRSPEP--------EVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHAD 52
MIIRSPEP EVKILVDRDPIKTSFE+WAKPGHFSRTIAKGPDTTTWIWNLHAD
Sbjct: 1 MIIRSPEPKVQILADPEVKILVDRDPIKTSFEQWAKPGHFSRTIAKGPDTTTWIWNLHAD 60
Query: 53 AHDFDSHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSA 112
AHDFDSHTSDLE+ISRK+FSAHFGQLSIIFLWLSGMYFHGARFSNYEAWL+DPTHIRPSA
Sbjct: 61 AHDFDSHTSDLEEISRKVFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLNDPTHIRPSA 120
Query: 113 QVVWPIVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLF 172
QVVWPIVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIG LVFAALMLF
Sbjct: 121 QVVWPIVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGALVFAALMLF 180
Query: 173 AGWFHYHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPK 232
AGWFHYHKAAPKL WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVD
Sbjct: 181 AGWFHYHKAAPKLVWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVD-- 238
Query: 233 EIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIA 292
LLAQLYPSFAEGATPFFTLNW+KYADFLTFRGGLDP+TGGLWLTDIA
Sbjct: 239 -------------LLAQLYPSFAEGATPFFTLNWSKYADFLTFRGGLDPLTGGLWLTDIA 285
Query: 293 HHHLAIAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKG--------------- 337
HHHLAIAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKG
Sbjct: 286 HHHLAIAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGPFTGQGHKGPFTGQG 345
Query: 338 ---LYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWI 394
LYEILTTSWHAQLSINLAMLGSLTII AHHM +MPP PYLATDYGTQLSLFTHHMWI
Sbjct: 346 HKGLYEILTTSWHAQLSINLAMLGSLTIIAAHHMIAMPPIPYLATDYGTQLSLFTHHMWI 405
Query: 395 GGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYI 454
GGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYI
Sbjct: 406 GGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYI 465
Query: 455 HNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVA 514
HNDTMSALGRPQDMFSDTAIQLQP+FAQWIQNTHALAPG TAPGATT TSL WGGGDLV+
Sbjct: 466 HNDTMSALGRPQDMFSDTAIQLQPVFAQWIQNTHALAPGTTAPGATTSTSLIWGGGDLVS 525
Query: 515 VGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCD 574
VG KVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCD
Sbjct: 526 VGGKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCD 585
Query: 575 GPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNF 634
GPGRGGTCQVSAW HVFLGLFWMYNAISVVIFHFSWKMQSDVWGSI+DQGVVTHITGG F
Sbjct: 586 GPGRGGTCQVSAWVHVFLGLFWMYNAISVVIFHFSWKMQSDVWGSINDQGVVTHITGGRF 645
Query: 635 AQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQE 694
AQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQE
Sbjct: 646 AQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQE 705
Query: 695 LIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
LIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG
Sbjct: 706 LIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 761
>gb|AAF29819.1| (AF180018) photosystem I P700 apoprotein A1 [Araucaria araucana]
Length = 716
Score = 1439 bits (3685), Expect = 0.0
Identities = 670/716 (93%), Positives = 698/716 (96%)
Query: 14 VDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKIFSA 73
V+RDPIKTSFE+WAKPGHFS+T++KGP+TTTWIWNLHADAHDFDSHT DLE+ISRK+FSA
Sbjct: 1 VERDPIKTSFEKWAKPGHFSKTLSKGPNTTTWIWNLHADAHDFDSHTDDLEEISRKVFSA 60
Query: 74 HFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGGFR 133
HFGQL+II +WLSGMYFHGARFSNYEAWLSDPTHI+PSAQVVWPIVGQEILNGDVGGGFR
Sbjct: 61 HFGQLAIILIWLSGMYFHGARFSNYEAWLSDPTHIKPSAQVVWPIVGQEILNGDVGGGFR 120
Query: 134 GIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDVES 193
GIQITSGFFQ+WRASGITSELQLYCTAIGGL+FA LMLFAGWFHYHKAAPKLAWFQDVES
Sbjct: 121 GIQITSGFFQLWRASGITSELQLYCTAIGGLIFAGLMLFAGWFHYHKAAPKLAWFQDVES 180
Query: 194 MLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPS 253
MLNHHLAGLLGLGSLSWAGHQVHVSLPINQ L+AGVDPKEIPLPHEFILNRDLLAQLYPS
Sbjct: 181 MLNHHLAGLLGLGSLSWAGHQVHVSLPINQLLDAGVDPKEIPLPHEFILNRDLLAQLYPS 240
Query: 254 FAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNW 313
FA+G TPFFTLNW++Y+DFLTFRGGL+PVTGGLWLTD HHHLAIAILFLIAGHMYRTNW
Sbjct: 241 FAKGLTPFFTLNWSEYSDFLTFRGGLNPVTGGLWLTDTVHHHLAIAILFLIAGHMYRTNW 300
Query: 314 GIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPP 373
IGH +K+ILEAHKGPFTG+GHKGLYEILTTSWHAQL++NLAMLGSLTI+VAHHMYSMPP
Sbjct: 301 SIGHSLKEILEAHKGPFTGEGHKGLYEILTTSWHAQLALNLAMLGSLTIVVAHHMYSMPP 360
Query: 374 YPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDA 433
YPYLA DYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTT+YN+LLDRVLRHRDA
Sbjct: 361 YPYLAIDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTQYNNLLDRVLRHRDA 420
Query: 434 IISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALAPG 493
IISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA AP
Sbjct: 421 IISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHASAPS 480
Query: 494 ITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVL 553
+TAP AT TSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVL
Sbjct: 481 LTAPDATASTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVL 540
Query: 554 FARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKMQ 613
FARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKMQ
Sbjct: 541 FARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKMQ 600
Query: 614 SDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFFLG 673
SDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGL FLG
Sbjct: 601 SDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLLFLG 660
Query: 674 AHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTH 729
AHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRALSIVQGRAVGV H
Sbjct: 661 AHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRALSIVQGRAVGVAH 716
>gb|AAF29813.1| (AF180012) photosystem I P700 apoprotein A1 [Sequoia sempervirens]
Length = 720
Score = 1439 bits (3684), Expect = 0.0
Identities = 667/720 (92%), Positives = 703/720 (97%)
Query: 10 VKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRK 69
+KI+V+RDPIKTSFE+WAKPGHFS+T+AKGP+TTTWIWNLHADAHDFDSHT+DLE+ISRK
Sbjct: 1 IKIVVERDPIKTSFEKWAKPGHFSKTLAKGPNTTTWIWNLHADAHDFDSHTNDLEEISRK 60
Query: 70 IFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVG 129
+FSAHFGQL+IIF+WLSGMYFHGARFSNYEAWL DPTHI+PSAQVVWPIVGQEILNGDVG
Sbjct: 61 VFSAHFGQLAIIFIWLSGMYFHGARFSNYEAWLGDPTHIKPSAQVVWPIVGQEILNGDVG 120
Query: 130 GGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQ 189
GGFRGIQITSGFFQIWRASGITSELQLYCTAIG L+FAALMLFAGWFHYHKAAPKLAWFQ
Sbjct: 121 GGFRGIQITSGFFQIWRASGITSELQLYCTAIGALIFAALMLFAGWFHYHKAAPKLAWFQ 180
Query: 190 DVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQ 249
DVESMLNHHLAGLLGLGSLSWAGHQ+HVSLPIN+ L+AGVDPKEIPLPHEFILNR+LLAQ
Sbjct: 181 DVESMLNHHLAGLLGLGSLSWAGHQIHVSLPINELLDAGVDPKEIPLPHEFILNRELLAQ 240
Query: 250 LYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMY 309
LYPSFA+G TPFFTLNW++Y++FLTFRGGL+PVTGGLWLTD AHHHLAIAILFLIAGHMY
Sbjct: 241 LYPSFAKGLTPFFTLNWSEYSEFLTFRGGLNPVTGGLWLTDTAHHHLAIAILFLIAGHMY 300
Query: 310 RTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMY 369
RTNW IGH +K+ILEAHKGPFTG+GH+GLYEILTTSWHAQL++NLAMLGSLTI+VAHHMY
Sbjct: 301 RTNWSIGHNLKEILEAHKGPFTGEGHRGLYEILTTSWHAQLALNLAMLGSLTIVVAHHMY 360
Query: 370 SMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLR 429
SMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTT+YN+LLDRVLR
Sbjct: 361 SMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTQYNNLLDRVLR 420
Query: 430 HRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA 489
HRDAI+SHLNW CIFLGFHSFGLYIHNDTMSALGRP+DMFSDTAIQLQPIFAQWIQNTHA
Sbjct: 421 HRDAIVSHLNWACIFLGFHSFGLYIHNDTMSALGRPKDMFSDTAIQLQPIFAQWIQNTHA 480
Query: 490 LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILL 549
LAP +TAP AT TSLTWGGGDLVAVG KVALLPIPLGTADFLVHHIHAFTIHVTVLILL
Sbjct: 481 LAPSLTAPDATASTSLTWGGGDLVAVGAKVALLPIPLGTADFLVHHIHAFTIHVTVLILL 540
Query: 550 KGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFS 609
KGVLFARSSRLIPDK NLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFS
Sbjct: 541 KGVLFARSSRLIPDKVNLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFS 600
Query: 610 WKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGL 669
WKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGL
Sbjct: 601 WKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGL 660
Query: 670 FFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTH 729
FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRALSIVQGRAVGV H
Sbjct: 661 LFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRALSIVQGRAVGVAH 720
>gb|AAF29826.1| (AF180025) photosystem I P700 apoprotein A1 [Torreya californica]
Length = 719
Score = 1436 bits (3676), Expect = 0.0
Identities = 668/718 (93%), Positives = 699/718 (97%)
Query: 10 VKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRK 69
+KI+V+RDPIKTSFE+WAKPGHFS+T+AKGP+TTTWIWNLHADAHDFDSHT+DLE+ISRK
Sbjct: 1 LKIVVERDPIKTSFEKWAKPGHFSKTLAKGPNTTTWIWNLHADAHDFDSHTNDLEEISRK 60
Query: 70 IFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVG 129
+FSAHFGQL+IIF+WLSGMYFHGARFSNYEAWL DPTHI+PSAQVVWPIVGQEILNGDVG
Sbjct: 61 VFSAHFGQLAIIFIWLSGMYFHGARFSNYEAWLGDPTHIKPSAQVVWPIVGQEILNGDVG 120
Query: 130 GGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQ 189
GGFRGIQITSGFFQIWRASGITSELQLYCTA G L+FAALMLFAGWFHYHKAAPKLAWFQ
Sbjct: 121 GGFRGIQITSGFFQIWRASGITSELQLYCTATGALIFAALMLFAGWFHYHKAAPKLAWFQ 180
Query: 190 DVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQ 249
DVESMLNHHLAGLLGLGSL WAGHQVHVSLPINQ L+AGVDPKEIPLPHEFI NRDLLAQ
Sbjct: 181 DVESMLNHHLAGLLGLGSLGWAGHQVHVSLPINQLLDAGVDPKEIPLPHEFISNRDLLAQ 240
Query: 250 LYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMY 309
LYPSFAEG TPFFTLNW++Y+DFLTFRGGL+PVTGGLWLTD AHHHLAIAILFLIAGHMY
Sbjct: 241 LYPSFAEGLTPFFTLNWSEYSDFLTFRGGLNPVTGGLWLTDTAHHHLAIAILFLIAGHMY 300
Query: 310 RTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMY 369
RTNWGIGH +K+ILEAHKGPFTG+GH+GLYEILTTSWHAQL++NLAMLGSLTI+VAHHMY
Sbjct: 301 RTNWGIGHNLKEILEAHKGPFTGEGHRGLYEILTTSWHAQLALNLAMLGSLTIVVAHHMY 360
Query: 370 SMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLR 429
SMPPYPYLA DYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTT+YN+LLDRVLR
Sbjct: 361 SMPPYPYLAIDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTQYNNLLDRVLR 420
Query: 430 HRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA 489
HRDAI+SHLNW CIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA
Sbjct: 421 HRDAIVSHLNWACIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA 480
Query: 490 LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILL 549
+P +TAP AT TSLTWGGGDLVAVG KVALLPIPLGTADFLVHHIHAFTIHVTVLILL
Sbjct: 481 SSPSLTAPDATASTSLTWGGGDLVAVGAKVALLPIPLGTADFLVHHIHAFTIHVTVLILL 540
Query: 550 KGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFS 609
KGVLFARSSRLIPDK NLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFS
Sbjct: 541 KGVLFARSSRLIPDKVNLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFS 600
Query: 610 WKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGL 669
WKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGL
Sbjct: 601 WKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGL 660
Query: 670 FFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGV 727
FFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRALSIVQGRAVGV
Sbjct: 661 FFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRALSIVQGRAVGV 718
>gb|AAF29812.1| (AF180011) photosystem I P700 apoprotein A1 [Encephalartos
lebomboensis]
Length = 719
Score = 1433 bits (3668), Expect = 0.0
Identities = 665/719 (92%), Positives = 699/719 (96%)
Query: 12 ILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKIF 71
I+V+RDPI+TSFE+WAKPGHFSRT+AKGP+TTTWIWNLHADAHDF SHT+DLE+ISRK+F
Sbjct: 1 IVVERDPIRTSFEKWAKPGHFSRTLAKGPNTTTWIWNLHADAHDFGSHTNDLEEISRKVF 60
Query: 72 SAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGG 131
SAHFGQL+IIF+WLSGMYFHGARFSNYEAWLSDPTHI+PSAQVVWPIVGQEILNGDVGGG
Sbjct: 61 SAHFGQLAIIFIWLSGMYFHGARFSNYEAWLSDPTHIKPSAQVVWPIVGQEILNGDVGGG 120
Query: 132 FRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDV 191
RGIQITSG FQ+WRASGITSELQLYCTAIG L+FAALMLFAGWFHYHKAAPKLAWFQDV
Sbjct: 121 SRGIQITSGLFQLWRASGITSELQLYCTAIGALIFAALMLFAGWFHYHKAAPKLAWFQDV 180
Query: 192 ESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLY 251
ESMLNHHLAGLLG+GSLSWAGHQVHVSLPINQ L+AGVDPKEIPLPHEFILNRDLLAQLY
Sbjct: 181 ESMLNHHLAGLLGIGSLSWAGHQVHVSLPINQLLDAGVDPKEIPLPHEFILNRDLLAQLY 240
Query: 252 PSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRT 311
PSFAEG TP FTLNW++Y++FLTFRGGL+PVTGGLWLTD AHHHLAIAILFLIAGHMYRT
Sbjct: 241 PSFAEGLTPLFTLNWSEYSEFLTFRGGLNPVTGGLWLTDTAHHHLAIAILFLIAGHMYRT 300
Query: 312 NWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSM 371
NWGIGH +K+ILEAHKGPFTG+GHKGLYEILTTSWHAQL++NLAMLGSLTI+VAHHMYSM
Sbjct: 301 NWGIGHSLKEILEAHKGPFTGEGHKGLYEILTTSWHAQLALNLAMLGSLTIVVAHHMYSM 360
Query: 372 PPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHR 431
PPYPYLA DYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTT+YN+LLDRVLRHR
Sbjct: 361 PPYPYLAIDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTQYNNLLDRVLRHR 420
Query: 432 DAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALA 491
DAI+SHLNW CIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQW+QNTHALA
Sbjct: 421 DAIVSHLNWACIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWVQNTHALA 480
Query: 492 PGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKG 551
PG TAP AT TS+TWGGGDLVAVG KVALLPIPLGTADFLVHHIHAFTIHVTVLILLKG
Sbjct: 481 PGSTAPDATASTSVTWGGGDLVAVGGKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKG 540
Query: 552 VLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWK 611
VLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWK
Sbjct: 541 VLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWK 600
Query: 612 MQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFF 671
MQSDVWGSISDQG+VTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGL F
Sbjct: 601 MQSDVWGSISDQGMVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLLF 660
Query: 672 LGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHY 730
LGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKL+VAP QPRALSIVQGRAVGV HY
Sbjct: 661 LGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLEVAPVIQPRALSIVQGRAVGVAHY 719
>gb|AAF29821.1| (AF180020) photosystem I P700 apoprotein A1 [Angiopteris evecta]
gb|AAF65218.1| (AF223226) photosystem protein [Ginkgo biloba]
Length = 721
Score = 1432 bits (3667), Expect = 0.0
Identities = 667/721 (92%), Positives = 699/721 (96%)
Query: 10 VKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRK 69
VKI+V+RDPIKTSFE+WAKPGHFSRT+AKGP+TTTWIWNLHADAHDFDSHT+DLE+ISRK
Sbjct: 1 VKIVVERDPIKTSFEKWAKPGHFSRTLAKGPNTTTWIWNLHADAHDFDSHTNDLEEISRK 60
Query: 70 IFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVG 129
+FSAHFGQL+II +WLSGMYFHGARFSNYEAWLSDPT I+PSAQVVWPIVGQEILNGDVG
Sbjct: 61 VFSAHFGQLAIILIWLSGMYFHGARFSNYEAWLSDPTRIKPSAQVVWPIVGQEILNGDVG 120
Query: 130 GGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQ 189
GGFRGIQITSGFFQIWRASGITSELQLYCTAIG L+FAALMLFAGWFHYHKAAPKLAWFQ
Sbjct: 121 GGFRGIQITSGFFQIWRASGITSELQLYCTAIGALIFAALMLFAGWFHYHKAAPKLAWFQ 180
Query: 190 DVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQ 249
DVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQ L+AGVDPKEIPLPHEFI NRDL+AQ
Sbjct: 181 DVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQLLDAGVDPKEIPLPHEFISNRDLVAQ 240
Query: 250 LYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMY 309
LYPSFA+G TPFF LNW++Y++FLTFRGGL+PVTGGLWLTD AHHHLAIAILFLIAGHMY
Sbjct: 241 LYPSFAKGLTPFFALNWSEYSEFLTFRGGLNPVTGGLWLTDTAHHHLAIAILFLIAGHMY 300
Query: 310 RTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMY 369
RTNWGIGH +K+ILEAHKGPFTG+GHKG+YEILTTSWHAQL++NLAMLGSLTI+VAHHMY
Sbjct: 301 RTNWGIGHSLKEILEAHKGPFTGEGHKGIYEILTTSWHAQLALNLAMLGSLTIVVAHHMY 360
Query: 370 SMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLR 429
SMPPYPYLA DYGTQLSLFTHHMWI GFLIVGAAAHAAIFMVRDYDPT +YN+LLDRVLR
Sbjct: 361 SMPPYPYLAIDYGTQLSLFTHHMWIRGFLIVGAAAHAAIFMVRDYDPTIQYNNLLDRVLR 420
Query: 430 HRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA 489
HRDAI+SHLNW CIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA
Sbjct: 421 HRDAIVSHLNWACIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA 480
Query: 490 LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILL 549
AP TAPGA TSLTWGGGDL+AVG KVALLPIPLGTADFLVHHIHAFTIHVTVLILL
Sbjct: 481 SAPSSTAPGAAASTSLTWGGGDLLAVGGKVALLPIPLGTADFLVHHIHAFTIHVTVLILL 540
Query: 550 KGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFS 609
KGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFS
Sbjct: 541 KGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFS 600
Query: 610 WKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGL 669
WKMQSDVWGSISD+GVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGL
Sbjct: 601 WKMQSDVWGSISDRGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGL 660
Query: 670 FFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTH 729
FFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRALSIVQGRAVGV H
Sbjct: 661 FFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRALSIVQGRAVGVAH 720
Query: 730 Y 730
Y
Sbjct: 721 Y 721
>gb|AAF29820.1| (AF180019) photosystem I P700 apoprotein A1 [Equisetum palustre]
Length = 719
Score = 1421 bits (3639), Expect = 0.0
Identities = 652/719 (90%), Positives = 701/719 (96%)
Query: 11 KILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKI 70
KI+V++DP++TSFE+WA+PGHFS+T+AKGP+TTTWIWNLHADAHDFDSHT+DLEDISRKI
Sbjct: 1 KIIVEKDPVRTSFEKWAQPGHFSKTLAKGPNTTTWIWNLHADAHDFDSHTNDLEDISRKI 60
Query: 71 FSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGG 130
FSAHFGQLSIIF+WLSGMYFHGARFSNYEAWLSDPTHI+PSAQVVWPIVGQEILNGDVGG
Sbjct: 61 FSAHFGQLSIIFIWLSGMYFHGARFSNYEAWLSDPTHIKPSAQVVWPIVGQEILNGDVGG 120
Query: 131 GFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQD 190
GF+GIQITSGFFQ+WRASGIT+ELQLYCTAIG L+FA LMLFAGWFHYHKAAPKLAWFQD
Sbjct: 121 GFQGIQITSGFFQLWRASGITNELQLYCTAIGALIFAGLMLFAGWFHYHKAAPKLAWFQD 180
Query: 191 VESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQL 250
VESMLNHHLAGLLGLGSL WAGHQVHVSLPINQ L++GVDP+EIPLPHEFILNRDLLAQL
Sbjct: 181 VESMLNHHLAGLLGLGSLGWAGHQVHVSLPINQLLDSGVDPREIPLPHEFILNRDLLAQL 240
Query: 251 YPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYR 310
YPSF+EG TPFF L W+KY+DFLTFRGGL+PVTGGLWLTD AHHH+AIA+LFLIAGHMYR
Sbjct: 241 YPSFSEGLTPFFNLEWSKYSDFLTFRGGLNPVTGGLWLTDTAHHHIAIAVLFLIAGHMYR 300
Query: 311 TNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYS 370
TNWGIGH IK+ILEAHKGPFTG+GH+GL+EILT+SWHAQL++NLAMLGSLTIIVAHHMY+
Sbjct: 301 TNWGIGHSIKEILEAHKGPFTGEGHRGLFEILTSSWHAQLAVNLAMLGSLTIIVAHHMYA 360
Query: 371 MPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRH 430
MPPYPYLATDYGTQLSLFTHHMWIGGFL+VGAAAHAAIFMVRDYDP+T+YN+LLDRV+RH
Sbjct: 361 MPPYPYLATDYGTQLSLFTHHMWIGGFLVVGAAAHAAIFMVRDYDPSTQYNNLLDRVIRH 420
Query: 431 RDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHAL 490
RDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHAL
Sbjct: 421 RDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHAL 480
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
AP +TAP AT TSLTWGGGDLV+VG +VALLPIPLGTADFLVHHIHAFTIHVTVLILLK
Sbjct: 481 APSLTAPNATASTSLTWGGGDLVSVGGRVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 540
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSW 610
GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+ISVVIFHFSW
Sbjct: 541 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSISVVIFHFSW 600
Query: 611 KMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLF 670
KMQSDVWGSI++QGV++HITGGNFAQS+ TINGWLRDFLWAQASQVIQSYGSSLSAYGL
Sbjct: 601 KMQSDVWGSINEQGVISHITGGNFAQSATTINGWLRDFLWAQASQVIQSYGSSLSAYGLL 660
Query: 671 FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTH 729
FLGAHFVWAFSLMFLFSGRGYWQELIESI+WAHNKLKVAPA QPRALSIVQGRAVGV H
Sbjct: 661 FLGAHFVWAFSLMFLFSGRGYWQELIESILWAHNKLKVAPAIQPRALSIVQGRAVGVAH 719
>gb|AAF29824.1| (AF180023) photosystem I P700 apoprotein A1 [Psilotum nudum]
Length = 719
Score = 1417 bits (3627), Expect = 0.0
Identities = 652/719 (90%), Positives = 695/719 (95%)
Query: 12 ILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKIF 71
ILV+RDP+KTS E+WA+PGHFSRT+AKGP+TTTWIWNLHAD+HDFDSHT+DLE+ISRK+F
Sbjct: 1 ILVERDPVKTSLEKWAQPGHFSRTLAKGPNTTTWIWNLHADSHDFDSHTNDLEEISRKVF 60
Query: 72 SAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGG 131
SAHFGQL+IIF+WLSGMYFHGARFSNYEAWLSDP HI+PSAQVVWPIVGQEILNGDVGGG
Sbjct: 61 SAHFGQLAIIFVWLSGMYFHGARFSNYEAWLSDPIHIKPSAQVVWPIVGQEILNGDVGGG 120
Query: 132 FRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDV 191
F+GIQITSGFFQIWRASGITSELQLYCTAIG L+FA LMLFAGWFHYHKAAPKLAWFQDV
Sbjct: 121 FQGIQITSGFFQIWRASGITSELQLYCTAIGALIFATLMLFAGWFHYHKAAPKLAWFQDV 180
Query: 192 ESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLY 251
ESMLNHHLAGLLGLGSL+WAGHQVHVSLPINQ L+AGVDPKEIPLPHEFILNRDLLAQLY
Sbjct: 181 ESMLNHHLAGLLGLGSLAWAGHQVHVSLPINQLLDAGVDPKEIPLPHEFILNRDLLAQLY 240
Query: 252 PSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRT 311
PSFA+G PFFTLNW++Y+DFLTFRGGL+PVTGGLWLTD HHHLAIA+LFL+AGHMYRT
Sbjct: 241 PSFAKGLIPFFTLNWSEYSDFLTFRGGLNPVTGGLWLTDTVHHHLAIAVLFLVAGHMYRT 300
Query: 312 NWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSM 371
NW IGH IK+ILEAH+GPFTG+GHKG++EILTTSWHAQL++NLAMLGSLTIIVAHHMYSM
Sbjct: 301 NWSIGHSIKEILEAHEGPFTGEGHKGIFEILTTSWHAQLALNLAMLGSLTIIVAHHMYSM 360
Query: 372 PPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHR 431
PPYPYLA DYGTQLSLFTHHMWIGGFL+VGAAAHAAIFMVRDYDPTT+YN+LLDRVLRHR
Sbjct: 361 PPYPYLAIDYGTQLSLFTHHMWIGGFLVVGAAAHAAIFMVRDYDPTTQYNNLLDRVLRHR 420
Query: 432 DAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALA 491
DAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQW+QNTHA A
Sbjct: 421 DAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWVQNTHASA 480
Query: 492 PGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKG 551
PG+TAP A TSLTWGG +L+AVG KVALLPIPLGTADFLVHHIHAFTIHVTVLILLKG
Sbjct: 481 PGLTAPNAIASTSLTWGGDNLIAVGGKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKG 540
Query: 552 VLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWK 611
VLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+IS+VIFHFSWK
Sbjct: 541 VLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSISIVIFHFSWK 600
Query: 612 MQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFF 671
MQSDVWGSI+DQGV+ HITGGNFAQSS TINGWLRDFLWAQASQVIQSYGSSLSAYGL F
Sbjct: 601 MQSDVWGSINDQGVINHITGGNFAQSSTTINGWLRDFLWAQASQVIQSYGSSLSAYGLLF 660
Query: 672 LGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHY 730
LGAHFVWAFSLMFLFSGRGYWQELIESI+WAHNKLKVAPA QPRALSIVQGRAVGV HY
Sbjct: 661 LGAHFVWAFSLMFLFSGRGYWQELIESIIWAHNKLKVAPAIQPRALSIVQGRAVGVAHY 719
>gb|AAF29816.1| (AF180015) photosystem I P700 apoprotein A1 [Cycas revoluta]
Length = 717
Score = 1414 bits (3619), Expect = 0.0
Identities = 659/716 (92%), Positives = 689/716 (96%)
Query: 12 ILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKIF 71
I+V+ DPIKTSFE+WAKPGHFS T+ KGP+TTTWIWNLHADAHDF SHT+DLE+ISRK+F
Sbjct: 1 IVVEMDPIKTSFEKWAKPGHFSXTLXKGPNTTTWIWNLHADAHDFGSHTNDLEEISRKVF 60
Query: 72 SAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGG 131
AHFGQL+II +WLSGMYFHGARFSNYEAWLSDPTHI+PSAQVVWPIVGQEILNGDVGGG
Sbjct: 61 RAHFGQLAIILIWLSGMYFHGARFSNYEAWLSDPTHIKPSAQVVWPIVGQEILNGDVGGG 120
Query: 132 FRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDV 191
RGIQITSG FQIWRASGITSELQLYCTAIG L+FAALMLFAGWFHYHKAAPKLAWFQDV
Sbjct: 121 SRGIQITSGLFQIWRASGITSELQLYCTAIGALIFAALMLFAGWFHYHKAAPKLAWFQDV 180
Query: 192 ESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLY 251
ESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQ L+AGVDPKEIPLPHEFI NRDLLAQLY
Sbjct: 181 ESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQLLDAGVDPKEIPLPHEFISNRDLLAQLY 240
Query: 252 PSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRT 311
PSFAEG TP FTLNW++Y++FLTFRGGL+PVTGGLWLTD AHHHLAIAILFLIAGHMYRT
Sbjct: 241 PSFAEGLTPLFTLNWSEYSEFLTFRGGLNPVTGGLWLTDTAHHHLAIAILFLIAGHMYRT 300
Query: 312 NWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSM 371
NWGIGH +K+ILE HKGPFTG+GHKGLYEILTTSWHAQL++NLAMLGSLTI+VAHHMYSM
Sbjct: 301 NWGIGHSLKEILETHKGPFTGEGHKGLYEILTTSWHAQLALNLAMLGSLTIVVAHHMYSM 360
Query: 372 PPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHR 431
PPYPYLA DYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTT+YN+LLDRVLRHR
Sbjct: 361 PPYPYLAIDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTQYNNLLDRVLRHR 420
Query: 432 DAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALA 491
DAI+SHLNW CIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQW+QNTHALA
Sbjct: 421 DAIVSHLNWACIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWVQNTHALA 480
Query: 492 PGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKG 551
PG TAP AT TSLTWGGGDLVAVG KVALLPIPLGTADFLVHHIHAFTIHVTVLILLKG
Sbjct: 481 PGSTAPDATASTSLTWGGGDLVAVGGKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKG 540
Query: 552 VLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWK 611
VLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWK
Sbjct: 541 VLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWK 600
Query: 612 MQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFF 671
MQSDVWGSISDQG+VTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGL F
Sbjct: 601 MQSDVWGSISDQGMVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLLF 660
Query: 672 LGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGV 727
LGAH VWAFSLMFLFSGRGYWQELIESIVWAHNKL+VAP QPRALSIVQGRAVGV
Sbjct: 661 LGAHXVWAFSLMFLFSGRGYWQELIESIVWAHNKLEVAPVIQPRALSIVQGRAVGV 716
>gb|AAF29825.1| (AF180024) photosystem I P700 apoprotein A1 [Huperzia squarrosa]
Length = 715
Score = 1412 bits (3616), Expect = 0.0
Identities = 655/714 (91%), Positives = 689/714 (95%)
Query: 11 KILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKI 70
KI V+RDP+KTSFE+WA+PGHFSRT+AKGP TTTWIWNLHADAHDFDSHT+DLEDISRK+
Sbjct: 1 KIAVERDPVKTSFEKWAQPGHFSRTLAKGPSTTTWIWNLHADAHDFDSHTNDLEDISRKV 60
Query: 71 FSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGG 130
FSAHFGQL+IIF+WLSGMYFHGARFSNYEAWLSDPTH++PSAQVVWPIVGQEILNGDVGG
Sbjct: 61 FSAHFGQLAIIFIWLSGMYFHGARFSNYEAWLSDPTHVKPSAQVVWPIVGQEILNGDVGG 120
Query: 131 GFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQD 190
GF+GIQITSGFFQIWRASGITSELQLY TAIGGL+FAALMLFAGWFHYHKAAPKL WFQD
Sbjct: 121 GFQGIQITSGFFQIWRASGITSELQLYSTAIGGLIFAALMLFAGWFHYHKAAPKLTWFQD 180
Query: 191 VESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQL 250
VESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQ L+AGVD KEIPLPHEFILNRDL+ QL
Sbjct: 181 VESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQLLDAGVDAKEIPLPHEFILNRDLMTQL 240
Query: 251 YPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYR 310
YPSFA+G TPFFTLNW++Y+DF TFRGGL+PVTGGLWLTD HHHLAIA+LFLIAGHMYR
Sbjct: 241 YPSFAKGLTPFFTLNWSEYSDFSTFRGGLNPVTGGLWLTDTVHHHLAIAVLFLIAGHMYR 300
Query: 311 TNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYS 370
TNWGIGH +K+ILEAHKGPFTG+GHKGLYEI TTSWHAQL++NLAMLGSLTIIVAHHMYS
Sbjct: 301 TNWGIGHSLKEILEAHKGPFTGEGHKGLYEIFTTSWHAQLALNLAMLGSLTIIVAHHMYS 360
Query: 371 MPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRH 430
MPPYPYLATDYGTQLSLFTHHMWIGGFL+VGAAAHAAIFMVRDYDPTT+YN+LLDRVLRH
Sbjct: 361 MPPYPYLATDYGTQLSLFTHHMWIGGFLVVGAAAHAAIFMVRDYDPTTQYNNLLDRVLRH 420
Query: 431 RDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHAL 490
RDAIISHLNW CIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQW+QNTHA+
Sbjct: 421 RDAIISHLNWACIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWVQNTHAV 480
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
AP TAP A TSLTWGG DLVAVG KVALLPIPLGTADFLVHHIHAFTIHVTVLILLK
Sbjct: 481 APFSTAPNAAASTSLTWGGIDLVAVGGKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 540
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSW 610
GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSW
Sbjct: 541 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSW 600
Query: 611 KMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLF 670
KMQSDVWGS+SDQ +VTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGL
Sbjct: 601 KMQSDVWGSVSDQKIVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLL 660
Query: 671 FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRA 724
FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRALSIVQGRA
Sbjct: 661 FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRALSIVQGRA 714
>gi|11467832 P700 apoprotein A1 of photosystem I [Nephroselmis olivacea]
gb|AAD54854.1|AF137379_77 (AF137379) P700 apoprotein A1 of photosystem I [Nephroselmis
olivacea]
Length = 751
Score = 1411 bits (3612), Expect = 0.0
Identities = 655/753 (86%), Positives = 707/753 (92%), Gaps = 5/753 (0%)
Query: 1 MIIRSPEPE---VKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFD 57
M I PE E VKI+VDR+P+ TSFE+WAKPGHFSRT+AKGP TTTWIW+LHADAHDFD
Sbjct: 1 MTISPPEREAKKVKIVVDRNPVVTSFEKWAKPGHFSRTLAKGPTTTTWIWDLHADAHDFD 60
Query: 58 SHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWP 117
SHT+DLEDIS KIFSAHFGQL +I +WLSGMYFHGARFSNYEAWLSDPTHI+PSAQVVWP
Sbjct: 61 SHTTDLEDISPKIFSAHFGQLGVILIWLSGMYFHGARFSNYEAWLSDPTHIKPSAQVVWP 120
Query: 118 IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFH 177
IVGQEILNGDVGGGF+GIQITSGFFQ+WRASGITSELQLY TAIGGL+ AA M FAGWFH
Sbjct: 121 IVGQEILNGDVGGGFQGIQITSGFFQLWRASGITSELQLYSTAIGGLLLAAAMFFAGWFH 180
Query: 178 YHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLP 237
YHKAAPKL WFQ+VESM+NHHL GLLGLGSL WAGHQ+HVSLP+N+ LNAGVDPKEIPLP
Sbjct: 181 YHKAAPKLEWFQNVESMMNHHLGGLLGLGSLGWAGHQIHVSLPVNKLLNAGVDPKEIPLP 240
Query: 238 HEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLA 297
HEF+LNRDL+AQLYPSFA+G TPFFTLNWA+Y DFLTFRGGL+PVTGGLWL+D AHHH+A
Sbjct: 241 HEFLLNRDLMAQLYPSFAKGLTPFFTLNWAEYGDFLTFRGGLNPVTGGLWLSDTAHHHVA 300
Query: 298 IAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAML 357
IA+LFL+AGHMYRTNWGIGH +K+ILEAHKGPFTG+GHKGLYEILTTSWHAQL+INLA+
Sbjct: 301 IAVLFLVAGHMYRTNWGIGHSMKEILEAHKGPFTGEGHKGLYEILTTSWHAQLAINLALF 360
Query: 358 GSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPT 417
GSL+I+VAHHMY+MPPYPYLATDYGTQLSLFTHHMWIGGF +VGAAAHAAIFMVRDYDPT
Sbjct: 361 GSLSIVVAHHMYAMPPYPYLATDYGTQLSLFTHHMWIGGFCVVGAAAHAAIFMVRDYDPT 420
Query: 418 TRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ 477
YN+LLDRV+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ
Sbjct: 421 NNYNNLLDRVIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ 480
Query: 478 PIFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIH 537
P+FAQWI THALAPG+TAP A TS +W GGD+VAVG KVA++PI LGTADFLVHHIH
Sbjct: 481 PVFAQWIHKTHALAPGLTAPNALASTSPSW-GGDVVAVGGKVAMMPISLGTADFLVHHIH 539
Query: 538 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 597
AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM
Sbjct: 540 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 599
Query: 598 YNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVI 657
YN+ISVVIFHFSWKMQSDVWG+++ QG V+HITGGNFA SS TINGWLRDFLWAQASQVI
Sbjct: 600 YNSISVVIFHFSWKMQSDVWGNVTAQG-VSHITGGNFALSSNTINGWLRDFLWAQASQVI 658
Query: 658 QSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRAL 717
QSYGS+LSAYGL FLGAHF+WAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP+ QPRAL
Sbjct: 659 QSYGSALSAYGLIFLGAHFIWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPSIQPRAL 718
Query: 718 SIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
SI QGRAVGV HYLLGGIATTWAFFLARIIAVG
Sbjct: 719 SITQGRAVGVAHYLLGGIATTWAFFLARIIAVG 751
>gb|AAF29814.1| (AF180013) photosystem I P700 apoprotein A1 [Welwitschia mirabilis]
Length = 720
Score = 1406 bits (3599), Expect = 0.0
Identities = 653/720 (90%), Positives = 689/720 (95%), Gaps = 1/720 (0%)
Query: 11 KILVDRDPIKTSFEEWAKPGHFSRTIAK-GPDTTTWIWNLHADAHDFDSHTSDLEDISRK 69
KI+V+RDPIKTSFE+WA PGHFS+T++K P+TTTWIWNLHADAHDFDSHT DLE+ISRK
Sbjct: 1 KIVVERDPIKTSFEKWANPGHFSKTLSKTDPETTTWIWNLHADAHDFDSHTEDLEEISRK 60
Query: 70 IFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVG 129
+FSAHFGQL+IIF WLSGMYFHGARFSNYEAWL+DPTHI+PSAQVVWPIVGQEILNGDVG
Sbjct: 61 VFSAHFGQLAIIFTWLSGMYFHGARFSNYEAWLADPTHIKPSAQVVWPIVGQEILNGDVG 120
Query: 130 GGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQ 189
GGFRGIQITSGFF IWRASGITSELQLYCTAIG L+FAALMLFAGWFHYHKAAPKLAWFQ
Sbjct: 121 GGFRGIQITSGFFPIWRASGITSELQLYCTAIGALIFAALMLFAGWFHYHKAAPKLAWFQ 180
Query: 190 DVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQ 249
VESMLNHHLAGLLGLGSLSWAGHQ+HVSLPINQ L+AGVDPKEIPLPHEFILNRDLL Q
Sbjct: 181 QVESMLNHHLAGLLGLGSLSWAGHQIHVSLPINQLLDAGVDPKEIPLPHEFILNRDLLNQ 240
Query: 250 LYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMY 309
LYPSFA+G PFFTLNW++Y++ LTFRGGL+PVTGGLWLTD AHHHLAIA+LFLIAGHMY
Sbjct: 241 LYPSFAQGLLPFFTLNWSEYSEILTFRGGLNPVTGGLWLTDTAHHHLAIAVLFLIAGHMY 300
Query: 310 RTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMY 369
+TNWGIGH +K+ILEAHKGPFTG+GHKGLYEILT SWHAQL++NLAMLGSLTIIVAHHMY
Sbjct: 301 KTNWGIGHSLKEILEAHKGPFTGEGHKGLYEILTNSWHAQLALNLAMLGSLTIIVAHHMY 360
Query: 370 SMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLR 429
SMPPYPYLA DY TQLSLFTHHMWIGGF+IVGAAAHAAIF+VRDYD TT YN+LLDRVLR
Sbjct: 361 SMPPYPYLAIDYSTQLSLFTHHMWIGGFIIVGAAAHAAIFLVRDYDSTTSYNNLLDRVLR 420
Query: 430 HRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA 489
HRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQW+QNTH
Sbjct: 421 HRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWLQNTHV 480
Query: 490 LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILL 549
AP TAP AT TSLTWGGGD++ VGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILL
Sbjct: 481 TAPSFTAPAATASTSLTWGGGDIITVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILL 540
Query: 550 KGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFS 609
KGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFS
Sbjct: 541 KGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFS 600
Query: 610 WKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGL 669
WKMQSDVWG+IS QGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGS+LSAYGL
Sbjct: 601 WKMQSDVWGNISKQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSALSAYGL 660
Query: 670 FFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTH 729
FFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRALSIVQGRAVGV H
Sbjct: 661 FFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRALSIVQGRAVGVAH 720
>gb|AAF29818.1| (AF180017) photosystem I P700 apoprotein A1 [Ephedra tweediana]
Length = 720
Score = 1403 bits (3591), Expect = 0.0
Identities = 651/721 (90%), Positives = 691/721 (95%), Gaps = 2/721 (0%)
Query: 10 VKILVDRDPIKTSFEEWAKPGHFSRTIAKG-PDTTTWIWNLHADAHDFDSHTSDLEDISR 68
+KI+V+RDPIKTSFE+WA PGHFSRT+AKG P+TTTWIWNLHADAHDFDSHT DLE+ISR
Sbjct: 1 IKIMVERDPIKTSFEKWANPGHFSRTLAKGDPETTTWIWNLHADAHDFDSHTEDLEEISR 60
Query: 69 KIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDV 128
KIFSAHFGQL+IIF+WLSGMYFHGARFSNYEAWL+DPTHI+PSAQVVWPIVGQEILNGDV
Sbjct: 61 KIFSAHFGQLAIIFIWLSGMYFHGARFSNYEAWLADPTHIKPSAQVVWPIVGQEILNGDV 120
Query: 129 GGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWF 188
GGGFRGIQITSGFF IWRASGITSELQLYCTAIGGL+FAALMLFAGWFHYHKAAPKL+WF
Sbjct: 121 GGGFRGIQITSGFFPIWRASGITSELQLYCTAIGGLIFAALMLFAGWFHYHKAAPKLSWF 180
Query: 189 QDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLA 248
Q VESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQ L+AGVDPKEIPLPHEFILNRDLL
Sbjct: 181 Q-VESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQLLDAGVDPKEIPLPHEFILNRDLLI 239
Query: 249 QLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
QLYPSF++G TPFFTLNW++Y++ LTFRGGL+P+TGGLWLTDI HHHLAIA++FLIAGHM
Sbjct: 240 QLYPSFSKGLTPFFTLNWSEYSEILTFRGGLNPITGGLWLTDIVHHHLAIAVIFLIAGHM 299
Query: 309 YRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHM 368
Y+TNWGIGH I++ILEAHKGPFTG+GHKGLYEILTTSWHAQL++NLA+LGSLTI+VAHHM
Sbjct: 300 YKTNWGIGHSIQEILEAHKGPFTGEGHKGLYEILTTSWHAQLALNLAILGSLTIVVAHHM 359
Query: 369 YSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVL 428
YSMPPYPYLA DYGTQLSLFTHHMWIGGF+I+GAAAHAAIF+VRDYD TT YN+L DRVL
Sbjct: 360 YSMPPYPYLAIDYGTQLSLFTHHMWIGGFVIIGAAAHAAIFLVRDYDSTTSYNNLFDRVL 419
Query: 429 RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTH 488
RHRDAIISHLNW CIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQW+QNTH
Sbjct: 420 RHRDAIISHLNWTCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWLQNTH 479
Query: 489 ALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLIL 548
AP TAP AT TS TWGG DLV VG KVALLPIPLGTADFLVHHIHAFTIHVTVLIL
Sbjct: 480 VTAPRFTAPAATASTSFTWGGNDLVTVGTKVALLPIPLGTADFLVHHIHAFTIHVTVLIL 539
Query: 549 LKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHF 608
LKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHF
Sbjct: 540 LKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHF 599
Query: 609 SWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYG 668
SWKMQSDVWG+IS QGVVTHITGGNFAQSS+TINGWLRDFLWAQASQVIQSYGS+LSAYG
Sbjct: 600 SWKMQSDVWGNISTQGVVTHITGGNFAQSSVTINGWLRDFLWAQASQVIQSYGSALSAYG 659
Query: 669 LFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVT 728
LFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRALSIVQGRAVGV
Sbjct: 660 LFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRALSIVQGRAVGVA 719
Query: 729 H 729
H
Sbjct: 720 H 720
>gi|11466389 P700 apoprotein A1 of photosystem I [Mesostigma viride]
gb|AAF43833.1|AF166114_45 (AF166114) P700 apoprotein A1 of photosystem I [Mesostigma viride]
Length = 750
Score = 1398 bits (3579), Expect = 0.0
Identities = 643/752 (85%), Positives = 703/752 (92%), Gaps = 4/752 (0%)
Query: 1 MIIRSPEPEV--KILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDS 58
M I PE E KI+VDRDP+KTSFE W KPGHFSR++AKGP+TTTWIWNLHADAHDFDS
Sbjct: 1 MTISPPEREANGKIVVDRDPVKTSFERWGKPGHFSRSLAKGPNTTTWIWNLHADAHDFDS 60
Query: 59 HTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPI 118
HT+DLEDISRK+FSAHFGQL++IF+WLSGMYFHGARFSNYEAWLSDPTHI+PSAQVVWPI
Sbjct: 61 HTNDLEDISRKVFSAHFGQLAVIFIWLSGMYFHGARFSNYEAWLSDPTHIKPSAQVVWPI 120
Query: 119 VGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHY 178
VGQEILNGDVGGGF+G+QITSGFFQ+WRASGI +E QLY TAIGGL+ A LM FAGWFHY
Sbjct: 121 VGQEILNGDVGGGFQGVQITSGFFQLWRASGIVNEQQLYTTAIGGLIAAGLMFFAGWFHY 180
Query: 179 HKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPH 238
HKAAPKL WFQ+ ESM+NHHLAGLLGLGSLSWAGHQ+HVSLP+NQ L+AGVDPKEIPLPH
Sbjct: 181 HKAAPKLEWFQNAESMMNHHLAGLLGLGSLSWAGHQIHVSLPVNQLLDAGVDPKEIPLPH 240
Query: 239 EFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAI 298
EF++NR+L+AQLYPSFA+G PFFTLNW +Y+DFLTFRGGL+PVTGGLWLTD HHH+AI
Sbjct: 241 EFVMNRELMAQLYPSFAKGLAPFFTLNWGEYSDFLTFRGGLNPVTGGLWLTDTVHHHVAI 300
Query: 299 AILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLG 358
A+LF++AGHMYRTNWGIGH +K+ILEAHKGPFTG+GHKGLYEILTTSWHAQL +NLA++G
Sbjct: 301 AVLFIVAGHMYRTNWGIGHSMKEILEAHKGPFTGEGHKGLYEILTTSWHAQLGLNLALMG 360
Query: 359 SLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTT 418
SL+IIVAHHMY+MPPYPYLATDYGTQLSLFTHHMWIGGF IVG AAHAAIFMVRDYDPT
Sbjct: 361 SLSIIVAHHMYAMPPYPYLATDYGTQLSLFTHHMWIGGFCIVGGAAHAAIFMVRDYDPTN 420
Query: 419 RYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP 478
YN+LLDRV+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP
Sbjct: 421 NYNNLLDRVIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP 480
Query: 479 IFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHA 538
+FAQ++QN + LAPG +AP A +S W GGD+VAVG KVA++PI LGT+DFLVHHIHA
Sbjct: 481 VFAQFVQNRNYLAPGFSAPNALASSSAVW-GGDVVAVGGKVAMMPIQLGTSDFLVHHIHA 539
Query: 539 FTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMY 598
FTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMY
Sbjct: 540 FTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMY 599
Query: 599 NAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQ 658
N +S+VIFHFSWKMQSDVWGS++ QG V+HITGGNFAQS+ TINGWLRDFLWAQASQVIQ
Sbjct: 600 NCLSIVIFHFSWKMQSDVWGSVTAQG-VSHITGGNFAQSANTINGWLRDFLWAQASQVIQ 658
Query: 659 SYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALS 718
SYGS+LSAYGL FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRALS
Sbjct: 659 SYGSALSAYGLMFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRALS 718
Query: 719 IVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
I QGRAVGV HYLLGGIATTWAFFLARIIAVG
Sbjct: 719 ITQGRAVGVAHYLLGGIATTWAFFLARIIAVG 750
>gb|AAF29823.1| (AF180022) photosystem I P700 apoprotein A1 [Adiantum
capillus-veneris]
Length = 719
Score = 1398 bits (3579), Expect = 0.0
Identities = 641/718 (89%), Positives = 688/718 (95%)
Query: 13 LVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKIFS 72
+V++DP+KTSFE+WA+PGHF+R +AKGP TTTWIWNLHADAHDFDSHT+DLEDISRKIFS
Sbjct: 2 MVEKDPVKTSFEKWAQPGHFARNLAKGPSTTTWIWNLHADAHDFDSHTNDLEDISRKIFS 61
Query: 73 AHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGGF 132
AHFGQL+IIF+ LSGMYFHGARFSNYE+WL+DPTH++PSAQVVWPIVGQEILNGDVGGGF
Sbjct: 62 AHFGQLAIIFIRLSGMYFHGARFSNYESWLNDPTHVKPSAQVVWPIVGQEILNGDVGGGF 121
Query: 133 RGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDVE 192
+G+QITSGFFQ+WRASGITSELQLYCTA+G LVFA LM FAGWFHYHKAAPKLAWFQ+VE
Sbjct: 122 QGVQITSGFFQLWRASGITSELQLYCTAVGALVFAGLMFFAGWFHYHKAAPKLAWFQNVE 181
Query: 193 SMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYP 252
SMLNHHLAGLLGLGSL WAGHQ+HVSLPINQ L+AGVDP+EIPLPHE ILNRDLLAQ YP
Sbjct: 182 SMLNHHLAGLLGLGSLGWAGHQIHVSLPINQLLDAGVDPEEIPLPHELILNRDLLAQTYP 241
Query: 253 SFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTN 312
SFA+G PFFTLNW++Y+DFLTFRGGL+P+TGGLWLTD AHHHLAIA+LFL+AGHMYRTN
Sbjct: 242 SFAKGLIPFFTLNWSEYSDFLTFRGGLNPITGGLWLTDTAHHHLAIAVLFLVAGHMYRTN 301
Query: 313 WGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMP 372
WGIGH K+ILEAHKGPFTG+GHKG+YEILT+SWHAQL+INLA LGSLTIIV+HHMY+MP
Sbjct: 302 WGIGHSTKEILEAHKGPFTGEGHKGIYEILTSSWHAQLAINLATLGSLTIIVSHHMYAMP 361
Query: 373 PYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRD 432
PYPYLATDY TQLSLFTHHMWIGGFLIVGAAAHAAIF+VRDYDPTT+YN+LLDRV+RHRD
Sbjct: 362 PYPYLATDYATQLSLFTHHMWIGGFLIVGAAAHAAIFVVRDYDPTTQYNNLLDRVIRHRD 421
Query: 433 AIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALAP 492
IISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALAP
Sbjct: 422 TIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALAP 481
Query: 493 GITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGV 552
G TAP AT GTSL WGG DLVAV KVAL PIPLGTADFLVHHIHAFTIHVTVLILLKGV
Sbjct: 482 GSTAPNATAGTSLAWGGSDLVAVAGKVALAPIPLGTADFLVHHIHAFTIHVTVLILLKGV 541
Query: 553 LFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKM 612
LFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+ISVVIFHFSWKM
Sbjct: 542 LFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSISVVIFHFSWKM 601
Query: 613 QSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFFL 672
QSDVWGS+S+QG V+HITGGNFAQSS TINGWLRDFLWAQASQVIQSYGSSLSAYGL FL
Sbjct: 602 QSDVWGSVSEQGAVSHITGGNFAQSSTTINGWLRDFLWAQASQVIQSYGSSLSAYGLLFL 661
Query: 673 GAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHY 730
GAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP TQPRALSI+QGRAVGVTHY
Sbjct: 662 GAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPVTQPRALSIIQGRAVGVTHY 719
>gb|AAF29822.1| (AF180021) photosystem I P700 apoprotein A1 [Asplenium nidus]
Length = 719
Score = 1393 bits (3565), Expect = 0.0
Identities = 635/717 (88%), Positives = 686/717 (95%)
Query: 13 LVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKIFS 72
+V++DP+KTSFE WA+PGHFS+ +AKGP TTTWIWNLHADAHDFDSHT+DLEDISRKIF
Sbjct: 2 IVEKDPVKTSFERWAQPGHFSKNLAKGPSTTTWIWNLHADAHDFDSHTNDLEDISRKIFG 61
Query: 73 AHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGGF 132
AHFGQL+IIF+WLSGMYFHGARFSNYEAWL+DPTH++PSAQVVWPIVGQEILNGDVGGGF
Sbjct: 62 AHFGQLAIIFIWLSGMYFHGARFSNYEAWLNDPTHVKPSAQVVWPIVGQEILNGDVGGGF 121
Query: 133 RGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDVE 192
+GIQITSGFFQ+WRASGIT+ELQLYCTA+G L+FA LM FAGWFHYHKAAPKLAWFQ+VE
Sbjct: 122 QGIQITSGFFQLWRASGITNELQLYCTAVGALIFAGLMFFAGWFHYHKAAPKLAWFQNVE 181
Query: 193 SMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYP 252
SMLNHHLAGLLGLGSL WAGHQ+HVSLPINQ L+AGVD KE+PLPHEFILNR++L Q YP
Sbjct: 182 SMLNHHLAGLLGLGSLGWAGHQIHVSLPINQLLDAGVDSKEVPLPHEFILNREILTQAYP 241
Query: 253 SFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTN 312
SFA+G PFFTL+W++Y+DFLTFRGGL+PVTGGLWLTD AHHHLAIA+LFL+AGHMYRTN
Sbjct: 242 SFAKGLIPFFTLDWSEYSDFLTFRGGLNPVTGGLWLTDTAHHHLAIAVLFLVAGHMYRTN 301
Query: 313 WGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMP 372
WGIGH ++ILEAHKGPFTG+GHKGLYEILT+SWHAQL+INLAMLGSLTIIV+HHMY+MP
Sbjct: 302 WGIGHSTREILEAHKGPFTGEGHKGLYEILTSSWHAQLAINLAMLGSLTIIVSHHMYAMP 361
Query: 373 PYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRD 432
PYPYLATDY TQLSLFTHHMWIGGFL+VGAAAHAAIFMVRDYDPTT+YN+LLDRV+RHRD
Sbjct: 362 PYPYLATDYATQLSLFTHHMWIGGFLVVGAAAHAAIFMVRDYDPTTQYNNLLDRVIRHRD 421
Query: 433 AIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALAP 492
AIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQW+QNTHALAP
Sbjct: 422 AIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWVQNTHALAP 481
Query: 493 GITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGV 552
G TAP A TSLTWGG +LVAVG K+A+ PI LGTADFLVHHIHAFTIHVTVLILLKGV
Sbjct: 482 GSTAPNAAAATSLTWGGSNLVAVGGKIAISPITLGTADFLVHHIHAFTIHVTVLILLKGV 541
Query: 553 LFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKM 612
LFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+ISVVIFHFSWKM
Sbjct: 542 LFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSISVVIFHFSWKM 601
Query: 613 QSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFFL 672
QSDVWGSIS+QGV+ HITGGNFAQSS TINGWLRDFLWAQASQVIQSYGSSLSAYGL FL
Sbjct: 602 QSDVWGSISEQGVINHITGGNFAQSSTTINGWLRDFLWAQASQVIQSYGSSLSAYGLLFL 661
Query: 673 GAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTH 729
GAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP TQPRALSI+QGRAVGVTH
Sbjct: 662 GAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPVTQPRALSIIQGRAVGVTH 718
>sp|P12154|PSAA_CHLRE PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||A28341 photosystem I P700 apoprotein A1 - Chlamydomonas reinhardtii
chloroplast
emb|CAA29286.1| (X05845) P700 chlorophyll a-apoprotein A2 [Chlamydomonas
reinhardtii]
prf||1310243A gene ps1A1 [Chlamydomonas reinhardtii]
Length = 751
Score = 1390 bits (3559), Expect = 0.0
Identities = 642/753 (85%), Positives = 705/753 (93%), Gaps = 5/753 (0%)
Query: 1 MIIRSPEPE---VKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFD 57
M I +PE E VKI VDR+P++TSFE+WAKPGHFSRT++KGP+TTTWIWNLHADAHDFD
Sbjct: 1 MTISTPEREAKKVKIAVDRNPVETSFEKWAKPGHFSRTLSKGPNTTTWIWNLHADAHDFD 60
Query: 58 SHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWP 117
SHTSDLE+ISRK+FSAHFGQL IIF+WLSGMYFHGARFSNYEAWLSDPTHI+PSAQVVWP
Sbjct: 61 SHTSDLEEISRKVFSAHFGQLGIIFIWLSGMYFHGARFSNYEAWLSDPTHIKPSAQVVWP 120
Query: 118 IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFH 177
IVGQEILNGDVGGGF+GIQITSGFFQ+WRASGITSELQLY TAIGGLV AA M FAGWFH
Sbjct: 121 IVGQEILNGDVGGGFQGIQITSGFFQLWRASGITSELQLYTTAIGGLVMAAAMFFAGWFH 180
Query: 178 YHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLP 237
YHKAAPKL WFQ+VESMLNHHL GLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLP
Sbjct: 181 YHKAAPKLEWFQNVESMLNHHLGGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLP 240
Query: 238 HEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLA 297
H+ +LNR ++A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+A
Sbjct: 241 HDLLLNRAIMADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVA 300
Query: 298 IAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAML 357
IA+LFL+AGHMYRTNWGIGH +K+ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+
Sbjct: 301 IAVLFLVAGHMYRTNWGIGHSMKEILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALF 360
Query: 358 GSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPT 417
GSL+IIVAHHMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT
Sbjct: 361 GSLSIIVAHHMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPT 420
Query: 418 TRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ 477
YN+LLDRV+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ
Sbjct: 421 NNYNNLLDRVIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ 480
Query: 478 PIFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIH 537
P+FAQWIQNTH LAP +TAP A TSLTW GG+L A G KVA++PI LGT+DF+VHHIH
Sbjct: 481 PVFAQWIQNTHFLAPQLTAPNALAATSLTW-GGELGAHGGKVAMMPISLGTSDFMVHHIH 539
Query: 538 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 597
AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM
Sbjct: 540 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 599
Query: 598 YNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVI 657
YN++S+VIFHFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVI
Sbjct: 600 YNSLSIVIFHFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVI 658
Query: 658 QSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRAL 717
QSYGS+LSAYGL FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRAL
Sbjct: 659 QSYGSALSAYGLIFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRAL 718
Query: 718 SIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
SI QGRAVGV HYLLGGIATTW+FFLARII+VG
Sbjct: 719 SITQGRAVGVAHYLLGGIATTWSFFLARIISVG 751
>gi|7524848 P700 apoprotein subunit Ia [Chlorella vulgaris]
sp|P56341|PSAA_CHLVU PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||T07278 photosystem I P700 apoprotein A1 - Chlorella vulgaris chloroplast
dbj|BAA57926.1| (AB001684) P700 apoprotein subunit Ia [Chlorella vulgaris]
Length = 751
Score = 1388 bits (3554), Expect = 0.0
Identities = 642/753 (85%), Positives = 702/753 (92%), Gaps = 5/753 (0%)
Query: 1 MIIRSPEPE---VKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFD 57
M I PE E VKI+VDR+P+ T+FE+WAKPGHFSRT++KGP TTTWIWNLHADAHDFD
Sbjct: 1 MTISPPEREAKKVKIVVDRNPVATNFEKWAKPGHFSRTLSKGPTTTTWIWNLHADAHDFD 60
Query: 58 SHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWP 117
+ TSDLE+ISRK+FSAHFGQL IIF+WLSGMYFHGARFSNYEAWL+DPTHI+PSAQVVWP
Sbjct: 61 TQTSDLEEISRKVFSAHFGQLGIIFIWLSGMYFHGARFSNYEAWLTDPTHIKPSAQVVWP 120
Query: 118 IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFH 177
IVGQEILN DVGGGF+G+QITSGFFQ+WRASGITSELQLY TAIGGLV AA M FAGWFH
Sbjct: 121 IVGQEILNADVGGGFQGLQITSGFFQLWRASGITSELQLYTTAIGGLVMAAAMFFAGWFH 180
Query: 178 YHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLP 237
YHKAAPKL WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLPIN+ L+AGVDPKEIPLP
Sbjct: 181 YHKAAPKLEWFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPINKLLDAGVDPKEIPLP 240
Query: 238 HEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLA 297
HEF+ N +L+AQLYPSFA+G PFFTL+WA+Y+DFLTF+GGL+PVTGGLWLTD HHHLA
Sbjct: 241 HEFLFNPELMAQLYPSFAKGLAPFFTLDWAQYSDFLTFQGGLNPVTGGLWLTDTVHHHLA 300
Query: 298 IAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAML 357
IA+LFL+AGH YRTNWGIG +K+ILEAHKGPFTG+GHKGLYEILTTSWHAQL+INLA+
Sbjct: 301 IAVLFLVAGHQYRTNWGIGSSLKEILEAHKGPFTGEGHKGLYEILTTSWHAQLAINLALF 360
Query: 358 GSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPT 417
GSL+IIV+HHMY+MPPYPYLATDYGTQLSLFTHHMWIGGF IVGA AHA IFMVRDYDPT
Sbjct: 361 GSLSIIVSHHMYAMPPYPYLATDYGTQLSLFTHHMWIGGFCIVGAGAHAGIFMVRDYDPT 420
Query: 418 TRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ 477
YN+LLDRVLRHRDA+ISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ
Sbjct: 421 NNYNNLLDRVLRHRDAMISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ 480
Query: 478 PIFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIH 537
P+FAQW+QNTH LAPG TAP A TS +W GGD+VAVG KVA++PI LGTADF+VHHIH
Sbjct: 481 PVFAQWVQNTHFLAPGFTAPNALASTSPSW-GGDVVAVGGKVAMMPISLGTADFMVHHIH 539
Query: 538 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 597
AFTIHVTVLILLKGVL+ARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM
Sbjct: 540 AFTIHVTVLILLKGVLYARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 599
Query: 598 YNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVI 657
YN+IS+VIFHFSWKMQSDVWG++S G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVI
Sbjct: 600 YNSISIVIFHFSWKMQSDVWGTVSANG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVI 658
Query: 658 QSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRAL 717
QSYGS+LSAYGL FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRAL
Sbjct: 659 QSYGSALSAYGLIFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRAL 718
Query: 718 SIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
SI QGRAVGV HYLLGGIATTW+FFLARI+AVG
Sbjct: 719 SITQGRAVGVAHYLLGGIATTWSFFLARILAVG 751
>sp|P25936|PSAA_SYNVU PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||S20922 photosystem I protein A1 - Synechococcus sp
dbj|BAA01759.1| (D10986) photosystem I core protein A [Synechococcus vulcanus]
Length = 755
Score = 1354 bits (3465), Expect = 0.0
Identities = 623/748 (83%), Positives = 681/748 (90%), Gaps = 5/748 (0%)
Query: 7 EPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDI 66
EP+V+++VD DP+ TSFE+WAKPGHF RT+A+GP TTTWIWNLHA AHDFD+HTSDLEDI
Sbjct: 9 EPKVRVVVDNDPVPTSFEKWAKPGHFDRTLARGPQTTTWIWNLHALAHDFDTHTSDLEDI 68
Query: 67 SRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNG 126
SRKIFSAHFG L+++F+WLSGMYFHGA+FSNYEAWL+DPT I+PSAQVVWPIVGQ ILNG
Sbjct: 69 SRKIFSAHFGHLAVVFIWLSGMYFHGAKFSNYEAWLADPTGIKPSAQVVWPIVGQGILNG 128
Query: 127 DVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLA 186
DVGGGF GIQITSG FQ+WRASGIT+E QLYCTAIGGLV A LMLFAGWFHYHK APKL
Sbjct: 129 DVGGGFHGIQITSGLFQLWRASGITNEFQLYCTAIGGLVMAGLMLFAGWFHYHKRAPKLE 188
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSLSWAGHQ+HVSLPIN+ L+AGV K+IPLPHEFILN L
Sbjct: 189 WFQNVESMLNHHLAGLLGLGSLSWAGHQIHVSLPINKLLDAGVAAKDIPLPHEFILNPSL 248
Query: 247 LAQLYPS----FAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILF 302
+A+LYP F G PFFT NWA Y+DFLTF GGL+PVTGGLWL+D AHHHLAIA+LF
Sbjct: 249 MAELYPKVDWGFFSGVIPFFTFNWAAYSDFLTFNGGLNPVTGGLWLSDTAHHHLAIAVLF 308
Query: 303 LIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTI 362
+IAGHMYRTNWGIGH +K+ILEAHKGPFTG GHKGLYE+LTTSWHAQL+INLAM+GSL+I
Sbjct: 309 IIAGHMYRTNWGIGHSLKEILEAHKGPFTGAGHKGLYEVLTTSWHAQLAINLAMMGSLSI 368
Query: 363 IVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYND 422
IVA HMY+MPPYPYLATDY TQLSLFTHHMWIGGFL+VG AAH AIFMVRDYDP N+
Sbjct: 369 IVAQHMYAMPPYPYLATDYPTQLSLFTHHMWIGGFLVVGGAAHGAIFMVRDYDPAMNQNN 428
Query: 423 LLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQ 482
+LDRVLRHRDAIISHLNWVCIFLGFHSFGLY+HNDTM A GRPQDMFSDT IQLQP+FAQ
Sbjct: 429 VLDRVLRHRDAIISHLNWVCIFLGFHSFGLYVHNDTMRAFGRPQDMFSDTGIQLQPVFAQ 488
Query: 483 WIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIH 542
W+QN H LAPG TAP A S+ +GG D+VAVG KVA++PI LGTADF+VHHIHAFTIH
Sbjct: 489 WVQNLHTLAPGGTAPNAAATASVAFGG-DVVAVGGKVAMMPIVLGTADFMVHHIHAFTIH 547
Query: 543 VTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAIS 602
VTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVS WDHVFLGLFWMYN IS
Sbjct: 548 VTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSGWDHVFLGLFWMYNCIS 607
Query: 603 VVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGS 662
VVIFHFSWKMQSDVWG+++ G V+HITGGNFAQS+ITINGWLRDFLWAQASQVI SYGS
Sbjct: 608 VVIFHFSWKMQSDVWGTVAPDGTVSHITGGNFAQSAITINGWLRDFLWAQASQVIGSYGS 667
Query: 663 SLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQG 722
+LSAYGL FLGAHF+WAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRALSI+QG
Sbjct: 668 ALSAYGLLFLGAHFIWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRALSIIQG 727
Query: 723 RAVGVTHYLLGGIATTWAFFLARIIAVG 750
RAVGV HYLLGGIATTWAFFLARII+VG
Sbjct: 728 RAVGVAHYLLGGIATTWAFFLARIISVG 755
>sp|P25896|PSAA_SYNEN PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
emb|CAA45304.1| (X63768) photosystem I subunit Ia [Synechococcus sp.]
Length = 755
Score = 1352 bits (3462), Expect = 0.0
Identities = 622/748 (83%), Positives = 681/748 (90%), Gaps = 5/748 (0%)
Query: 7 EPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDI 66
EP+V+++VD DP+ TSFE+WAKPGHF RT+A+GP TTTWIWNLHA AHDFD+HTSDLEDI
Sbjct: 9 EPKVRVVVDNDPVPTSFEKWAKPGHFDRTLARGPQTTTWIWNLHALAHDFDTHTSDLEDI 68
Query: 67 SRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNG 126
SRKIFSAHFG L+++F+WLSGMYFHGA+FSNYEAWL+DPT I+PSAQVVWPIVGQ ILNG
Sbjct: 69 SRKIFSAHFGHLAVVFIWLSGMYFHGAKFSNYEAWLADPTGIKPSAQVVWPIVGQGILNG 128
Query: 127 DVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLA 186
DVGGGF GIQITSG FQ+WRASGIT+E QLYCTAIGGLV A LMLFAGWFHYHK APKL
Sbjct: 129 DVGGGFHGIQITSGLFQLWRASGITNEFQLYCTAIGGLVMAGLMLFAGWFHYHKRAPKLE 188
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLPIN+ L+AGV K+IPLPHEFILN L
Sbjct: 189 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPINKLLDAGVAAKDIPLPHEFILNPSL 248
Query: 247 LAQLYPS----FAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILF 302
+A+LYP F G PFFT NWA Y+DFLTF GGL+PVTGGLWL+D AHHHLAIA+LF
Sbjct: 249 MAELYPKVDWGFFSGVIPFFTFNWAAYSDFLTFNGGLNPVTGGLWLSDTAHHHLAIAVLF 308
Query: 303 LIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTI 362
+IAGHMYRTNWGIGH +K+ILEAHKGPFTG GHKGLYE+LTTSWHAQL+INLAM+GSL+I
Sbjct: 309 IIAGHMYRTNWGIGHSLKEILEAHKGPFTGAGHKGLYEVLTTSWHAQLAINLAMMGSLSI 368
Query: 363 IVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYND 422
IVA HMY+MPPYPYLATDY TQLSLFTHHMWIGGFL+VG AAH AIFMVRDYDP N+
Sbjct: 369 IVAQHMYAMPPYPYLATDYPTQLSLFTHHMWIGGFLVVGGAAHGAIFMVRDYDPAMNQNN 428
Query: 423 LLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQ 482
+LDRVLRHRDAIISHLNWVCIFLGFHSFGLY+HNDTM A GRPQDMFSDT IQLQP+FAQ
Sbjct: 429 VLDRVLRHRDAIISHLNWVCIFLGFHSFGLYVHNDTMRAFGRPQDMFSDTGIQLQPVFAQ 488
Query: 483 WIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIH 542
W+QN H LAPG TAP A S+ +GG D+VAVG KVA++PI LGTADF+VHHIHAFTIH
Sbjct: 489 WVQNLHTLAPGGTAPNAAATASVAFGG-DVVAVGGKVAMMPIVLGTADFMVHHIHAFTIH 547
Query: 543 VTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAIS 602
VTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVS WDHVFLGLFWMYN IS
Sbjct: 548 VTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSGWDHVFLGLFWMYNCIS 607
Query: 603 VVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGS 662
VVIFHFSWKMQSDVWG+++ G V+HITGGNFAQS+ITINGWLRDFLWAQASQVI SYGS
Sbjct: 608 VVIFHFSWKMQSDVWGTVAPDGTVSHITGGNFAQSAITINGWLRDFLWAQASQVIGSYGS 667
Query: 663 SLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQG 722
+LSAYGL FLGAHF+WAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRALSI+QG
Sbjct: 668 ALSAYGLLFLGAHFIWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRALSIIQG 727
Query: 723 RAVGVTHYLLGGIATTWAFFLARIIAVG 750
RAVGV HYLLGGIATTWAFFLARII+VG
Sbjct: 728 RAVGVAHYLLGGIATTWAFFLARIISVG 755
>gi|11465750 Photosystem I p700 chlorophyll A apoprotein A1 [Porphyra purpurea]
sp|P51284|PSAA_PORPU PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||S73205 photosystem I protein A1 - red alga (Porphyra purpurea) chloroplast
gb|AAC08170.1| (U38804) Photosystem I p700 chlorophyll A apoprotein A1 [Porphyra
purpurea]
Length = 752
Score = 1352 bits (3461), Expect = 0.0
Identities = 619/753 (82%), Positives = 693/753 (91%), Gaps = 4/753 (0%)
Query: 1 MIIRSPEPE---VKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFD 57
M I S E E VKI VD++P+ TSFE+WA+PGHFSRT+AKGP TTTWIWNLHADAHDFD
Sbjct: 1 MAISSKEQETKKVKISVDKNPVDTSFEKWAQPGHFSRTLAKGPKTTTWIWNLHADAHDFD 60
Query: 58 SHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWP 117
S TS LE++SRKIFSAHFGQL++IFLWLSGMYFHGARFSNY AWLS+PT I+PSAQVVWP
Sbjct: 61 SQTSSLEEVSRKIFSAHFGQLAVIFLWLSGMYFHGARFSNYVAWLSNPTGIKPSAQVVWP 120
Query: 118 IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFH 177
IVGQEILNGDVGGGF+G+Q+TSG+FQ+WRASGIT+E QLYCTAIGGL AALMLFAGWFH
Sbjct: 121 IVGQEILNGDVGGGFQGVQVTSGWFQLWRASGITTEFQLYCTAIGGLGMAALMLFAGWFH 180
Query: 178 YHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLP 237
YHKAAPKL WFQ+VESM+NHHLAGLLGLG L WAGHQ+H+SLPIN+ L++GV P+EIPLP
Sbjct: 181 YHKAAPKLEWFQNVESMMNHHLAGLLGLGCLGWAGHQIHLSLPINKLLDSGVSPQEIPLP 240
Query: 238 HEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLA 297
HEF++NR+L+AQLYPSF++G PFFTLNWA+Y+DFLTF+GGL+PVTGGLWL+D AHHHLA
Sbjct: 241 HEFLINRELMAQLYPSFSKGLVPFFTLNWAEYSDFLTFKGGLNPVTGGLWLSDTAHHHLA 300
Query: 298 IAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAML 357
+A+LFL AGHMYRTNWGIGH +K+ILEAHKGPFTG GH+GLYEILTTSWHAQL+INLAM+
Sbjct: 301 LAVLFLAAGHMYRTNWGIGHSMKEILEAHKGPFTGNGHEGLYEILTTSWHAQLAINLAMM 360
Query: 358 GSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPT 417
GSL+IIVAHHMY+MPPYPY+ATDY TQLSLFTHHMWIGGF +VGA AHA+IFMVRDY+P
Sbjct: 361 GSLSIIVAHHMYAMPPYPYIATDYPTQLSLFTHHMWIGGFCVVGAGAHASIFMVRDYNPA 420
Query: 418 TRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ 477
YN+LLDR++RHRDAI+SHLNWVCIFLGFHSFGLYIHNDTM ALGR QDMFSDTAIQLQ
Sbjct: 421 ENYNNLLDRIIRHRDAIVSHLNWVCIFLGFHSFGLYIHNDTMRALGRSQDMFSDTAIQLQ 480
Query: 478 PIFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIH 537
PIFAQW+Q+ H LAPG TAP A S + GG++V+VGNKVA++PI LGTADF+VHHIH
Sbjct: 481 PIFAQWVQSIHTLAPGNTAPNALATASYAF-GGEVVSVGNKVAMMPISLGTADFMVHHIH 539
Query: 538 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 597
AFTIHVTVLIL+KG LF+R+SRLIPDKANLGFRFPCDGPGRGGTCQVS WDHVFLGLFWM
Sbjct: 540 AFTIHVTVLILVKGFLFSRNSRLIPDKANLGFRFPCDGPGRGGTCQVSGWDHVFLGLFWM 599
Query: 598 YNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVI 657
YN++SV IFHFSWKMQSDVWGS+S G V+HITGGNFAQS+ITINGWLRDFLWAQASQVI
Sbjct: 600 YNSLSVAIFHFSWKMQSDVWGSVSPSGNVSHITGGNFAQSAITINGWLRDFLWAQASQVI 659
Query: 658 QSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRAL 717
QSYGS+LSAYGL FL AHFVWAFSLMFLFSGRGYWQELIESIVWAHNK+KVAPA QPRAL
Sbjct: 660 QSYGSALSAYGLIFLAAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKIKVAPAIQPRAL 719
Query: 718 SIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
SI QGRAVGV HYLLGGI TTWAFFLARII+VG
Sbjct: 720 SITQGRAVGVAHYLLGGIGTTWAFFLARIISVG 752
>gi|11467293 PsaA subunit of photosystem I reaction center [Cyanophora paradoxa]
sp|P48112|PSAA_CYAPA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||T06838 probable photosystem I protein A1 - Cyanophora paradoxa cyanelle
gb|AAA81181.1| (U30821) PsaA subunit of photosystem I reaction center [Cyanophora
paradoxa]
Length = 752
Score = 1349 bits (3453), Expect = 0.0
Identities = 619/753 (82%), Positives = 687/753 (91%), Gaps = 4/753 (0%)
Query: 1 MIIRSPEPE---VKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFD 57
M I PE E VKI++D+DP+ TSF++WA PGHFSRT+AKGP TTTWIWNLHAD HDFD
Sbjct: 1 MRISPPEREAKKVKIVIDKDPVSTSFDKWAVPGHFSRTLAKGPKTTTWIWNLHADVHDFD 60
Query: 58 SHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWP 117
S+TSDLED+SRKIFSAHFG L+++F+WLSG YFHGARFSNYEAWLS+PT I+PSAQVVWP
Sbjct: 61 SYTSDLEDVSRKIFSAHFGHLAVVFIWLSGAYFHGARFSNYEAWLSNPTTIKPSAQVVWP 120
Query: 118 IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFH 177
IVGQEILNGDVGGGF+GIQITSG FQ+WRASGIT+ELQLY TAIG LV AALMLFAGWFH
Sbjct: 121 IVGQEILNGDVGGGFQGIQITSGLFQMWRASGITTELQLYVTAIGALVMAALMLFAGWFH 180
Query: 178 YHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLP 237
YHKAAPKL WFQ+ ESM+NHHLAGL GLGSLSWAGHQ+HVSLP+N+ L++GV P+EIPLP
Sbjct: 181 YHKAAPKLEWFQNAESMMNHHLAGLFGLGSLSWAGHQIHVSLPVNKLLDSGVSPQEIPLP 240
Query: 238 HEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLA 297
HEFILN+DL+AQLYPSF +G TPFFTLNW +Y+DFLTF+GGL+PVTGGLWL+D AHHHLA
Sbjct: 241 HEFILNKDLIAQLYPSFGQGLTPFFTLNWNEYSDFLTFKGGLNPVTGGLWLSDRAHHHLA 300
Query: 298 IAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAML 357
IA+LF++AGHMYRTNWGIGH +K++LE HKGPFTG+GHKGLYEI T SWHAQLS+NLA+
Sbjct: 301 IAVLFIVAGHMYRTNWGIGHSMKEMLETHKGPFTGEGHKGLYEIFTNSWHAQLSLNLALF 360
Query: 358 GSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPT 417
GSL+IIVAHHMYSMPPYPYLATDY T L LFTHH+WIGGFLIVGA AHAAIFMVRDYDP
Sbjct: 361 GSLSIIVAHHMYSMPPYPYLATDYATSLCLFTHHVWIGGFLIVGAGAHAAIFMVRDYDPA 420
Query: 418 TRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ 477
YN+LLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTM ALGRPQDMFSD AIQLQ
Sbjct: 421 QNYNNLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMRALGRPQDMFSDAAIQLQ 480
Query: 478 PIFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIH 537
P+FAQW+Q ++ A G TAP A S + GGD+V+VG KVA++PI LGTADFLVHHIH
Sbjct: 481 PVFAQWVQGVNSAAAGNTAPNALRNASYAF-GGDIVSVGEKVAMMPISLGTADFLVHHIH 539
Query: 538 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 597
AFTIHVTVLILLKGVLFAR+SRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM
Sbjct: 540 AFTIHVTVLILLKGVLFARNSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 599
Query: 598 YNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVI 657
YN++SVV+FHFSWKMQSDVWG+++ G V+HITG NFAQSSITINGWLRDFLWAQASQVI
Sbjct: 600 YNSLSVVLFHFSWKMQSDVWGNVTADGAVSHITGNNFAQSSITINGWLRDFLWAQASQVI 659
Query: 658 QSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRAL 717
QSYGS+LSAYGL FLGAHF+WAFSLMFLFSGRGYWQELIESIVWAHNKLK AP+ QPRAL
Sbjct: 660 QSYGSALSAYGLMFLGAHFIWAFSLMFLFSGRGYWQELIESIVWAHNKLKFAPSIQPRAL 719
Query: 718 SIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
SI QGRAVGV HYLLGGIATTW+FF ARII+VG
Sbjct: 720 SITQGRAVGVAHYLLGGIATTWSFFHARIISVG 752
>gi|11467713 PSI P700 apoprotein A1 [Guillardia theta]
sp|O78508|PSAA_GUITH PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
gb|AAC35699.1| (AF041468) PSI P700 apoprotein A1 [Guillardia theta]
Length = 752
Score = 1335 bits (3416), Expect = 0.0
Identities = 607/753 (80%), Positives = 685/753 (90%), Gaps = 4/753 (0%)
Query: 1 MIIRSPEPEVK---ILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFD 57
M I S E E K ++VDR+P+ TSFE+W +PGHFSRT+AKGP TTTWIWNLHADAHDFD
Sbjct: 1 MTISSTEQEAKKVNVVVDRNPVSTSFEKWGQPGHFSRTLAKGPKTTTWIWNLHADAHDFD 60
Query: 58 SHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWP 117
SHTS LEDISRKIFSAHFGQLSIIFLW+SGM+FHGARFSNY AWLS+PT ++PSAQVVWP
Sbjct: 61 SHTSSLEDISRKIFSAHFGQLSIIFLWISGMHFHGARFSNYSAWLSNPTTVKPSAQVVWP 120
Query: 118 IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFH 177
IVGQEILNGDVGGGF+G+Q+TSGFFQIWRA GITSE++LY A+ GL+ + LM+FAGWFH
Sbjct: 121 IVGQEILNGDVGGGFQGVQVTSGFFQIWRAEGITSEVELYWCAVAGLLMSGLMIFAGWFH 180
Query: 178 YHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLP 237
YHKAAPKL WFQ+ ESMLNHHL+GLLGLG LSWAGHQ+HV LPIN+ L+AGV P+EIPLP
Sbjct: 181 YHKAAPKLEWFQNAESMLNHHLSGLLGLGCLSWAGHQIHVGLPINKLLDAGVAPQEIPLP 240
Query: 238 HEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLA 297
HEF++NRDL+AQLYPSF +G PFF+LNW++Y+DFLTF+GGL+PVTGGLWL+D AHHHLA
Sbjct: 241 HEFLMNRDLMAQLYPSFNKGLIPFFSLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHLA 300
Query: 298 IAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAML 357
+A+LF++AGHMYRTNWGIGH +K+ILEAHKGPFTG+GHKGLYEIL TSWH+QL+INLAM+
Sbjct: 301 LAVLFIVAGHMYRTNWGIGHSMKEILEAHKGPFTGEGHKGLYEILITSWHSQLAINLAMM 360
Query: 358 GSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPT 417
GSL+IIVAHHMY+MP YP++ATDY TQLS+FTHHMWIGGF I GAAAHA IFMVRDY+P
Sbjct: 361 GSLSIIVAHHMYAMPAYPFIATDYPTQLSIFTHHMWIGGFCICGAAAHAGIFMVRDYNPA 420
Query: 418 TRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ 477
YN+LLDRV+RHRDAIISHLNW+CIFLGFHSFGLYIHNDTM ALGR QDMFSDTAIQL+
Sbjct: 421 QNYNNLLDRVIRHRDAIISHLNWICIFLGFHSFGLYIHNDTMRALGRTQDMFSDTAIQLK 480
Query: 478 PIFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIH 537
P+FAQW+QN H +APG T P A S + GGD+++VGNKVA++PI LGTADF+VHHIH
Sbjct: 481 PVFAQWVQNIHTIAPGNTTPNALATASYAF-GGDVISVGNKVAMMPISLGTADFMVHHIH 539
Query: 538 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 597
AFTIHVTVLILLKGVLF+R+SRLIPDKANLGFRFPCDGPGRGGTCQ SAWD VFLGLFWM
Sbjct: 540 AFTIHVTVLILLKGVLFSRNSRLIPDKANLGFRFPCDGPGRGGTCQSSAWDSVFLGLFWM 599
Query: 598 YNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVI 657
YN ISVVIFHFSWKMQSDVWG++ + G VTHITGGNFAQSSITINGWLRDFLWA+ASQVI
Sbjct: 600 YNCISVVIFHFSWKMQSDVWGTVQNDGTVTHITGGNFAQSSITINGWLRDFLWAEASQVI 659
Query: 658 QSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRAL 717
QSYGS+LSAYGL FLGAHF+WAFSLMFLFSGRGYWQELIESIVWAHNKL APA QPRAL
Sbjct: 660 QSYGSALSAYGLIFLGAHFIWAFSLMFLFSGRGYWQELIESIVWAHNKLHFAPAIQPRAL 719
Query: 718 SIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
SI QGRAVG+ HYLLGGI TTWAFFLARII+VG
Sbjct: 720 SITQGRAVGLAHYLLGGIGTTWAFFLARIISVG 752
>sp|O52474|PSAA_MASLA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
gb|AAC12866.1| (AF038558) photosystem I PsaA [Fischerella PCC7605]
Length = 752
Score = 1322 bits (3384), Expect = 0.0
Identities = 610/753 (81%), Positives = 668/753 (88%), Gaps = 4/753 (0%)
Query: 1 MIIRSPEPEVK---ILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFD 57
M I PE E K ++VD DP+ TSFE WAKPGHF RT+A+GP TTTWIWNLHA AHDFD
Sbjct: 1 MTISPPEREEKKARVIVDNDPVPTSFELWAKPGHFDRTLARGPKTTTWIWNLHALAHDFD 60
Query: 58 SHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWP 117
+HTSDLEDISRKIF+AHFG L+++ LWLSGM FHGARFSNYEAWLSDP H++PSAQVVWP
Sbjct: 61 THTSDLEDISRKIFAAHFGHLAVVTLWLSGMIFHGARFSNYEAWLSDPLHVKPSAQVVWP 120
Query: 118 IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFH 177
IVGQ+ILNGDVGGGF GIQITSG FQIWR GIT+ QLY TAIGGLV A L LFAGWFH
Sbjct: 121 IVGQDILNGDVGGGFHGIQITSGLFQIWRGWGITNSFQLYVTAIGGLVLAGLFLFAGWFH 180
Query: 178 YHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLP 237
YHK APKL WFQ+VESMLNHHLA LLG GSL WAGH +HVS P N+ L+AGV K+IPLP
Sbjct: 181 YHKRAPKLEWFQNVESMLNHHLAVLLGCGSLGWAGHLIHVSAPTNKLLDAGVSVKDIPLP 240
Query: 238 HEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLA 297
HEFILN+ LL +LYP FA G TPFFTLNW +YADFLTF+GGL+PVTGGLWLTDI+HHHLA
Sbjct: 241 HEFILNKGLLTELYPGFASGITPFFTLNWGQYADFLTFKGGLNPVTGGLWLTDISHHHLA 300
Query: 298 IAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAML 357
IA+LF+IAGHMYRTNWGIGH IK+ILE HKGPFTG+GHKGLYE +TTSWHAQL+ NLA L
Sbjct: 301 IAVLFIIAGHMYRTNWGIGHSIKEILENHKGPFTGEGHKGLYENMTTSWHAQLATNLAFL 360
Query: 358 GSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPT 417
GSLTIIVAHHMY+MPPYPYLATDY TQL +FTHHMWIGGFLIVG AAHA IFMVRDYDP
Sbjct: 361 GSLTIIVAHHMYAMPPYPYLATDYATQLCIFTHHMWIGGFLIVGGAAHATIFMVRDYDPV 420
Query: 418 TRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ 477
N++LDRV+RHRDAIISHLNWVCIFLGFHSFGLY+HNDTM ALGRPQDMFSDTAIQLQ
Sbjct: 421 VNQNNVLDRVIRHRDAIISHLNWVCIFLGFHSFGLYVHNDTMRALGRPQDMFSDTAIQLQ 480
Query: 478 PIFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIH 537
P+FAQW+QN H LAPG TAP A S +GGG +VAVG KVA++PI LGTADFLVHHIH
Sbjct: 481 PVFAQWVQNIHTLAPGSTAPNALEPASYAFGGG-IVAVGGKVAMMPIALGTADFLVHHIH 539
Query: 538 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 597
AFTIHVTVLILLKG LFAR+SRLIPDKANLGFRFPCDGPGRGGTCQVS WDHVFLGLFWM
Sbjct: 540 AFTIHVTVLILLKGFLFARNSRLIPDKANLGFRFPCDGPGRGGTCQVSGWDHVFLGLFWM 599
Query: 598 YNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVI 657
YN+IS+VIFHFSWKMQSDVWG++ G VTHITGGN+AQS++TINGWLRDFLWAQ+ QVI
Sbjct: 600 YNSISIVIFHFSWKMQSDVWGTVDAAGNVTHITGGNWAQSALTINGWLRDFLWAQSVQVI 659
Query: 658 QSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRAL 717
SYGS+LSAYGL FLGAHF+WAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRAL
Sbjct: 660 NSYGSALSAYGLMFLGAHFIWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRAL 719
Query: 718 SIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
SI+QGRAVGV HYLLGGIATTWAFF A I+++G
Sbjct: 720 SIIQGRAVGVAHYLLGGIATTWAFFHAHILSLG 752
>sp|Q44550|PSAA_ANAVA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||I39615 psaA protein - Anabaena variabilis
gb|AAA18488.1| (L26326) PsaA [Anabaena variabilis]
Length = 752
Score = 1322 bits (3383), Expect = 0.0
Identities = 609/753 (80%), Positives = 671/753 (88%), Gaps = 4/753 (0%)
Query: 1 MIIRSPEPEVK---ILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFD 57
M I PE E K ++VD+DP+ TSFE+WA+PGHF RT+A+GP TTTWIWNLHA AHDFD
Sbjct: 1 MTISPPEREEKKARVIVDKDPVPTSFEKWAQPGHFDRTLARGPKTTTWIWNLHALAHDFD 60
Query: 58 SHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWP 117
+HTSDLEDISRKIF+AHFG L+++ +WLSGM FHGA+FSNYEAWLSDP ++RPSAQVVWP
Sbjct: 61 THTSDLEDISRKIFAAHFGHLAVVTIWLSGMIFHGAKFSNYEAWLSDPLNVRPSAQVVWP 120
Query: 118 IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFH 177
IVGQ+ILNGDVGGGF GIQITSG FQ+WR GIT+ QLYCTAIGGLV A L LFAGWFH
Sbjct: 121 IVGQDILNGDVGGGFHGIQITSGLFQVWRGWGITNSFQLYCTAIGGLVLAGLFLFAGWFH 180
Query: 178 YHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLP 237
YHK APKL WFQ+VESMLNHHL LLG GSL WAGH +HVS PIN+ L+AGV K+IPLP
Sbjct: 181 YHKRAPKLEWFQNVESMLNHHLQVLLGCGSLGWAGHLIHVSAPINKLLDAGVAVKDIPLP 240
Query: 238 HEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLA 297
HEFILN+ +L L+P FA G TPFFTLNW +YADFLTF+GGL+PVTGGLWLTDI+HHHLA
Sbjct: 241 HEFILNKSVLIDLFPGFAAGLTPFFTLNWGQYADFLTFKGGLNPVTGGLWLTDISHHHLA 300
Query: 298 IAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAML 357
IA+LF+IAGH YRTNWGIGH IK+ILE HKGPFTG+GHKGLYE LTTSWHAQL+ NLA L
Sbjct: 301 IAVLFIIAGHQYRTNWGIGHSIKEILENHKGPFTGEGHKGLYENLTTSWHAQLATNLAFL 360
Query: 358 GSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPT 417
GSLTII+AHHMY+MPPYPYLATDY TQL +FTHH+WIGGFLIVG AAHAAIFMVRDYDP
Sbjct: 361 GSLTIIIAHHMYAMPPYPYLATDYATQLCIFTHHIWIGGFLIVGGAAHAAIFMVRDYDPV 420
Query: 418 TRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ 477
N++LDRV+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTM ALGRPQDMFSDTAIQLQ
Sbjct: 421 VNQNNVLDRVIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMRALGRPQDMFSDTAIQLQ 480
Query: 478 PIFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIH 537
P+FAQW+QN H LAPG TAP A S +GGG ++AVG KVA++PI LGTADFL+HHIH
Sbjct: 481 PVFAQWVQNLHTLAPGGTAPNALEPVSYAFGGG-VLAVGGKVAMMPIALGTADFLIHHIH 539
Query: 538 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 597
AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVS WDHVFLGLFWM
Sbjct: 540 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSGWDHVFLGLFWM 599
Query: 598 YNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVI 657
YN++S+VIFHFSWKMQSDVWG++ G V+HITGGNFAQS+ITINGWLRDFLWAQASQVI
Sbjct: 600 YNSLSIVIFHFSWKMQSDVWGTVDAAGNVSHITGGNFAQSAITINGWLRDFLWAQASQVI 659
Query: 658 QSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRAL 717
SYGS+LSAYGL FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA QPRAL
Sbjct: 660 NSYGSALSAYGLMFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAIQPRAL 719
Query: 718 SIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
SI QGRAVGV HYLLGGIATTWAFF A I++VG
Sbjct: 720 SITQGRAVGVAHYLLGGIATTWAFFHAHILSVG 752
>dbj|BAA17437.1| (D90906) P700 apoprotein subunit Ia [Synechocystis sp.]
Length = 751
Score = 1321 bits (3381), Expect = 0.0
Identities = 606/752 (80%), Positives = 670/752 (88%), Gaps = 3/752 (0%)
Query: 1 MIIRSPEPEVK--ILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDS 58
M I PE E K + VD +P+ TSFE+W KPGHF RT+A+GP TTTWIWNLHA+AHDFDS
Sbjct: 1 MTISPPEREAKAKVSVDNNPVPTSFEKWGKPGHFDRTLARGPKTTTWIWNLHANAHDFDS 60
Query: 59 HTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPI 118
TSDLED+SRKIFSAHFG L+++F+WLSGMYFHGA+FSNYE WL+DPTHI+PSAQVVWPI
Sbjct: 61 QTSDLEDVSRKIFSAHFGHLAVVFVWLSGMYFHGAKFSNYEGWLADPTHIKPSAQVVWPI 120
Query: 119 VGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHY 178
VGQ ILNGDVGGGF GIQITSG F +WRASG T QLYCTAIGGLV AALMLFAGWFHY
Sbjct: 121 VGQGILNGDVGGGFHGIQITSGLFYLWRASGFTDSYQLYCTAIGGLVMAALMLFAGWFHY 180
Query: 179 HKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPH 238
H APKL WFQ+VESM+NHHLAGLLGLGSL WAGHQ+HVS+PIN+ L+AGV PK+IPLPH
Sbjct: 181 HVKAPKLEWFQNVESMMNHHLAGLLGLGSLGWAGHQIHVSMPINKLLDAGVAPKDIPLPH 240
Query: 239 EFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAI 298
EFIL +A+LYPSFA+G TPFFTLNW Y+DFLTF+GGL+PVTGGLWL+D AHHHLAI
Sbjct: 241 EFILEPSKMAELYPSFAQGLTPFFTLNWGVYSDFLTFKGGLNPVTGGLWLSDTAHHHLAI 300
Query: 299 AILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLG 358
A+LF+IAGHMYRTNWGIGH +K+ILEAHKGPFTG+GHKGLYEILTTSWHAQL+INLA+LG
Sbjct: 301 AVLFIIAGHMYRTNWGIGHSMKEILEAHKGPFTGEGHKGLYEILTTSWHAQLAINLALLG 360
Query: 359 SLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTT 418
SLTIIVA HMY+MPPYPY A DY TQLSLFTHHMWIGGFLIVGA AH AIFMVRDYDP
Sbjct: 361 SLTIIVAQHMYAMPPYPYQAIDYATQLSLFTHHMWIGGFLIVGAGAHGAIFMVRDYDPAK 420
Query: 419 RYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP 478
N+LLDR+LRHRDAIISHLNWVCIFLGFHSFGLYIHNDTM ALGRPQDMFSDTAIQLQP
Sbjct: 421 NVNNLLDRMLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMRALGRPQDMFSDTAIQLQP 480
Query: 479 IFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHA 538
IFAQW+Q+ H LAPG TAP A S + GG+ +AV KVA++PI LGTADF+VHHIHA
Sbjct: 481 IFAQWVQHLHTLAPGATAPNALATASYAF-GGETIAVAGKVAMMPITLGTADFMVHHIHA 539
Query: 539 FTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMY 598
FTIHVT LILLKGVL+ARSSRL+PDKANLGFRFPCDGPGRGGTCQVS WDHVFLGLFWMY
Sbjct: 540 FTIHVTALILLKGVLYARSSRLVPDKANLGFRFPCDGPGRGGTCQVSGWDHVFLGLFWMY 599
Query: 599 NAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQ 658
N++SVVIFHFSWKMQSDVWG++S G VTH+T GNFAQS+ITINGWLRDFLWAQA+ VI
Sbjct: 600 NSLSVVIFHFSWKMQSDVWGTVSPDGSVTHVTLGNFAQSAITINGWLRDFLWAQAANVIN 659
Query: 659 SYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALS 718
SYGS+LSAYG+ FL HFV+AFSLMFLFSGRGYWQELIESIVWAHNKL VAPA QPRALS
Sbjct: 660 SYGSALSAYGIMFLAGHFVFAFSLMFLFSGRGYWQELIESIVWAHNKLNVAPAIQPRALS 719
Query: 719 IVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
I+QGRAVGV HYLLGGI TTWAFFLAR +++G
Sbjct: 720 IIQGRAVGVAHYLLGGIVTTWAFFLARSLSIG 751
>sp|P29254|PSAA_SYNY3 PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||S18242 photosystem I protein A1 - Synechocystis sp. (strain PCC 6803)
emb|CAA41629.1| (X58825) P700 apoprotein subunit Ia (PSA-A) [Synechocystis
PCC6803]
Length = 751
Score = 1321 bits (3380), Expect = 0.0
Identities = 605/752 (80%), Positives = 670/752 (88%), Gaps = 3/752 (0%)
Query: 1 MIIRSPEPEVK--ILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDS 58
M I PE E K + VD +P+ TSFE+W KPGHF RT+A+GP TTTWIWNLHA+AHDFDS
Sbjct: 1 MTISPPEREAKAKVSVDNNPVPTSFEKWGKPGHFDRTLARGPKTTTWIWNLHANAHDFDS 60
Query: 59 HTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPI 118
TSDLED+SRKIFSAHFG L+++F+WLSGMYFHGA+FSNYE WL+DPTHI+PSAQVVWPI
Sbjct: 61 QTSDLEDVSRKIFSAHFGHLAVVFVWLSGMYFHGAKFSNYEGWLADPTHIKPSAQVVWPI 120
Query: 119 VGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHY 178
VGQ ILNGDVGGGF GIQITSG F +WRASG T QLYCTAIGGLV AALMLFAGWFHY
Sbjct: 121 VGQGILNGDVGGGFHGIQITSGLFYLWRASGFTDSYQLYCTAIGGLVMAALMLFAGWFHY 180
Query: 179 HKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPH 238
H APKL WFQ+VESM+NHHLAGLLGLGSL WAGHQ+HVS+PIN+ L+AGV PK+IPLPH
Sbjct: 181 HVKAPKLEWFQNVESMMNHHLAGLLGLGSLGWAGHQIHVSMPINKLLDAGVAPKDIPLPH 240
Query: 239 EFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAI 298
EFIL +A+LYPSFA+G TPFFTLNW Y+DFLTF+GGL+PVTGGLWL+D AHHHLAI
Sbjct: 241 EFILEPSKMAELYPSFAQGLTPFFTLNWGVYSDFLTFKGGLNPVTGGLWLSDTAHHHLAI 300
Query: 299 AILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLG 358
A+LF+IAGHMYRTNWGIGH +K+ILEAHKGPFTG+GHKGLYEILTTSWHAQL+INLA+LG
Sbjct: 301 AVLFIIAGHMYRTNWGIGHSMKEILEAHKGPFTGEGHKGLYEILTTSWHAQLAINLALLG 360
Query: 359 SLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTT 418
SLTIIVA HMY+MPPYPY A DY TQLSLFTHHMWIGGFLIVGA AH AIFMVRDYDP
Sbjct: 361 SLTIIVAQHMYAMPPYPYQAIDYATQLSLFTHHMWIGGFLIVGAGAHGAIFMVRDYDPAK 420
Query: 419 RYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP 478
N+LLDR+LRHRDAIISHLNWVCIFLGFHSFGLYIHNDTM ALGRPQDMFSDTAIQLQP
Sbjct: 421 NVNNLLDRMLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMRALGRPQDMFSDTAIQLQP 480
Query: 479 IFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHA 538
IFAQW+Q+ H LAPG TAP A S + GG+ +AV KVA++PI LGTADF+VHHIHA
Sbjct: 481 IFAQWVQHLHTLAPGATAPNALATASYAF-GGETIAVAGKVAMMPITLGTADFMVHHIHA 539
Query: 539 FTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMY 598
FTIHVT LILLKGVL+ARSSRL+PDKANLGFRFPCDGPGRGGTCQVS WDHVFLGLFWMY
Sbjct: 540 FTIHVTALILLKGVLYARSSRLVPDKANLGFRFPCDGPGRGGTCQVSGWDHVFLGLFWMY 599
Query: 599 NAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQ 658
N++S+VIFHFSWKMQSDVWG++S G VTH+T GNFAQS+ITINGWLRDFLWAQA+ VI
Sbjct: 600 NSLSIVIFHFSWKMQSDVWGTVSPDGSVTHVTLGNFAQSAITINGWLRDFLWAQAANVIN 659
Query: 659 SYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALS 718
SYGS+LSAYG+ FL HFV+AFSLMFLFSGRGYWQELIESIVWAHNKL VAPA QPRALS
Sbjct: 660 SYGSALSAYGIMFLAGHFVFAFSLMFLFSGRGYWQELIESIVWAHNKLNVAPAIQPRALS 719
Query: 719 IVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
I+QGRAVGV HYLLGGI TTWAFFLAR +++G
Sbjct: 720 IIQGRAVGVAHYLLGGIVTTWAFFLARSLSIG 751
>gi|11467572 PSI, P700 apoprotein A1 [Odontella sinensis]
sp|P49479|PSAA_ODOSI PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||S78377 photosystem I P700 apoprotein A1 - Odontella sinensis chloroplast
emb|CAA91750.1| (Z67753) PSI, P700 apoprotein A1 [Odontella sinensis]
Length = 752
Score = 1307 bits (3345), Expect = 0.0
Identities = 592/740 (80%), Positives = 672/740 (90%), Gaps = 1/740 (0%)
Query: 10 VKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRK 69
V+I V++D ++TSF +WA+PGHFSRT+AKGP TTTWIWNLHADAHDFDS TS LE++SRK
Sbjct: 13 VQIFVEKDAVETSFAKWAQPGHFSRTLAKGPKTTTWIWNLHADAHDFDSQTSSLEEVSRK 72
Query: 70 IFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVG 129
IFSAHFGQL++IFLW+SGM+FHGA FSNY AWLSDP I+ S+QVVWPIVGQEILN DVG
Sbjct: 73 IFSAHFGQLAVIFLWISGMHFHGAYFSNYSAWLSDPISIKQSSQVVWPIVGQEILNADVG 132
Query: 130 GGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQ 189
G F+G+Q TSG+FQ+WRA GITSE++LY AIGGL+ +ALMLFAGWFHYHKAAPKL WFQ
Sbjct: 133 GNFQGVQTTSGWFQMWRAEGITSEVELYWIAIGGLIMSALMLFAGWFHYHKAAPKLEWFQ 192
Query: 190 DVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQ 249
+ ESM+NHHLAGLLGLG LSW+GHQ+H++LPIN+ L+AGV P+EIPLPHEF++NR+L+AQ
Sbjct: 193 NAESMMNHHLAGLLGLGCLSWSGHQIHIALPINKLLDAGVAPQEIPLPHEFLINRELMAQ 252
Query: 250 LYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMY 309
LYPSF +G PFF+ W +Y+DFLTF+GGL+PVTGGLWL+DIAHHHLA+++LF+ AGHMY
Sbjct: 253 LYPSFNKGLAPFFSGQWGEYSDFLTFKGGLNPVTGGLWLSDIAHHHLALSVLFIFAGHMY 312
Query: 310 RTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMY 369
RTNWGIGH +K+ILEAHKGPFTG+GHKGLYEILTTSWHAQL+INLAM+GSL+IIVAHHMY
Sbjct: 313 RTNWGIGHSMKEILEAHKGPFTGEGHKGLYEILTTSWHAQLAINLAMMGSLSIIVAHHMY 372
Query: 370 SMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLR 429
+MPPYPY+ATDY TQLSLFTHHMWIGGF +VG AAH AIFMVRDY P YN+LLDRVLR
Sbjct: 373 AMPPYPYIATDYATQLSLFTHHMWIGGFCVVGGAAHGAIFMVRDYTPANNYNNLLDRVLR 432
Query: 430 HRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA 489
HRDAIISHLNWVCIFLG HSFGLYIHNDTM ALGRPQDMFSD AIQLQPIFAQW+QN H
Sbjct: 433 HRDAIISHLNWVCIFLGCHSFGLYIHNDTMRALGRPQDMFSDKAIQLQPIFAQWVQNIHL 492
Query: 490 LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILL 549
LAPG TAP A TS + GGD+V VG K+A++PI LGTADF+VHHIHAFTIHVTVLILL
Sbjct: 493 LAPGTTAPNALATTSYAF-GGDVVEVGGKIAMMPIKLGTADFMVHHIHAFTIHVTVLILL 551
Query: 550 KGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFS 609
KGVL+ARSS+LIPDKANLGFRFPCDGPGRGGTCQ S+WDHVFLGLFWMYN+ISVVIFHFS
Sbjct: 552 KGVLYARSSKLIPDKANLGFRFPCDGPGRGGTCQSSSWDHVFLGLFWMYNSISVVIFHFS 611
Query: 610 WKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGL 669
WKMQSDVWG+I+ G ++HITGGNFAQSSITINGWLRDFLW+QASQVIQSYGS+ SAYGL
Sbjct: 612 WKMQSDVWGTITPDGNISHITGGNFAQSSITINGWLRDFLWSQASQVIQSYGSASSAYGL 671
Query: 670 FFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTH 729
FLGAHF+WAFSLMFLFSGRGYWQELIESIVWAHNKL APA QPRALSI QGRAVG+ H
Sbjct: 672 IFLGAHFIWAFSLMFLFSGRGYWQELIESIVWAHNKLNFAPAIQPRALSITQGRAVGLAH 731
Query: 730 YLLGGIATTWAFFLARIIAV 749
YLLGGI TTW+FFLAR I++
Sbjct: 732 YLLGGIGTTWSFFLARAISI 751
>gi|11465395 unknown; Photosystem I p700 chlorophyll A apoprotein A1 [Cyanidium
caldarium]
gb|AAF12880.1|AF022186_2 (AF022186) unknown; Photosystem I p700 chlorophyll A apoprotein A1
[Cyanidium caldarium]
Length = 752
Score = 1306 bits (3342), Expect = 0.0
Identities = 591/740 (79%), Positives = 670/740 (89%), Gaps = 1/740 (0%)
Query: 11 KILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKI 70
KILVDR+P+ TSFE+W PGHFSR++AKGP TTTWIWNLHAD HDFDSHTS LEDISRKI
Sbjct: 14 KILVDRNPVPTSFEKWGVPGHFSRSLAKGPKTTTWIWNLHADVHDFDSHTSSLEDISRKI 73
Query: 71 FSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGG 130
FSAHFGQLS+IF+WLSGMYFHGARFSNY WLS+P+ I+PSAQ+VWPIVGQEILN D+GG
Sbjct: 74 FSAHFGQLSLIFIWLSGMYFHGARFSNYTIWLSNPSLIKPSAQIVWPIVGQEILNADLGG 133
Query: 131 GFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQD 190
G +GIQITSGFF +WRASGIT+EL+LY TA+GGL A LM F GWFHYHKAAPKL WFQ+
Sbjct: 134 GSQGIQITSGFFHLWRASGITNELELYVTALGGLFMAGLMAFGGWFHYHKAAPKLEWFQN 193
Query: 191 VESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQL 250
VESMLNHHLAGLLGLGSLSWAGHQ+HVSLPIN+ L+AGVDPK IPLPHEFILNR+L++QL
Sbjct: 194 VESMLNHHLAGLLGLGSLSWAGHQIHVSLPINKLLDAGVDPKTIPLPHEFILNRELMSQL 253
Query: 251 YPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYR 310
YPSF +G PFF+LNW Y+DFLTFRGGL+P+TG LW++DIAHHHLAI++LF++AGHMYR
Sbjct: 254 YPSFEKGLWPFFSLNWGAYSDFLTFRGGLNPITGSLWMSDIAHHHLAISVLFIVAGHMYR 313
Query: 311 TNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYS 370
TNWGIGH IK+IL+AH+GP TG GH+GLYE LTTSWHA L+INLA+ GSL+IIVAHHMY+
Sbjct: 314 TNWGIGHSIKEILDAHRGPLTGSGHRGLYEALTTSWHANLAINLALFGSLSIIVAHHMYA 373
Query: 371 MPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRH 430
MPPYPY++ DY TQLSLFTHH WIGGF IVGA+AH AIFM+RDY P+ YN++LDR++RH
Sbjct: 374 MPPYPYISIDYPTQLSLFTHHTWIGGFCIVGASAHGAIFMIRDYVPSQHYNNVLDRLIRH 433
Query: 431 RDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHAL 490
RDA+ISHLNWVCIFLG HSFGLYIHNDTM ALGR QDMFSDTAIQLQPIFAQWIQ+ H L
Sbjct: 434 RDALISHLNWVCIFLGTHSFGLYIHNDTMRALGRSQDMFSDTAIQLQPIFAQWIQSLHTL 493
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
AP TAP A TS + GG++VAV NK+A++P+ LGTADF+VHHIHAFTIHVT+LILLK
Sbjct: 494 APANTAPNALATTSYVF-GGEVVAVANKIAMMPMKLGTADFMVHHIHAFTIHVTLLILLK 552
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSW 610
GVLFAR+SRLIPDKANLGFRFPCDGPGRGGTCQVS+WDHVFLGLFWMYN ISVVIFHFSW
Sbjct: 553 GVLFARNSRLIPDKANLGFRFPCDGPGRGGTCQVSSWDHVFLGLFWMYNCISVVIFHFSW 612
Query: 611 KMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLF 670
KMQSDVWG++S +V+H+ GGNF+QS+ITINGWLRDFLWAQASQVIQSYGSS+SAYGL
Sbjct: 613 KMQSDVWGTVSQNSLVSHVVGGNFSQSAITINGWLRDFLWAQASQVIQSYGSSISAYGLM 672
Query: 671 FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHY 730
FL AHF+WAFSLMFLFSGRGYWQELIESIVWAH+KLK+AP QPRALSI QGRAVG HY
Sbjct: 673 FLAAHFIWAFSLMFLFSGRGYWQELIESIVWAHSKLKIAPTIQPRALSITQGRAVGAAHY 732
Query: 731 LLGGIATTWAFFLARIIAVG 750
LLGGIATTWAFFL+R I++G
Sbjct: 733 LLGGIATTWAFFLSRAISIG 752
>gi|11466999 PSI P700 apoprotein, subunit 1a [Euglena gracilis]
sp|P19430|PSAA_EUGGR PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||S26071 photosystem I protein A1 - Euglena gracilis chloroplast
emb|CAA77910.1| (Z11874) PSI P700 chl. a apoprotein [Euglena gracilis]
emb|CAA50093.1| (X70810) PSI P700 apoprotein, subunit 1a [Euglena gracilis]
Length = 751
Score = 1303 bits (3336), Expect = 0.0
Identities = 595/753 (79%), Positives = 681/753 (90%), Gaps = 5/753 (0%)
Query: 1 MIIRSPEPEVK---ILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFD 57
M I PE +VK + +P++TSFE+W++PGHFSR ++KGP+TTTWIWNLHADAHDFD
Sbjct: 1 MTITPPEQQVKRVRVAFVSNPVETSFEKWSRPGHFSRLLSKGPNTTTWIWNLHADAHDFD 60
Query: 58 SHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWP 117
+HT+DLEDISRK+FSAHFGQL+II +WLSGM+FHGARFSNYEAWL DP H++PSAQ+VWP
Sbjct: 61 NHTTDLEDISRKVFSAHFGQLAIIEIWLSGMFFHGARFSNYEAWLLDPIHVKPSAQIVWP 120
Query: 118 IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFH 177
IVGQEILNGDVGG F+GIQITSG FQ+WR+ GITSE QLY TA+ GL+F+A++ FAGWFH
Sbjct: 121 IVGQEILNGDVGGNFQGIQITSGLFQLWRSCGITSEFQLYITALTGLIFSAVLFFAGWFH 180
Query: 178 YHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLP 237
YHKAAPKL WFQ+VESMLNHHL+GLLGLG LSWAGHQ+HVSLPIN+ L++GV+P E+PLP
Sbjct: 181 YHKAAPKLEWFQNVESMLNHHLSGLLGLGCLSWAGHQIHVSLPINKLLDSGVNPAELPLP 240
Query: 238 HEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLA 297
H+FIL++ L++QLYPSF++G PFFT +W++Y+DFLTFRGGL+ VTGGLWLTD+AHHHLA
Sbjct: 241 HDFILDKSLISQLYPSFSKGLAPFFTFHWSEYSDFLTFRGGLNNVTGGLWLTDVAHHHLA 300
Query: 298 IAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAML 357
+A+LF++AGHMY+TNW IGH IK +LE+H GPFTGQGHKGLYEI T SWHAQLS+NLAM+
Sbjct: 301 LAVLFILAGHMYKTNWKIGHDIKGLLESHTGPFTGQGHKGLYEIFTNSWHAQLSLNLAMM 360
Query: 358 GSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPT 417
GSL+IIVA HMYSMPPYPY+A DYGT+LSLFTHH WIGGF IVGAAAHAAIFMVRDYDP
Sbjct: 361 GSLSIIVAQHMYSMPPYPYIAIDYGTELSLFTHHYWIGGFCIVGAAAHAAIFMVRDYDPA 420
Query: 418 TRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ 477
+N+LLDRVL HRDAIISHLNWVCIFLG HSFGLYIHNDT+SALGRPQDMFSD+AIQLQ
Sbjct: 421 LNFNNLLDRVLLHRDAIISHLNWVCIFLGLHSFGLYIHNDTLSALGRPQDMFSDSAIQLQ 480
Query: 478 PIFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIH 537
P+FAQWIQNTH LAP +TA + T+ W GGD+V++ KVA++PI LGTADFLVHHIH
Sbjct: 481 PVFAQWIQNTHYLAPTLTAFNLVSPTTPVW-GGDVVSISGKVAMMPIKLGTADFLVHHIH 539
Query: 538 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 597
AFTIHVTVLILLKGVLF+RSSRLIPDKA+LGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM
Sbjct: 540 AFTIHVTVLILLKGVLFSRSSRLIPDKASLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 599
Query: 598 YNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVI 657
YN+ISV IFHFSWKMQSDVWG++ V+HITGGNF+Q S+TINGWLRDFLWAQ+SQVI
Sbjct: 600 YNSISVAIFHFSWKMQSDVWGTVL-ANKVSHITGGNFSQGSLTINGWLRDFLWAQSSQVI 658
Query: 658 QSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRAL 717
QSYGS LSAYGL FLGAHFVWAF+LMFLFSGRGYWQELIESIVWAHNKLKVAP QPRAL
Sbjct: 659 QSYGSPLSAYGLMFLGAHFVWAFTLMFLFSGRGYWQELIESIVWAHNKLKVAPNIQPRAL 718
Query: 718 SIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
SI QGRAVGV HYLLGGIATTW+FFLARII+VG
Sbjct: 719 SITQGRAVGVAHYLLGGIATTWSFFLARIISVG 751
>sp|P17154|PSAA_SYNP2 PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A1
pir||S06397 photosystem I protein A1 - Synechococcus sp. (PCC 7002)
gb|AAA88632.1| (M18165) chlorophyll a/b apoprotein [Synechococcus sp.]
prf||1410329A P700 chlorophyll a 81.7kD protein [Synechococcus sp.]
Length = 739
Score = 1291 bits (3305), Expect = 0.0
Identities = 586/739 (79%), Positives = 665/739 (89%), Gaps = 2/739 (0%)
Query: 12 ILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKIF 71
+ VD+DP+ TSFE+W KPGHF RT+A+GP TTTWIWNLHADAHDFDS TSDLEDISRKIF
Sbjct: 3 VKVDKDPVPTSFEKWGKPGHFDRTLARGPKTTTWIWNLHADAHDFDSQTSDLEDISRKIF 62
Query: 72 SAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGG 131
SAHFG L+++F+WLSGMYFHGA+FSNYEAWL++PT I+PSAQVVWP+VGQ ILNGDVGGG
Sbjct: 63 SAHFGHLAVVFVWLSGMYFHGAKFSNYEAWLNNPTVIKPSAQVVWPVVGQGILNGDVGGG 122
Query: 132 FRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDV 191
F GIQITSG F +WRA+G T+ QLY TAIGGLV AALM+FAGWFHYHKAAPKL WFQ+
Sbjct: 123 FSGIQITSGLFYLWRAAGFTNSYQLYVTAIGGLVMAALMVFAGWFHYHKAAPKLEWFQNA 182
Query: 192 ESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLY 251
E+M+NHHL+ LLG GSL W GH +HVSLP+N+ L++GV K+IPLPHEF + ++A+LY
Sbjct: 183 EAMMNHHLSVLLGCGSLGWTGHLIHVSLPVNKLLDSGVAAKDIPLPHEFF-DASVMAELY 241
Query: 252 PSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRT 311
PSFA+G PFFTL+WA Y+DFLTF+GGL+P TG LWL+D+AHHHLAIA+LF+IAGHMYRT
Sbjct: 242 PSFAQGLKPFFTLDWAAYSDFLTFKGGLNPTTGSLWLSDVAHHHLAIAVLFIIAGHMYRT 301
Query: 312 NWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSM 371
+GIGH +K+ILEAHKGPFTG+GHKGLYEILTTSWHAQL++NLA+LGSLTI++AHHMYSM
Sbjct: 302 GFGIGHSMKEILEAHKGPFTGEGHKGLYEILTTSWHAQLAVNLALLGSLTIVIAHHMYSM 361
Query: 372 PPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHR 431
PPYPY+ATDYGTQLSLFTHH WIGGFLIVGA AH AIFMVRDYDP N+LLDRV+RHR
Sbjct: 362 PPYPYMATDYGTQLSLFTHHTWIGGFLIVGAGAHGAIFMVRDYDPAKNVNNLLDRVIRHR 421
Query: 432 DAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALA 491
DAIISHLNWVCIFLGFHSFGLYIHNDTM ALGRPQDMFSD+AIQLQPIFAQWIQN H LA
Sbjct: 422 DAIISHLNWVCIFLGFHSFGLYIHNDTMRALGRPQDMFSDSAIQLQPIFAQWIQNIHTLA 481
Query: 492 PGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKG 551
PG TAP A S + GGD+VA+GNKVAL+PI LGTADF+VHHIHAFTIHVTVLILLKG
Sbjct: 482 PGGTAPNAIASASQVF-GGDVVAIGNKVALMPITLGTADFMVHHIHAFTIHVTVLILLKG 540
Query: 552 VLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWK 611
+L++RSSRL+PDK LGFRFPCDGPGRGGTCQVS WDHVFLGLFWMYN++S+VIFHFSWK
Sbjct: 541 LLYSRSSRLVPDKGQLGFRFPCDGPGRGGTCQVSGWDHVFLGLFWMYNSLSIVIFHFSWK 600
Query: 612 MQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFF 671
MQSDVWG+I G V+H+TGGNFA S+ITINGWLRDFLWAQ+SQVI SYGS+LSAYG+ F
Sbjct: 601 MQSDVWGTILPDGSVSHVTGGNFATSAITINGWLRDFLWAQSSQVINSYGSALSAYGIMF 660
Query: 672 LGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHYL 731
L HFV+AFSLMFLFSGRGYWQELIESIVWAHNKLK+APA QPRALSIVQGRAVGV HYL
Sbjct: 661 LAGHFVFAFSLMFLFSGRGYWQELIESIVWAHNKLKLAPAIQPRALSIVQGRAVGVAHYL 720
Query: 732 LGGIATTWAFFLARIIAVG 750
LGGI TTWAFFLAR +++G
Sbjct: 721 LGGIVTTWAFFLARSLSIG 739
>gb|AAF29815.1| (AF180014) photosystem I P700 apoprotein A1 [Marsilea botrycarpa]
Length = 720
Score = 1291 bits (3304), Expect = 0.0
Identities = 600/719 (83%), Positives = 641/719 (88%)
Query: 12 ILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKIF 71
I+V++DP+KTSFE+WA+PGHFS+ +AKGP TTTWIWNLHADAHDFDSHT+DLEDISRKIF
Sbjct: 2 IMVEKDPVKTSFEKWAQPGHFSKALAKGPSTTTWIWNLHADAHDFDSHTNDLEDISRKIF 61
Query: 72 SAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGG 131
SAHFGQL IIF+WLSGMYFHGARFSNYEAWL+DPTH++PSAQVVWPIVGQEILNGDVGGG
Sbjct: 62 SAHFGQLGIIFIWLSGMYFHGARFSNYEAWLNDPTHVKPSAQVVWPIVGQEILNGDVGGG 121
Query: 132 FRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDV 191
F+GIQITSGFFQ+WRASGITSELQLYCTAIGGL+FA LM FAGWFHYHKAAPKLAWFQ+V
Sbjct: 122 FQGIQITSGFFQLWRASGITSELQLYCTAIGGLIFAGLMFFAGWFHYHKAAPKLAWFQNV 181
Query: 192 ESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLY 251
ESMLNHHLAGLLGLGSL+WAGHQ+HVSLPINQ L+AGVDPKEIPLPHEFILNR+L+AQ +
Sbjct: 182 ESMLNHHLAGLLGLGSLAWAGHQIHVSLPINQLLDAGVDPKEIPLPHEFILNRELMAQTF 241
Query: 252 PSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRT 311
PSFA+G PFFTL+W++Y+DFLTFRGGL+PVTGGLWLTD AHHHLAIA+LFL+AGHMYRT
Sbjct: 242 PSFAKGLIPFFTLDWSEYSDFLTFRGGLNPVTGGLWLTDTAHHHLAIAVLFLVAGHMYRT 301
Query: 312 NWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSM 371
AHHMY+M
Sbjct: 302 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXAHHMYAM 361
Query: 372 PPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHR 431
PPYPYLATDY TQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTT+YN+LLDRV+RHR
Sbjct: 362 PPYPYLATDYATQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTQYNNLLDRVIRHR 421
Query: 432 DAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALA 491
DAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQW+QNTHALA
Sbjct: 422 DAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWVQNTHALA 481
Query: 492 PGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKG 551
PG TAP A TS TWGG DLVAVG KVAL PIPLGTADFLVHHIHAFTIHVTVLILLKG
Sbjct: 482 PGSTAPNAAASTSPTWGGSDLVAVGGKVALAPIPLGTADFLVHHIHAFTIHVTVLILLKG 541
Query: 552 VLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWK 611
VLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+ISVVIFHFSWK
Sbjct: 542 VLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSISVVIFHFSWK 601
Query: 612 MQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFF 671
MQSDVWGSIS++GVV HITGGNFAQSS TINGWLRDFLWAQASQVIQSYGSSLSAYGL F
Sbjct: 602 MQSDVWGSISEEGVVNHITGGNFAQSSTTINGWLRDFLWAQASQVIQSYGSSLSAYGLLF 661
Query: 672 LGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHY 730
LGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP TQPRALSI+QGRAVGV HY
Sbjct: 662 LGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPVTQPRALSIIQGRAVGVAHY 720
>gb|AAA84451.1| (M37526) alpha-apoprotein [Euglena gracilis]
Length = 711
Score = 1209 bits (3095), Expect = 0.0
Identities = 561/753 (74%), Positives = 643/753 (84%), Gaps = 45/753 (5%)
Query: 1 MIIRSPEPEVK---ILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFD 57
M I PE +VK + +P++TSFE+W++PGHFSR ++KGP+TTTWIWNLHADAHDFD
Sbjct: 1 MTITPPEQQVKRVRVAFVSNPVETSFEKWSRPGHFSRLLSKGPNTTTWIWNLHADAHDFD 60
Query: 58 SHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWP 117
+HT+DL DP H++PSAQ+VWP
Sbjct: 61 NHTTDL----------------------------------------DPIHVKPSAQIVWP 80
Query: 118 IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFH 177
IVGQEILNGDVGG F+GIQITSG FQ+WR+ GITSE QLY TA+ GL+F+A++ FAGWFH
Sbjct: 81 IVGQEILNGDVGGNFQGIQITSGLFQLWRSCGITSEFQLYITALTGLIFSAVLFFAGWFH 140
Query: 178 YHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLP 237
YHKAAPKL WFQ+VESMLNHHL+GLLGLG LSWAGHQ+HVSLPIN+ L++GV+P E+PLP
Sbjct: 141 YHKAAPKLEWFQNVESMLNHHLSGLLGLGCLSWAGHQIHVSLPINKLLDSGVNPAELPLP 200
Query: 238 HEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLA 297
H+FIL++ L++QLYPSF++G PFFT +W++Y+DFLTFRGGL+ VTGGLWLTD+AHHHLA
Sbjct: 201 HDFILDKSLISQLYPSFSKGLAPFFTFHWSEYSDFLTFRGGLNNVTGGLWLTDVAHHHLA 260
Query: 298 IAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAML 357
+A+LF++AGHMY+TNW IGH IK +LE+H GPFTGQGHKGLYEI T SWHAQLS+NLAM+
Sbjct: 261 LAVLFILAGHMYKTNWKIGHDIKGLLESHTGPFTGQGHKGLYEIFTNSWHAQLSLNLAMM 320
Query: 358 GSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPT 417
GSL+IIVA HMYSMPPYPY+A DYGT+LSLFTHH WIGGF IVGAAAHAAIFMVRDYDP
Sbjct: 321 GSLSIIVAQHMYSMPPYPYIAIDYGTELSLFTHHYWIGGFCIVGAAAHAAIFMVRDYDPA 380
Query: 418 TRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQ 477
+N+LLDRVL HRDAIISHLNWVCIFLG HSFGLYIHNDT+SALGRPQDMFSD+AIQLQ
Sbjct: 381 LNFNNLLDRVLLHRDAIISHLNWVCIFLGLHSFGLYIHNDTLSALGRPQDMFSDSAIQLQ 440
Query: 478 PIFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIH 537
P+FAQWIQNTH LAP +TA + T+ W GGD+V++ KVA++PI LGTADFLVHHIH
Sbjct: 441 PVFAQWIQNTHYLAPTLTAFNLVSPTTPVW-GGDVVSISGKVAMMPIKLGTADFLVHHIH 499
Query: 538 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 597
AFTIHVTVLILLKGVLF+RSSRLIPDKA+LGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM
Sbjct: 500 AFTIHVTVLILLKGVLFSRSSRLIPDKASLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 559
Query: 598 YNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVI 657
YN+ISV IFHFSWKMQSDVWG++ V+HITGGNF+Q S+TINGWLRDFLWAQ+SQVI
Sbjct: 560 YNSISVAIFHFSWKMQSDVWGTVL-ANKVSHITGGNFSQGSLTINGWLRDFLWAQSSQVI 618
Query: 658 QSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRAL 717
QSYGS LSAYGL FLGAHFVWAF+LMFLFSGRGYWQELIESIVWAHNKLKVAP QPRAL
Sbjct: 619 QSYGSPLSAYGLMFLGAHFVWAFTLMFLFSGRGYWQELIESIVWAHNKLKVAPNIQPRAL 678
Query: 718 SIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
SI QGRAVGV HYLLGGIATTW+FFLARII+VG
Sbjct: 679 SITQGRAVGVAHYLLGGIATTWSFFLARIISVG 711
>emb|CAB64208.1| (AJ133190) photosystem I subunit PsaA [Synechococcus sp. WH7803]
Length = 767
Score = 1185 bits (3031), Expect = 0.0
Identities = 561/771 (72%), Positives = 637/771 (81%), Gaps = 25/771 (3%)
Query: 1 MIIRSPE--PEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDS 58
M I PE K V++ +F KPGHF R +AKGP TTTW+WNLHA+AHDFDS
Sbjct: 1 MTISPPERGSTAKTQVEKVDNPATFGAVRKPGHFDRALAKGPKTTTWVWNLHANAHDFDS 60
Query: 59 HTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPI 118
HTSDLE++SRKIFSAHFG L++IF+WLSG +FHGARFSN+ WL+DPTH++PSAQVVWP+
Sbjct: 61 HTSDLEEVSRKIFSAHFGHLAVIFIWLSGAFFHGARFSNFSGWLADPTHVKPSAQVVWPV 120
Query: 119 VGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHY 178
GQEILNGD+G GF+GIQITSG F +WRA GIT+E QL AIG LV A LML AG FHY
Sbjct: 121 FGQEILNGDMGAGFQGIQITSGLFHVWRAWGITNETQLMSLAIGALVMAGLMLNAGVFHY 180
Query: 179 HKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDP------- 231
HKAAPKL WFQ+VESMLNHHLAGLLGLGSLSW GH +HVSLP + ++A +D
Sbjct: 181 HKAAPKLEWFQNVESMLNHHLAGLLGLGSLSWTGHLLHVSLPTTKLMDA-IDAGQPLVLN 239
Query: 232 -------KEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTG 284
+IPLPHEF N+D +AQLYP F G FF+ NWA Y+DFLTF+GGL+PVTG
Sbjct: 240 GKTIASVADIPLPHEFF-NQDFVAQLYPGFGAGIGAFFSGNWAAYSDFLTFKGGLNPVTG 298
Query: 285 GLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIKDILEAHKG-----PFTGQGHKGLY 339
+W++DIAHHHLAIA+LF++AGHMYRTNWGIGH IK+ILE KG P T +GH GL+
Sbjct: 299 SMWMSDIAHHHLAIAVLFIVAGHMYRTNWGIGHSIKEILEGQKGDPLLFPAT-KGHDGLF 357
Query: 340 EILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLI 399
E +TTSWHAQL++NLAMLGSL+IIVA HMY+MPPY Y++ DY TQ+ LFTHHMWIGGFLI
Sbjct: 358 EFMTTSWHAQLAVNLAMLGSLSIIVAQHMYAMPPYAYMSIDYPTQIGLFTHHMWIGGFLI 417
Query: 400 VGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTM 459
VGAAAHA M+RDYDP +++LDRVL+ RDA+ISHLNWVCI+LGFHSFGLYIHNDTM
Sbjct: 418 VGAAAHADTAMIRDYDPAKHVDNVLDRVLKARDALISHLNWVCIWLGFHSFGLYIHNDTM 477
Query: 460 SALGRPQDMFSDTAIQLQPIFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKV 519
ALGRPQDMFSD+AIQL+P+FAQWIQ HA A G TAP A G S + G +VAVG KV
Sbjct: 478 RALGRPQDMFSDSAIQLKPVFAQWIQGLHAAAAGSTAPNALAGVSEVFNGS-VVAVGGKV 536
Query: 520 ALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRG 579
A PIPLGTADF+VHHIHAFTIHVTVLILLKGVL+AR+SRLIPDKANLGFRFPCDGPGRG
Sbjct: 537 AAAPIPLGTADFMVHHIHAFTIHVTVLILLKGVLYARNSRLIPDKANLGFRFPCDGPGRG 596
Query: 580 GTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSI 639
GTCQVSAWDHVFLGLFWMYN++S+VIFHFSWKMQSDVWG+++ G V HIT GNFA S+I
Sbjct: 597 GTCQVSAWDHVFLGLFWMYNSLSIVIFHFSWKMQSDVWGTVNADGSVQHITNGNFANSAI 656
Query: 640 TINGWLRDFLWAQASQVIQSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESI 699
TINGWLRDFLWAQA+QVI SYGS+ SAYGL FLGAHFVW FSLMFLFSGRGYWQELIESI
Sbjct: 657 TINGWLRDFLWAQAAQVINSYGSNTSAYGLMFLGAHFVWPFSLMFLFSGRGYWQELIESI 716
Query: 700 VWAHNKLKVAPATQPRALSIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
VWAHNKLKVAPA QPRALSI QGRAVGV HYLLGGIATTWAFF A I+ VG
Sbjct: 717 VWAHNKLKVAPAIQPRALSITQGRAVGVAHYLLGGIATTWAFFHAHILVVG 767
>gb|AAK08970.1| (AY026898) photosystem I P700 protein subunit A [Prochlorothrix
hollandica]
Length = 699
Score = 1140 bits (2916), Expect = 0.0
Identities = 540/701 (77%), Positives = 599/701 (85%), Gaps = 8/701 (1%)
Query: 56 FDSHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVV 115
FDSHTSDLED+SRKIFSAHFG L++IF+WLSG YFHGARFSNY AWLSDP I+PSAQVV
Sbjct: 1 FDSHTSDLEDVSRKIFSAHFGHLAVIFIWLSGAYFHGARFSNYTAWLSDPISIKPSAQVV 60
Query: 116 WP---IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLF 172
WP I GQEILNGDVGGGF GIQITSG F +WRA GIT +LY TAIG LV A LMLF
Sbjct: 61 WPFRSIFGQEILNGDVGGGFHGIQITSGLFHLWRACGITHSSELYATAIGALVMAGLMLF 120
Query: 173 AGWFHYHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPK 232
AGWFHYHKAAPKL WFQ+VESMLNHHL+ LLG GSL WAGH +H+SLP+N L+AGV P
Sbjct: 121 AGWFHYHKAAPKLEWFQNVESMLNHHLSVLLGCGSLGWAGHLIHISLPVNALLDAGVSPA 180
Query: 233 EIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIA 292
+IPL +++L+ +A+ + G PFFTLNW Y+DFLTF+GGL+P TG LWLTDIA
Sbjct: 181 DIPLAKDYVLDAGYMAK-FSQLCRGLNPFFTLNWGVYSDFLTFKGGLNPQTGSLWLTDIA 239
Query: 293 HHHLAIAILFLIAGHMYRTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQL 350
HH LAIA+LF+IAGHMYRTNWGIGH +K +L+ HKGP G GH GLYEILTTSWHAQL
Sbjct: 240 HHQLAIAVLFIIAGHMYRTNWGIGHDMKALLDGHKGPVGEVGTGHAGLYEILTTSWHAQL 299
Query: 351 SINLAMLGSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFM 410
+INLA+LGSL+IIVAHHMY+MPPYPYLA DY TQLSLFTHH+WIGGFLIVGA AHAAIFM
Sbjct: 300 AINLALLGSLSIIVAHHMYAMPPYPYLAIDYPTQLSLFTHHVWIGGFLIVGAGAHAAIFM 359
Query: 411 VRDYDPTTRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFS 470
+RDYDP ++LLDRV+RHRDAIISHLNWVCI+LGFHSFGLYIHNDTM ALGRPQDMFS
Sbjct: 360 IRDYDPAKNVDNLLDRVIRHRDAIISHLNWVCIWLGFHSFGLYIHNDTMRALGRPQDMFS 419
Query: 471 DTAIQLQPIFAQWIQNTHALAPGIT-APGATTGTSLTWGGGDLVAVGNKVALLPIPLGTA 529
D+AIQLQPIFAQ IQ+ A G AP TS WGG D +AVG KVA+ IPLGTA
Sbjct: 420 DSAIQLQPIFAQGIQSIQAAVAGSAQAPWVGAATSPVWGG-DTIAVGGKVAMSAIPLGTA 478
Query: 530 DFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDH 589
DF+VHHIHAFTIHVTVLILLKGVL+AR+SRL+PDKA LGF FPCDGPGRGGTCQVSAWDH
Sbjct: 479 DFMVHHIHAFTIHVTVLILLKGVLYARNSRLVPDKAELGFAFPCDGPGRGGTCQVSAWDH 538
Query: 590 VFLGLFWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFL 649
VFLGLFWMYN++S+VIFHFSWKMQSDVWG++ G V +IT GNFA+S++TING LRDFL
Sbjct: 539 VFLGLFWMYNSLSIVIFHFSWKMQSDVWGTVYPDGSVLNITVGNFAESALTINGRLRDFL 598
Query: 650 WAQASQVIQSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVA 709
WAQA+ VI SYGS+LSAYGL FL A FVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVA
Sbjct: 599 WAQAASVINSYGSALSAYGLMFLAADFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVA 658
Query: 710 PATQPRALSIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
PA QPRALSI QGRAVGV HYLLGGIATTWAFFLARII+VG
Sbjct: 659 PAIQPRALSITQGRAVGVAHYLLGGIATTWAFFLARIISVG 699
>emb|CAB64200.1| (AJ133191) photosystem I subunit PsaA [Prochlorococcus sp.
CCMP1378]
Length = 767
Score = 1080 bits (2763), Expect = 0.0
Identities = 510/772 (66%), Positives = 613/772 (79%), Gaps = 38/772 (4%)
Query: 5 SPEPEVKIL---VDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTS 61
S E + KIL V DP F + KPG +S ++KGP TTTWIWNLHADAHDFD HT
Sbjct: 8 SGEKDKKILESPVKADPRPIDFAKLDKPGFWSSKLSKGPKTTTWIWNLHADAHDFDIHTG 67
Query: 62 DLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQ 121
D E+ +RKIFSAHFG L++IF+W+S +FHGARFSNY WL+DPTH++P AQ VW IVGQ
Sbjct: 68 DAEEATRKIFSAHFGHLAVIFIWMSAAFFHGARFSNYSGWLADPTHVKPGAQQVWAIVGQ 127
Query: 122 EILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKA 181
E+LNGD+G + GIQI+SG F +WRA GIT+E +L AIG +V AALML AG FHYHKA
Sbjct: 128 EMLNGDLGANYNGIQISSGVFHMWRAWGITNESELMALAIGAVVMAALMLHAGIFHYHKA 187
Query: 182 APKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNA-------GVDPKEI 234
APK+ WFQ++ESMLNHH+AGL+GLGSL+WAGH +H+ P L+A ++ KEI
Sbjct: 188 APKMEWFQNIESMLNHHIAGLVGLGSLAWAGHCIHIGAPTAALLDAIDAGTPLVINGKEI 247
Query: 235 ------PLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWL 288
P+PH+ + + ++AQ++P A G FF+LNW ++DFLTF+GGL+PVTG LW+
Sbjct: 248 ATIADMPMPHQ-LCDPQIIAQIFPGLASGTGNFFSLNWLAFSDFLTFKGGLNPVTGSLWM 306
Query: 289 TDIAHHHLAIAILFLIAGHMYRTNWGIGHGIKDILEAHKG-PF---TGQGHKGLYEILTT 344
TD++HHHLA ++ +I GHMYRTN+GIGH +K+IL++ +G P +GH+GL+E +
Sbjct: 307 TDVSHHHLAFGVIAIIGGHMYRTNYGIGHSMKEILDSQQGDPILFPAPKGHQGLFEFMAE 366
Query: 345 SWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAA 404
S HAQLS+NLAMLGSL+I+++HHMY+MPPYPY+ATDY T L LFTHHMWIGG IVGA A
Sbjct: 367 SRHAQLSVNLAMLGSLSILISHHMYAMPPYPYIATDYMTVLGLFTHHMWIGGLFIVGAGA 426
Query: 405 HAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGR 464
HA I MVRDYDP +++LDR+L+ RDA+ISHLNWVC++LGFHSFGLYIHNDTM ALGR
Sbjct: 427 HAGIAMVRDYDPAKHIDNVLDRILKARDALISHLNWVCMWLGFHSFGLYIHNDTMRALGR 486
Query: 465 PQDMFSDTAIQLQPIFAQWIQNTHALAPGITAPGATTGTSLTWG------GGDLVAVGNK 518
PQDMFSD+AIQLQPIFAQW+Q+ A A G T+ A T +L G LV VG K
Sbjct: 487 PQDMFSDSAIQLQPIFAQWVQSIQASAVG-TSILAGTAEALPHKAISEVFNGSLVEVGGK 545
Query: 519 VALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGR 578
VA+ PIPLGTAD ++HHIHAF IHVTVLILLKGVL+ARSSRLIPDKA+LGFRFPCDGPGR
Sbjct: 546 VAIAPIPLGTADLMIHHIHAFQIHVTVLILLKGVLYARSSRLIPDKASLGFRFPCDGPGR 605
Query: 579 GGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSS 638
GGTCQVS+WDHVFLGLFWMYN +S+VIFHFSWKMQSDVWG +TGGNF+QS+
Sbjct: 606 GGTCQVSSWDHVFLGLFWMYNCLSIVIFHFSWKMQSDVWG----------LTGGNFSQSA 655
Query: 639 ITINGWLRDFLWAQASQVIQSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIES 698
ITINGWLRDFLWAQ+SQV+ SYGS++S YGL FLGAHF+WAFSLMFLFSGRGYWQEL ES
Sbjct: 656 ITINGWLRDFLWAQSSQVLTSYGSAISMYGLMFLGAHFIWAFSLMFLFSGRGYWQELFES 715
Query: 699 IVWAHNKLKVAPATQPRALSIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
IVWAHNKLKVAP QPRALSI QGRAVGVTH+L+GGIATTWAFF AR+ +G
Sbjct: 716 IVWAHNKLKVAPTIQPRALSITQGRAVGVTHFLVGGIATTWAFFHARLFGLG 767
>emb|CAB64198.2| (AJ133192) photosystem I subunit PsaA [Prochlorococcus marinus]
Length = 773
Score = 1044 bits (2671), Expect = 0.0
Identities = 489/763 (64%), Positives = 588/763 (76%), Gaps = 39/763 (5%)
Query: 14 VDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKIFSA 73
VDRD +E+ KPG +S ++KGP TTTWIWNLHADAHDFD+H DLE+ SRKIFSA
Sbjct: 24 VDRDHAPIDYEKLNKPGFWSSKLSKGPKTTTWIWNLHADAHDFDTHLGDLEETSRKIFSA 83
Query: 74 HFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGGFR 133
HFG L+++F+W+S +FHGARFSNY WL+DPT+++P AQVVWP+VGQEILN D+GG ++
Sbjct: 84 HFGHLAVVFIWMSAAFFHGARFSNYTGWLADPTNVKPGAQVVWPVVGQEILNADLGGNYQ 143
Query: 134 GIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDVES 193
G+QITSG FQ+WRA GITSE+QL AIGG++ AALML G +HYHKAAPKL WF+ +E
Sbjct: 144 GLQITSGIFQMWRAWGITSEVQLMALAIGGVIMAALMLHGGIYHYHKAAPKLEWFRKIEP 203
Query: 194 MLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNA----------GV---DPKEIPLPHEF 240
ML HH L+GLGS++WAGH +H+ P+ L+A GV ++
Sbjct: 204 MLQHHQIALIGLGSIAWAGHLIHIGAPVAALLDAIDAGNPLVVDGVSICSAADVTNLAPR 263
Query: 241 ILNRDLLAQLYPSFA-EGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIA 299
+ + + +Q++PS A FFTLNW + D LT +GGL+PVTG LW+TDI+HHHLA
Sbjct: 264 LCDPAVASQIFPSLAGRTVENFFTLNWWAFTDILTNKGGLNPVTGSLWMTDISHHHLAFG 323
Query: 300 ILFLIAGHMYRTN-WGIGHGIKDILEAHKG-PF---TGQGHKGLYEILTTSWHAQLSINL 354
+ + GHM+R N G+GH +K+I++ HKG P +GH+G++E L+ SWH QLSINL
Sbjct: 324 VFAIFGGHMWRNNVHGVGHSMKEIMDVHKGDPILFPAPKGHQGIFEFLSNSWHGQLSINL 383
Query: 355 AMLGSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDY 414
AM+GS +I+VAHHMY++PPYPY+A DY T L LFTHHMWIGG I GAAAHA I M+RDY
Sbjct: 384 AMVGSASIVVAHHMYALPPYPYIAIDYPTVLGLFTHHMWIGGLFICGAAAHAGIAMIRDY 443
Query: 415 DPTTRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAI 474
DP +++LDR+L+ RDAIISHLNWVC++LGFHSFGLYIHND M ALGRP+DMFSDT I
Sbjct: 444 DPAVHIDNVLDRILKARDAIISHLNWVCMWLGFHSFGLYIHNDVMRALGRPKDMFSDTGI 503
Query: 475 QLQPIFAQWIQNTHALAPG-------ITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLG 527
QLQP AQW+QN A G + PG+ G++V VG KVA+ PIPLG
Sbjct: 504 QLQPFLAQWVQNLQQSAVGTGDLVGVVILPGSVLSEVF---NGNVVEVGGKVAIAPIPLG 560
Query: 528 TADFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAW 587
TAD ++HH+HAFTIHVT+LILLKGVL+ARSSRLIPDKA LGFRFPCDGPGRGGTCQVS+W
Sbjct: 561 TADLMIHHVHAFTIHVTLLILLKGVLYARSSRLIPDKAQLGFRFPCDGPGRGGTCQVSSW 620
Query: 588 DHVFLGLFWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRD 647
DHVFLGLFWMYN++SVVIFHFSWKMQSDVWG +TGGNFAQSSITINGWLRD
Sbjct: 621 DHVFLGLFWMYNSLSVVIFHFSWKMQSDVWG----------LTGGNFAQSSITINGWLRD 670
Query: 648 FLWAQASQVIQSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLK 707
FLWAQ+SQV+ SYG +S YGL FLGAHFVWAFSLMFLFSGRGYWQEL ESI+WAHNKLK
Sbjct: 671 FLWAQSSQVLTSYGQPISMYGLMFLGAHFVWAFSLMFLFSGRGYWQELFESIIWAHNKLK 730
Query: 708 VAPATQPRALSIVQGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
VAP QPRALSI QGRAVGV H+LLGGIATTWAFF AR+I +G
Sbjct: 731 VAPTIQPRALSITQGRAVGVAHFLLGGIATTWAFFHARLIGLG 773
>dbj|BAB18852.1| (AB044231) P700 chlorophyll a-protein A1 [Pandorina morum]
Length = 498
Score = 936 bits (2394), Expect = 0.0
Identities = 427/499 (85%), Positives = 471/499 (93%), Gaps = 1/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ ILNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLILNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH IK+ILEAHKGPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIV+H
Sbjct: 121 HMYRTNWGIGHSIKEILEAHKGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVSH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHHMWIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHMWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHH+HAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHVHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+ISVVIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSISVVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ +V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTSNNLVSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 479
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 480 YGLIFLGAHFVWAFSLMFL 498
>dbj|BAB18347.1| (AB044421) photosystem I P700 chlorophyll a apoprotein A1
[Lobomonas monstruosa]
Length = 497
Score = 936 bits (2393), Expect = 0.0
Identities = 430/499 (86%), Positives = 470/499 (94%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHL GLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLGGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLLLNRGI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGLAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH +K+ILEAHKGPFTG+GH GLYEILTTSWHAQL+INLAM GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSMKEILEAHKGPFTGEGHVGLYEILTTSWHAQLAINLAMFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMYSMPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYSMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAPG+TAP A TSLTW GGDLVAVG KVA++PI LGTADF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPGLTAPNALGATSLTW-GGDLVAVGGKVAMMPISLGTADFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWGS++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGSSLSA
Sbjct: 420 HFSWKMQSDVWGSVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSSLSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18850.1| (AB044227) P700 chlorophyll a-protein A1 [Pandorina morum]
Length = 497
Score = 933 bits (2385), Expect = 0.0
Identities = 428/499 (85%), Positives = 471/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ ILNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLILNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH IK+ILEAHKGPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIV+H
Sbjct: 121 HMYRTNWGIGHSIKEILEAHKGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVSH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHHMWIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHMWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHH+HAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHVHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+ISVVIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSISVVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18849.1| (AB044226) P700 chlorophyll a-protein A1 [Pandorina morum]
Length = 497
Score = 932 bits (2382), Expect = 0.0
Identities = 427/499 (85%), Positives = 471/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ ILNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLILNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH +K+ILEAHKGPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIV+H
Sbjct: 121 HMYRTNWGIGHSMKEILEAHKGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVSH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHHMWIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHMWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHH+HAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHVHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+ISVVIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSISVVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18346.1| (AB044420) photosystem I P700 chlorophyll a apoprotein A1
[Vitreochlamys ordinata]
Length = 497
Score = 930 bits (2377), Expect = 0.0
Identities = 427/499 (85%), Positives = 469/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHL GLLGLGSL+WAGHQ+HVSLPIN+ L+AGVDPKEIPLPH+ ILNR +
Sbjct: 1 WFQNVESMLNHHLGGLLGLGSLAWAGHQIHVSLPINKLLDAGVDPKEIPLPHDLILNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMYSMPPYPYLATDYGTQLSLFTHH WIGGF IVGAAAHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYSMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAAAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP TAP A TSLTW GGDLVAVG KVA++PI LGT+DFLVHHIHAFTIHVTVL
Sbjct: 301 THFLAPQFTAPNALAATSLTW-GGDLVAVGGKVAIMPISLGTSDFLVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDH+FLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHIFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18848.1| (AB044225) P700 chlorophyll a-protein A1 [Volvulina boldii]
Length = 497
Score = 929 bits (2376), Expect = 0.0
Identities = 425/499 (85%), Positives = 471/499 (94%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ ILNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLILNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH+WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHVWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18846.1| (AB044223) P700 chlorophyll a-protein A1 [Volvulina steinii]
dbj|BAB18847.1| (AB044224) P700 chlorophyll a-protein A1 [Volvulina steinii]
Length = 497
Score = 929 bits (2376), Expect = 0.0
Identities = 424/499 (84%), Positives = 470/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHL GLLGLGSL+WAGHQ+HVSLP+N+ L+AG+DPKEIPLPH+F+LNR +
Sbjct: 1 WFQNVESMLNHHLGGLLGLGSLAWAGHQIHVSLPVNKLLDAGIDPKEIPLPHDFLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH +++IL+AH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSMREILDAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMYSMPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYSMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DFLVHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFLVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDH+FLGLFWMYNAIS+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHIFLGLFWMYNAISIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLW+Q+SQVIQSYGSSLSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWSQSSQVIQSYGSSLSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18844.1| (AB044216) P700 chlorophyll a-protein A1 [Yamagishiella unicocca]
Length = 497
Score = 929 bits (2375), Expect = 0.0
Identities = 425/499 (85%), Positives = 470/499 (94%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AG+DPKEIPLPH+ ILNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGIDPKEIPLPHDLILNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGLAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DFLVHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFLVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18342.1| (AB044416) photosystem I P700 chlorophyll a apoprotein A1
[Basichlamys sacculifera]
Length = 497
Score = 929 bits (2375), Expect = 0.0
Identities = 424/499 (84%), Positives = 471/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ ILNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLILNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGLAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH +++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIV+H
Sbjct: 121 HMYRTNWGIGHSMREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVSH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHHMWIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHMWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DFLVHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFLVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHF+WAFSLMFL
Sbjct: 479 YGLIFLGAHFIWAFSLMFL 497
>dbj|BAB18881.1| (AB044230) P700 chlorophyll a-protein A1 [Pandorina morum]
Length = 497
Score = 929 bits (2374), Expect = 0.0
Identities = 425/499 (85%), Positives = 470/499 (94%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGLAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DFLVHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFLVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18841.1| (AB044199) P700 chlorophyll a-protein A1 [Eudorina elegans]
dbj|BAB18842.1| (AB044210) P700 chlorophyll a-protein A1 [Eudorina cylindrica]
dbj|BAB18873.1| (AB044201) P700 chlorophyll a-protein A1 [Eudorina elegans]
Length = 497
Score = 929 bits (2374), Expect = 0.0
Identities = 425/499 (85%), Positives = 470/499 (94%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ ILNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLILNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18856.1| (AB044242) P700 chlorophyll a-protein A1 [Gonium pectorale]
Length = 497
Score = 928 bits (2373), Expect = 0.0
Identities = 424/499 (84%), Positives = 470/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AG+DPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGIDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DFLVHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFLVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18840.1| (AB044198) P700 chlorophyll a-protein A1 [Eudorina illinoisensis]
dbj|BAB18874.1| (AB044203) P700 chlorophyll a-protein A [Eudorina elegans]
dbj|BAB18870.1| (AB044192) P700 chlorophyll a-protein A1 [Pleodorina californica]
dbj|BAB18871.1| (AB044194) P700 chlorophyll a-protein A1 [Pleodorina japonica]
dbj|BAB18875.1| (AB044206) P700 chlorophyll a-protein A1 [Eudorina unicocca]
dbj|BAB18876.1| (AB044209) P700 chlorophyll a-protein A1 [Eudorina unicocca]
Length = 497
Score = 928 bits (2372), Expect = 0.0
Identities = 424/499 (84%), Positives = 470/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18835.1| (AB044182) P700 chlorophyll a-protein A1 [Volvox aureus]
Length = 497
Score = 928 bits (2372), Expect = 0.0
Identities = 424/499 (84%), Positives = 469/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y DFLTF+GGL+PVTGGLWL+D AHHHLAIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYGDFLTFKGGLNPVTGGLWLSDTAHHHLAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAI+SHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAILSHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18877.1| (AB044212) P700 chlorophyll a-protein A1 [Platydorina caudata]
Length = 497
Score = 927 bits (2371), Expect = 0.0
Identities = 424/499 (84%), Positives = 470/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ ILNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLILNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH +++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSMREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18855.1| (AB044241) P700 chlorophyll a-protein A1 [Gonium octonarium]
Length = 497
Score = 927 bits (2371), Expect = 0.0
Identities = 423/499 (84%), Positives = 470/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AG+DPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGIDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18853.1| (AB044232) P700 chlorophyll a-protein A1 [Pandorina colemaniae]
Length = 497
Score = 927 bits (2371), Expect = 0.0
Identities = 424/499 (84%), Positives = 470/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSF++G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFSKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DFLVHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFLVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18851.1| (AB044228) P700 chlorophyll a-protein A1 [Pandorina morum]
Length = 497
Score = 927 bits (2371), Expect = 0.0
Identities = 424/499 (84%), Positives = 470/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSF++G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFSKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DFLVHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFLVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTANG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18341.1| (AB044415) photosystem I P700 chlorophyll a apoprotein A1
[Tetrabaena socialis]
Length = 497
Score = 927 bits (2371), Expect = 0.0
Identities = 424/499 (84%), Positives = 470/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGLAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH +++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSMREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DFLVHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFLVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18869.1| (AB044185) P700 chlorophyll a-protein A1 [Volvox carteri f.
kawasakiensis]
Length = 497
Score = 927 bits (2370), Expect = 0.0
Identities = 423/499 (84%), Positives = 470/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ ++NR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLLINRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDL+AVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLIAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18878.1| (AB044215) unnamed protein product [Yamagishiella unicocca]
Length = 497
Score = 927 bits (2369), Expect = 0.0
Identities = 424/499 (84%), Positives = 469/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL WAGHQ+HVSLPIN+ L+AGVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLGWAGHQIHVSLPINKLLDAGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGLAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH +++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSMREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18843.1| (AB044213) P700 chlorophyll a-protein A1 [Yamagishiella unicocca]
Length = 497
Score = 927 bits (2369), Expect = 0.0
Identities = 423/499 (84%), Positives = 470/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGLAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH +++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSMREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18838.1| (AB044187) P700 chlorophyll a-protein A1 [Volvox globator]
Length = 497
Score = 927 bits (2369), Expect = 0.0
Identities = 424/499 (84%), Positives = 470/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPIFLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18837.1| (AB044186) P700 chlorophyll a-protein A1 [Volvox barberi]
dbj|BAB18839.1| (AB044188) P700 chlorophyll a-protein A1 [Volvox rousseletii]
Length = 497
Score = 927 bits (2369), Expect = 0.0
Identities = 424/499 (84%), Positives = 470/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPKLTAPNALAATSLTW-GGDLVAVGGKVAMMPIFLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18836.1| (AB044183) P700 chlorophyll a-protein A1 [Volvox dissipatrix]
Length = 497
Score = 927 bits (2369), Expect = 0.0
Identities = 423/499 (84%), Positives = 470/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHNIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH W+GGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWMGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18345.1| (AB044419) photosystem I P700 chlorophyll a apoprotein A1
[Chlamydomonas reinhardtii]
Length = 497
Score = 926 bits (2368), Expect = 0.0
Identities = 423/499 (84%), Positives = 469/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHL GLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLGGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH +K+ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSMKEILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18872.1| (AB044197) P700 chlorophyll a-protein A1 [Pleodorina indica]
Length = 497
Score = 925 bits (2365), Expect = 0.0
Identities = 422/499 (84%), Positives = 469/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++IL+AH+GPFTG+GH GLYEILTTSWHAQL+INL + GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILDAHRGPFTGEGHVGLYEILTTSWHAQLAINLTLFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18884.1| (AB044240) P700 chlorophyll a-protein A1 [Gonium multicoccum]
Length = 497
Score = 924 bits (2363), Expect = 0.0
Identities = 420/499 (84%), Positives = 470/499 (94%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AG+DPKEIPLPH+F+LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGIDPKEIPLPHDFLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH +++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSMREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLW+Q++ VIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWSQSASVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18882.1| (AB044234) P700 chlorophyll a-protein A1 [Astrephomene
gubernaculifera]
Length = 497
Score = 923 bits (2360), Expect = 0.0
Identities = 419/499 (83%), Positives = 468/499 (92%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AG+DPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGIDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFNGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILE+H+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILESHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHHMWIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHMWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP TAP A TSLTW GGDLVAVG KVA++PI LGT+DF++HHIHAFTIHVTVL
Sbjct: 301 THFLAPEFTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMIHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ++ VIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSANVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHF+WAFSLMFL
Sbjct: 479 YGLIFLGAHFIWAFSLMFL 497
>dbj|BAB18854.1| (AB044235) P700 chlorophyll a-protein A1 [Astrephomene
gubernaculifera]
Length = 497
Score = 922 bits (2358), Expect = 0.0
Identities = 420/499 (84%), Positives = 468/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L++GVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDSGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW +Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWNEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILE+H+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILESHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHHMWIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHMWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQFTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ++ VIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSANVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHF+WAFSLMFL
Sbjct: 479 YGLIFLGAHFIWAFSLMFL 497
>dbj|BAB18879.1| (AB044219) P700 chlorophyll a-protein A1 [Volvulina compacta]
Length = 497
Score = 922 bits (2357), Expect = 0.0
Identities = 421/499 (84%), Positives = 468/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+ GVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDYGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HIT GNFAQS+ TINGWLRDFLW+Q+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITNGNFAQSANTINGWLRDFLWSQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18858.1| (AB044244) P700 chlorophyll a-protein A1 [Gonium viridistellatum]
Length = 497
Score = 922 bits (2357), Expect = 0.0
Identities = 419/499 (83%), Positives = 469/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AG+DPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGIDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH +++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSMREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLW+Q++ VIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWSQSASVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18845.1| (AB044220) P700 chlorophyll a-protein A1 [Volvulina pringsheimii]
Length = 497
Score = 922 bits (2357), Expect = 0.0
Identities = 421/499 (84%), Positives = 467/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL WAGHQ+HVSLP+N+ L+ GVDPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLGWAGHQIHVSLPVNKLLDYGVDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGLAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHHIHAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHIHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWAQ++ VIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAQSASVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18857.1| (AB044243) P700 chlorophyll a-protein A1 [Gonium quadratum]
Length = 497
Score = 922 bits (2356), Expect = 0.0
Identities = 419/499 (83%), Positives = 469/499 (93%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AG+DPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGIDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGILPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++IL+AH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSIREILDAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQW+QN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWVQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHH+HAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHVHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+ISVVIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSISVVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLW+Q++ VIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWSQSAAVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHFVWAFSLMFL
Sbjct: 479 YGLIFLGAHFVWAFSLMFL 497
>dbj|BAB18883.1| (AB044238) P700 chlorophyll a-protein A1 [Astrephomene perforata]
Length = 497
Score = 917 bits (2344), Expect = 0.0
Identities = 417/499 (83%), Positives = 465/499 (92%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHLAGLLGLGSL+WAGHQ+HVSLP+N+ L+AG+DPKEIPLPH+ +LNR +
Sbjct: 1 WFQNVESMLNHHLAGLLGLGSLAWAGHQIHVSLPVNKLLDAGIDPKEIPLPHDLLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFNGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH I++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHNIREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPYPYLATDYGTQLSLFTHH+WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYAMPPYPYLATDYGTQLSLFTHHVWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP TAP A TSLTW GGDLVAVG KVA++PI LGT+DF+VHH+HAFTIHVTVL
Sbjct: 301 THLLAPQFTAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFMVHHVHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN++S+VIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSLSIVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRD LW+ +SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDMLWSHSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHF WA SLMFL
Sbjct: 479 YGLIFLGAHFCWALSLMFL 497
>dbj|BAB18880.1| (AB044222) P700 chlorophyll a-protein A1 [Volvulina steinii]
Length = 497
Score = 917 bits (2343), Expect = 0.0
Identities = 419/499 (83%), Positives = 466/499 (92%), Gaps = 2/499 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHL GLLGLGSL+WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+F+LNR +
Sbjct: 1 WFQNVESMLNHHLGGLLGLGSLAWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDFLLNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIMPFFTLNWSEYSDFLTFNGGLNPVTGGLWLSDQAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH +++ILEAH+GPFTG+GH GLYEILTTSWHAQL+INLA GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHNMREILEAHRGPFTGEGHVGLYEILTTSWHAQLAINLASCGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMYSMPPYPYLATDYGTQLSLFTHH WIGGF IVGA AHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYSMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAGAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
TH LAP +TAP A TSLTW GG+LV+VG KVA++PI LGT+DF+VHH+HAFTIHVTVL
Sbjct: 301 THFLAPQLTAPNALAATSLTW-GGNLVSVGGKVAMMPISLGTSDFMVHHVHAFTIHVTVL 359
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFL LFW YN+ISVVIF
Sbjct: 360 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLALFWAYNSISVVIF 419
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSA 666
HFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLW+Q+SQVIQSYGS+LSA
Sbjct: 420 HFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWSQSSQVIQSYGSALSA 478
Query: 667 YGLFFLGAHFVWAFSLMFL 685
YGL FLGAHF+WAFSLMFL
Sbjct: 479 YGLIFLGAHFIWAFSLMFL 497
>gb|AAD44698.1| (AF130031) PSI P700 apoprotein A1 [Heterocapsa triquetra]
Length = 732
Score = 809 bits (2066), Expect = 0.0
Identities = 399/751 (53%), Positives = 525/751 (69%), Gaps = 61/751 (8%)
Query: 26 WAKPG--HFSRTIAKGPDTTTWIWNLHADAHDFDSHTSD--LEDISRKIFSAHFGQLSII 81
W++ G HF++ +AKGP TTTWIW+LHADAHDF +S+ + K+FS+ SI+
Sbjct: 13 WSQAGSSHFNKALAKGPKTTTWIWSLHADAHDFQQSSSESTAASVGAKVFSSGLAHFSIV 72
Query: 82 FLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGGFRGIQITSGF 141
F WL GM+FHGA FSNY AWL DP P +Q+VW +VGQ+ILN D+GG F+ I++TSGF
Sbjct: 73 FFWLGGMHFHGAYFSNYSAWLKDPK--TPGSQLVWSLVGQDILNQDLGGYFQSIRVTSGF 130
Query: 142 FQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDVESM--LNHHL 199
FQ+WRA GI +++ L A L+ + L+A +FH H ++W + +M L+ +
Sbjct: 131 FQLWRAEGIVTQVHLKYAAAAALIGSIATLWAAYFHMH-----ISWSSSLRTMGSLSSYN 185
Query: 200 AG----LLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFA 255
AG L GLGS+SWAGHQ+H++LPIN+ L++GVDP ++P P + +L +DL+ ++P F
Sbjct: 186 AGQLAILAGLGSISWAGHQIHIALPINRLLDSGVDPSQLPSPQD-LLFKDLMQVIFPGFG 244
Query: 256 EGATPFFTLNWAKYADFLTFRG-----GLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYR 310
G F++ +L +G GL+P TG ++L IA HH + I +I+G +
Sbjct: 245 VGPAVDFSI-------YLNQKGAASEVGLNPSTGSIYLGQIASHHFFVGITCIISGIIAL 297
Query: 311 TNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYS 370
+ + G F SWH++LSINLA+ GSL+I AHH+Y+
Sbjct: 298 L----------VKRSKAGSFQDAA------AFNNSWHSRLSINLAIAGSLSITFAHHIYA 341
Query: 371 MPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYD-----PTTR-----Y 420
MP YP+ +DY T LSLF HHMWIGGF IVGA AHA+IFM+RD P+ +
Sbjct: 342 MPVYPFCGSDYATVLSLFVHHMWIGGFFIVGAGAHASIFMIRDEGVPAGIPSAKGRLYSV 401
Query: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
N ++ ++L HRD I+ HL +V I LG H+FG+YIHNDT+ ALGRP+D+FSD +IQL+P+F
Sbjct: 402 NSIIQQLLAHRDIIMGHLIYVTIALGMHAFGIYIHNDTLQALGRPEDIFSDNSIQLKPLF 461
Query: 481 AQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 540
A W+Q+ +L T G T + G L + KV + LGTADF+VHHIHAFT
Sbjct: 462 AVWVQSLPSLFLLNTLSGDAAVTGIP--GFGLEVLDGKVVTMTQELGTADFMVHHIHAFT 519
Query: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
IH T+LIL+KGVL++RSSRL+ DK LGFR+PCDGPGRGGTCQ+S WDHVFLG+FWMYN+
Sbjct: 520 IHCTLLILMKGVLYSRSSRLVSDKLELGFRYPCDGPGRGGTCQISPWDHVFLGVFWMYNS 579
Query: 601 ISVVIFHFSWKMQSDVWG--SISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQ 658
+S+VIFHF WKMQSDVWG +S Q +V H+TGG+F+ +SITINGWLR+FLW+QA++VIQ
Sbjct: 580 LSIVIFHFFWKMQSDVWGVYDVSTQKIV-HLTGGDFSVNSITINGWLRNFLWSQAAEVIQ 638
Query: 659 SYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALS 718
SYGS LS YGL FLGAHF WAFSLMFL+SGRGYWQELIESI+WAH+KLK+ P QPRALS
Sbjct: 639 SYGSGLSGYGLIFLGAHFTWAFSLMFLWSGRGYWQELIESILWAHHKLKIVPNIQPRALS 698
Query: 719 IVQGRAVGVTHYLLGGIATTWAFFLARIIAV 749
I QGRAVG+ HY+LGGI TTWAFFLAR++A+
Sbjct: 699 ITQGRAVGLVHYMLGGIGTTWAFFLARVVAL 729
>pir||S71178 photosystem I protein A1 - Arabidopsis thaliana chloroplast
(fragment)
gb|AAB67985.1| (L36246) anoxia-induced protein [Arabidopsis thaliana]
Length = 347
Score = 714 bits (1824), Expect = 0.0
Identities = 334/345 (96%), Positives = 339/345 (97%)
Query: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT
Sbjct: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
Query: 61 SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVG 120
SDLE+ISRK+FS HFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHI PSAQVVWPIVG
Sbjct: 61 SDLEEISRKVFSGHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIGPSAQVVWPIVG 120
Query: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHK 180
QEILNG VGGGFRGIQITSGFFQIWRASGITSELQLYCTAIG LVFAALMLFAGWFHYHK
Sbjct: 121 QEILNGVVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGALVFAALMLFAGWFHYHK 180
Query: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 240
AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS PINQFLNAGVDPKEIPLPHEF
Sbjct: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSYPINQFLNAGVDPKEIPLPHEF 240
Query: 241 ILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
ILNRDLLAQLYPSFAEGATPFFTLNW+KY++FLTFRGGLDPVTGGLWLTDIAHHHL IAI
Sbjct: 241 ILNRDLLAQLYPSFAEGATPFFTLNWSKYSEFLTFRGGLDPVTGGLWLTDIAHHHLRIAI 300
Query: 301 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTS 345
LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTS
Sbjct: 301 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTS 345
>dbj|BAA86297.1| (AB027235) P700 chlorophyll a apoprotein [Gymnodinium mikimotoi]
Length = 449
Score = 684 bits (1745), Expect = 0.0
Identities = 317/449 (70%), Positives = 359/449 (79%), Gaps = 5/449 (1%)
Query: 173 AGWFHYHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPK 232
AGWFHYHK+AP L WFQ+V++ML HHL GLLGLG LSWAGHQ+H+SLPIN L+AGVD +
Sbjct: 1 AGWFHYHKSAPSLDWFQNVQAMLAHHLTGLLGLGCLSWAGHQIHISLPINFLLDAGVDLR 60
Query: 233 EIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIA 292
IP P + NR + LYPSF +G PFFT WA Y+DFLTF+G L+PVTGGLWLTDIA
Sbjct: 61 VIPTPASLLFNRTYIENLYPSFTDGLWPFFTGAWANYSDFLTFKGRLNPVTGGLWLTDIA 120
Query: 293 HHHLAIAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSI 352
HHHLA++I+FL AGHMYRT W IGH ++++L AHKGP TG GHK L ILT+SWHAQL+I
Sbjct: 121 HHHLALSIIFLFAGHMYRTAWPIGHNMQNLLAAHKGPLTGAGHKALDTILTSSWHAQLAI 180
Query: 353 NLAMLGSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVR 412
NLAM+GSL+I+VAHHMY+MPPYP+LA DY TQLSLF HHMWIG FLIVGA AHAA+FM R
Sbjct: 181 NLAMVGSLSILVAHHMYAMPPYPFLAIDYATQLSLFVHHMWIGSFLIVGAGAHAALFMTR 240
Query: 413 DYDPTT-----RYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQD 467
DY P +LLDRVL HR +I+HLNWVCIFLG HSFGLY HNDTM ALGRPQD
Sbjct: 241 DYTPNAGDVNFTATNLLDRVLLHRTTLIAHLNWVCIFLGTHSFGLYAHNDTMRALGRPQD 300
Query: 468 MFSDTAIQLQPIFAQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLG 527
FSD AI L PIFA+WIQ HA APG TAP A +SL + +AV ++A+ PI LG
Sbjct: 301 TFSDGAISLSPIFAKWIQALHAEAPGTTAPNALAVSSLCFLDSGTIAVNGRIAIQPIFLG 360
Query: 528 TADFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAW 587
TADF++HHIHAF IHV LILLKGVLFARSS LIPDK LGFRFPCDGPGRGGTCQVS W
Sbjct: 361 TADFMIHHIHAFQIHVATLILLKGVLFARSSALIPDKHRLGFRFPCDGPGRGGTCQVSPW 420
Query: 588 DHVFLGLFWMYNAISVVIFHFSWKMQSDV 616
DHVFLGLFWMYN IS+ IFHF WKMQSDV
Sbjct: 421 DHVFLGLFWMYNCISITIFHFYWKMQSDV 449
>gb|AAA84232.1| (M17309) P700 chlorophyll a-protein [Euglena gracilis]
prf||1401246A trnD/trnK/psaA gene [Euglena gracilis]
Length = 398
Score = 680 bits (1735), Expect = 0.0
Identities = 297/398 (74%), Positives = 358/398 (89%), Gaps = 3/398 (0%)
Query: 1 MIIRSPEPEVK---ILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFD 57
M I PE +VK + +P++TSFE+W++PGHFSR ++KGP+TTTWIWNLHADAHDFD
Sbjct: 1 MTITPPEQQVKRVRVAFVSNPVETSFEKWSRPGHFSRLLSKGPNTTTWIWNLHADAHDFD 60
Query: 58 SHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWP 117
+HT+DLEDISRK+FSAHFGQL+II +WLSGM+FHGARFSNYEAWL DP H++PSAQ+VWP
Sbjct: 61 NHTTDLEDISRKVFSAHFGQLAIIEIWLSGMFFHGARFSNYEAWLLDPIHVKPSAQIVWP 120
Query: 118 IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFH 177
IVGQEILNGDVGG F+GIQITSG FQ+WR+ GITSE QLY TA+ GL+F+A++ FAGWFH
Sbjct: 121 IVGQEILNGDVGGNFQGIQITSGLFQLWRSCGITSEFQLYITALTGLIFSAVLFFAGWFH 180
Query: 178 YHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLP 237
YHKAAPKL WFQ+VESMLNHHL+GLLGLG LSWAGHQ+HVSLPIN+ L++GV+P E+PLP
Sbjct: 181 YHKAAPKLEWFQNVESMLNHHLSGLLGLGCLSWAGHQIHVSLPINKLLDSGVNPAELPLP 240
Query: 238 HEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLA 297
H+FIL++ L++QLYPSF++G PFFT +W++Y+DFLTFRGGL+ VTGGLWLTD+AHHHLA
Sbjct: 241 HDFILDKSLISQLYPSFSKGLAPFFTFHWSEYSDFLTFRGGLNNVTGGLWLTDVAHHHLA 300
Query: 298 IAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAML 357
+A+LF++AGHMY+TNW IGH IK +LE+H GPFTGQGHKGLYEI T SWHAQLS+NLAM+
Sbjct: 301 LAVLFILAGHMYKTNWKIGHDIKGLLESHTGPFTGQGHKGLYEIFTNSWHAQLSLNLAMM 360
Query: 358 GSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIG 395
GSL+IIVA HMYSMPPYPY+A DYGT+LSLFTHH WIG
Sbjct: 361 GSLSIIVAQHMYSMPPYPYIAIDYGTELSLFTHHYWIG 398
>dbj|BAB18349.1| (AB044423) photosystem I P700 chlorophyll a apoprotein A1
[Paulschulzia pseudovolvox]
Length = 355
Score = 673 bits (1717), Expect = 0.0
Identities = 316/357 (88%), Positives = 336/357 (93%), Gaps = 2/357 (0%)
Query: 329 PFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLATDYGTQLSLF 388
PFTG+GH GLYEILTTSWHAQL+INLA+ GSL+IIVAHHMYSMPPYPYLATDYGTQLSLF
Sbjct: 1 PFTGEGHVGLYEILTTSWHAQLAINLALFGSLSIIVAHHMYSMPPYPYLATDYGTQLSLF 60
Query: 389 THHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWVCIFLGFH 448
THH WIGGF IVGAAAHAAIFMVRDYDPT YN+LLDR++RHRDAIISHLNWVCIFLGFH
Sbjct: 61 THHTWIGGFCIVGAAAHAAIFMVRDYDPTNNYNNLLDRMIRHRDAIISHLNWVCIFLGFH 120
Query: 449 SFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALAPGITAPGATTGTSLTWG 508
SFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQNTH LAP TAP A TSL+W
Sbjct: 121 SFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQNTHFLAPQFTAPNALAATSLSW- 179
Query: 509 GGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLG 568
GGDLVAVG KVA++PI LGT+DFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLG
Sbjct: 180 GGDLVAVGGKVAMMPISLGTSDFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLG 239
Query: 569 FRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTH 628
FRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+ISVVIFHFSWKMQSDVWGS++ G V+H
Sbjct: 240 FRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSISVVIFHFSWKMQSDVWGSVTASG-VSH 298
Query: 629 ITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFFLGAHFVWAFSLMFL 685
ITGGNFAQS+ TINGWLRDFLWAQ+SQVIQSYGS+LSAYGL FLGAHF+WAFSLMFL
Sbjct: 299 ITGGNFAQSANTINGWLRDFLWAQSSQVIQSYGSALSAYGLIFLGAHFIWAFSLMFL 355
>emb|CAB75844.1| (AJ250264) photosystem I P700 chlorophyll a apoprotein A1
[Amphidinium operculatum]
Length = 671
Score = 661 bits (1688), Expect = 0.0
Identities = 356/734 (48%), Positives = 455/734 (61%), Gaps = 90/734 (12%)
Query: 19 IKTSFEEWAKPGHFSRTIAKGPD-TTTWIWNLHADAHDFDSHTSDLEDISRKIFSAHFGQ 77
+ T F + A G + +AK TTT IWNLHADAHDF S+ +S+++F+A+
Sbjct: 21 LNTDFSK-AAFGSATGLVAKAASQTTTAIWNLHADAHDF----SNSSYLSKQVFAANLAH 75
Query: 78 LSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGGFRGIQI 137
+ ++F+WLSGM+FHGA FSNY WL DP+ I P+AQ V I Q +LN I++
Sbjct: 76 IGVVFIWLSGMHFHGAYFSNYLDWLQDPS-IAPTAQQVSNIANQSVLNP--------IRV 126
Query: 138 TSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQD-VESMLN 196
TSGFF +W A GITS QL A GL+ +AL +FH H + + S+
Sbjct: 127 TSGFFNLWLAEGITSTYQLKVIAAFGLIASALCFLGSYFHMHSSTSFTRVLNTKLTSLST 186
Query: 197 HHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAE 256
HHL GLLGLGSL+WAGH +H+SLP+N LNAGV +P PH + ++ +
Sbjct: 187 HHLVGLLGLGSLAWAGHLIHISLPVNILLNAGV---AVPSPHSLLSSKAV---------- 233
Query: 257 GATPFFTLNWAKYADFLTFRGGLDPVTGGLW--LTDIAHHHLAIAILFLIAGHMYRTNWG 314
A + L+F L G +W L A HH A+A++ ++ +
Sbjct: 234 ----------ATIVEQLSF-SALTSSDGYVWQPLVYSAMHHFALALVLIVGSVL------ 276
Query: 315 IGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPY 374
GP + + + + +SWH L + L + G+ +++ A + P Y
Sbjct: 277 -------------GPLSTASNPLMSFTVGSSWHLVLGVQLFVTGTASVLYAQMSNAYPVY 323
Query: 375 PYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAI 434
PYL TD+ T +SLF HHMWIGGF +VGA AH +I +VRD P + VL RD I
Sbjct: 324 PYLLTDHPTVVSLFVHHMWIGGFFLVGAFAHLSIGLVRDTLPQS-----FSVVLTQRDII 378
Query: 435 ISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALAPGI 494
+ HL WV FLG HSFGLY+HNDTM ALGRP DMFSD AI L P+FA+W
Sbjct: 379 LGHLTWVVAFLGVHSFGLYVHNDTMQALGRPDDMFSDNAISLLPVFARW----------- 427
Query: 495 TAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLF 554
++LT L + G+ V++L + L TADF+V HIHAFTIH TVLIL+KG L+
Sbjct: 428 --------STLT-----LNSTGSAVSVLGVELSTADFMVTHIHAFTIHTTVLILVKGFLY 474
Query: 555 ARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKMQS 614
ARSSRL+ DK L FR+PCDGPGRGGTCQ+S WDHVFLGLFWMYN+ISVVIFHF W+ QS
Sbjct: 475 ARSSRLVNDKYKLDFRYPCDGPGRGGTCQISPWDHVFLGLFWMYNSISVVIFHFFWEYQS 534
Query: 615 DVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFFLGA 674
++ + G +F +SI NGWLR+FLW+ A+QVIQSYGS L+AYGL FL +
Sbjct: 535 NLASIKASAGGSIRALASDFELNSINTNGWLRNFLWSGAAQVIQSYGSPLAAYGLTFLAS 594
Query: 675 HFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHYLLGG 734
HFVWA SLMFLFSGRGYWQELIES++WAH+KL V P QPRALSI GRAVG+THYLLGG
Sbjct: 595 HFVWALSLMFLFSGRGYWQELIESVLWAHHKLYVVPHIQPRALSITSGRAVGLTHYLLGG 654
Query: 735 IATTWAFFLARIIA 748
I TTW+FFLARI+A
Sbjct: 655 IGTTWSFFLARIVA 668
>gi|7524850 P700 apoprotein subunit Ib [Chlorella vulgaris]
sp|P56342|PSAB_CHLVU PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||T07280 photosystem I P700 apoprotein 1B - Chlorella vulgaris chloroplast
dbj|BAA57928.1| (AB001684) P700 apoprotein subunit Ib [Chlorella vulgaris]
Length = 734
Score = 654 bits (1668), Expect = 0.0
Identities = 352/754 (46%), Positives = 456/754 (59%), Gaps = 75/754 (9%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT +W A AHDF+SH E+ + +KIF++HFGQL+IIFLW SG F
Sbjct: 8 FSQALAQDP-TTRRLWFGIATAHDFESHDGMTEERLYQKIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRA 147
H A N+E W+ DP HIRP A +W P GQ + GG G + I TSG +Q W
Sbjct: 67 HVAWQGNFEQWVQDPLHIRPIAHAIWDPHFGQAAVEAFTRGGASGPVNISTSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYHKA-APKLAWFQDVESMLNHHLAGLLGLG 206
GI + +LY +I LV A L LFAGW H + P L+WF++ ES LNHHLAGL G+
Sbjct: 127 IGIRTNQELYVGSIFLLVLAGLFLFAGWLHLQPSFQPALSWFKNAESRLNHHLAGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+W GH VHV++P ++ + G D LPH G TPFFT NW
Sbjct: 187 SLAWTGHLVHVAIPESRGQHVGWDNFLTVLPHP----------------AGLTPFFTGNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
A YA+ LTF GG P T LWLTD+AHHHLAIA++F++AGHM
Sbjct: 231 AAYAENPDSLSQLFGTGEGSGTAILTFLGGFHPQTQSLWLTDMAHHHLAIAVVFILAGHM 290
Query: 309 YRTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
YRT +GIGH +++ILEA P G GHKGLY+ + S H QL + LA +G++ +VA
Sbjct: 291 YRTIFGIGHSMREILEAQTPPSGRLGAGHKGLYDTVNNSLHFQLGLALASVGTICSLVAQ 350
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMYS+PPY +LA D+ TQ SL+THH +I GF++ GA AH AIF VRDYDP ++L R
Sbjct: 351 HMYSLPPYAFLAQDFTTQASLYTHHQYIAGFIMCGAFAHGAIFFVRDYDPEANRGNVLAR 410
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
VL H++AIISHL+WV +FLGFH+ GLY+HND + A G P+ I ++P+FAQWIQ
Sbjct: 411 VLDHKEAIISHLSWVSLFLGFHTLGLYVHNDVVQAFGTPEKQ-----ILIEPVFAQWIQA 465
Query: 487 THA--------LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHA 538
H L T+ + G SL W G L + + L + +G DFLVHH A
Sbjct: 466 AHGKTVYGFDFLLSSATSAPSLAGQSL-WLPGWLQGINSDTNSLFLTIGPGDFLVHHAIA 524
Query: 539 FTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMY 598
+H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM
Sbjct: 525 LGLHTTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWML 584
Query: 599 NAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQ 658
N I V F+F WK +W QG V F +SS + GWLRD+LW +SQ+I
Sbjct: 585 NTIGWVTFYFHWK-HLGIW-----QGNV-----NQFNESSTYLMGWLRDYLWLNSSQLIN 633
Query: 659 SYG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAT-- 712
Y +SLS + FL H ++A MFL S RGYWQELIE++ WAH + +A
Sbjct: 634 GYNPFGMNSLSVWAWMFLFGHLIYATGFMFLISWRGYWQELIETLAWAHERTPLANLVRW 693
Query: 713 --QPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+TH+ +G + T AF +A
Sbjct: 694 RDKPVALSIVQARLVGLTHFSVGYVLTYAAFLIA 727
>sp|P29255|PSAB_SYNY3 PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||S18243 photosystem I protein A2 - Synechocystis sp. (strain PCC 6803)
dbj|BAA17438.1| (D90906) P700 apoprotein subunit Ib [Synechocystis sp.]
Length = 731
Score = 651 bits (1660), Expect = 0.0
Identities = 346/753 (45%), Positives = 455/753 (59%), Gaps = 76/753 (10%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF++H E+ + +KIF++HFG ++IIFLW SG F
Sbjct: 8 FSQDLAQDP-TTRRIWYGIATAHDFETHDGMTEENLYQKIFASHFGHIAIIFLWTSGTLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDV-GGGFRGIQIT-SGFFQIWRA 147
H A N+E W+ DP +IRP A +W P G+ +N G + I SG + +
Sbjct: 67 HVAWQGNFEQWIKDPLNIRPIAHAIWDPHFGEGAVNAFTQAGASNPVNIAYSGVYHWFYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+T+ +LY A+ LV A+L LFAGW H K P LAWF++ ES LNHHLAGL G+
Sbjct: 127 IGMTTNQELYSGAVFLLVLASLFLFAGWLHLQPKFRPSLAWFKNAESRLNHHLAGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+WAGH VHV++P + + G D PH G PFFT NW
Sbjct: 187 SLAWAGHLVHVAIPEARGQHVGWDNFLSTPPHP----------------AGLMPFFTGNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
YA LTF GG P T LWLTDIAHHHLAIA++F+IAGHM
Sbjct: 231 GVYAADPDTAGHIFGTSEGAGTAILTFLGGFHPQTESLWLTDIAHHHLAIAVIFIIAGHM 290
Query: 309 YRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHM 368
YRTNWGIGH IK+IL AHKGP TG GH LY+ + S H QL + LA LG +T +VA HM
Sbjct: 291 YRTNWGIGHSIKEILNAHKGPLTGAGHTNLYDTINNSLHFQLGLALASLGVITSLVAQHM 350
Query: 369 YSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVL 428
YS+P Y ++A D+ TQ +L+THH +I GFL+VGA AH AIF VRDYDP +++L R+L
Sbjct: 351 YSLPSYAFIAQDHTTQAALYTHHQYIAGFLMVGAFAHGAIFFVRDYDPVANKDNVLARML 410
Query: 429 RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNT- 487
H++A+ISHL+WV +FLGFH+ GLY+HND + A G P+ I ++P+FAQWIQ T
Sbjct: 411 EHKEALISHLSWVSLFLGFHTLGLYVHNDVVVAFGTPEKQ-----ILIEPVFAQWIQATS 465
Query: 488 -------HALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 540
L + +TTG + W G L A+ + L + +G DFLVHH A
Sbjct: 466 GKALYGFDVLLSNPDSIASTTGAA--WLPGWLDAINSGTNSLFLTIGPGDFLVHHAIALG 523
Query: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
+H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N
Sbjct: 524 LHTTALILIKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAMFWMLNT 583
Query: 601 ISVVIFHFSWKMQSDVW-GSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQS 659
+ + F++ WK VW G+++ F ++S + GW RD+LWA ++Q+I
Sbjct: 584 LGWLTFYWHWK-HLGVWSGNVA-----------QFNENSTYLMGWFRDYLWANSAQLING 631
Query: 660 YG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAT--- 712
Y ++LS + FL H VWA MFL S RGYWQELIE+IVWAH + +A
Sbjct: 632 YNPYGVNNLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETIVWAHERTPLANLVRWK 691
Query: 713 -QPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+ H+ +G + T AF +A
Sbjct: 692 DKPVALSIVQARLVGLAHFTVGYVLTYAAFLIA 724
>emb|CAA41630.1| (X58825) P700 apoprotein subunit Ib (PSA-B) [Synechocystis
PCC6803]
Length = 731
Score = 649 bits (1657), Expect = 0.0
Identities = 345/753 (45%), Positives = 455/753 (59%), Gaps = 76/753 (10%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF++H E+ + +KIF++HFG ++IIFLW SG F
Sbjct: 8 FSQDLAQDP-TTRRIWYGIATAHDFETHDGMTEENLYQKIFASHFGHIAIIFLWTSGTLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDV-GGGFRGIQIT-SGFFQIWRA 147
H A N+E W+ DP +IRP A +W P G+ +N G + I SG + +
Sbjct: 67 HVAWQGNFEQWIKDPLNIRPIAHAIWDPHFGEGAVNAFTQAGASNPVNIAYSGVYHWFYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+T+ +LY A+ LV A+L LFAGW H K P LAWF++ ES LNHHLAGL G+
Sbjct: 127 IGMTTNQELYSGAVFLLVLASLFLFAGWLHLQPKFRPSLAWFKNAESRLNHHLAGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+WAGH VHV++P + + G D PH G PFFT NW
Sbjct: 187 SLAWAGHLVHVAIPEARGQHVGWDNFLSTPPHP----------------AGLMPFFTGNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
YA LTF GG P T LWLTDIAHHHLAIA++F+IAGHM
Sbjct: 231 GVYAADPDTAGHIFGTSEGAGTAILTFLGGFHPQTESLWLTDIAHHHLAIAVIFIIAGHM 290
Query: 309 YRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHM 368
YRTNWGIGH IK+IL AHKGP TG GH LY+ + S H QL + LA LG +T +VA HM
Sbjct: 291 YRTNWGIGHSIKEILNAHKGPLTGAGHTNLYDTINNSLHFQLGLALASLGVITSLVAQHM 350
Query: 369 YSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVL 428
YS+P Y ++A D+ TQ +L+THH +I GFL+VGA AH AIF VRDYDP +++L R+L
Sbjct: 351 YSLPSYAFIAQDHTTQAALYTHHQYIAGFLMVGAFAHGAIFFVRDYDPVANKDNVLARML 410
Query: 429 RHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNT- 487
H++A+ISHL+WV +FLGFH+ GLY+HND + A G P+ I ++P+FAQWIQ T
Sbjct: 411 EHKEALISHLSWVSLFLGFHTLGLYVHNDVVVAFGTPEKQ-----ILIEPVFAQWIQATS 465
Query: 488 -------HALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 540
L + +TTG + W G L A+ + L + +G DFLVHH A
Sbjct: 466 GKALYGFDVLLSNPDSIASTTGAA--WLPGWLDAINSGTNSLFLTIGPGDFLVHHAIALG 523
Query: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
+H T LIL+KG L +R S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N
Sbjct: 524 LHTTALILIKGALESRGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAMFWMLNT 583
Query: 601 ISVVIFHFSWKMQSDVW-GSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQS 659
+ + F++ WK VW G+++ F ++S + GW RD+LWA ++Q+I
Sbjct: 584 LGWLTFYWHWK-HLGVWSGNVA-----------QFNENSTYLMGWFRDYLWANSAQLING 631
Query: 660 YG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAT--- 712
Y ++LS + FL H VWA MFL S RGYWQELIE+IVWAH + +A
Sbjct: 632 YNPYGVNNLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETIVWAHERTPLANLVRWK 691
Query: 713 -QPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+ H+ +G + T AF +A
Sbjct: 692 DKPVALSIVQARLVGLAHFTVGYVLTYAAFLIA 724
>gi|11467833 P700 apoprotein A2 of photosystem I [Nephroselmis olivacea]
gb|AAD54855.1|AF137379_78 (AF137379) P700 apoprotein A2 of photosystem I [Nephroselmis
olivacea]
Length = 734
Score = 647 bits (1650), Expect = 0.0
Identities = 345/754 (45%), Positives = 455/754 (59%), Gaps = 75/754 (9%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + +KIF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAEDP-TTRRIWYGIATAHDFESHDGMTEESLYQKIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E W DP H+RP A +W P GQ + GG G + I+ SG +Q W
Sbjct: 67 HVAWQGNFEQWGQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGAAGPVNISYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ S LY A+ LV AA++LFAGW H K P L+WF++ ES LNHHLAGL G+
Sbjct: 127 IGMRSSNDLYTGALFLLVGAAILLFAGWLHLQPKFQPSLSWFKNAESRLNHHLAGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+W GH VHV++P ++ + G D LPH G PFF NW
Sbjct: 187 SLAWTGHLVHVAIPESRGQHVGWDNFLTTLPHP----------------AGLAPFFNGNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
+ YA LTF GG P T LWLTD+AHHHLAIA++F++AGHM
Sbjct: 231 SVYAQNPDSASHLFGTSTGAGTAILTFLGGFHPQTQSLWLTDMAHHHLAIAVVFILAGHM 290
Query: 309 YRTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
YRTN+GIGH +++ILEAH+ P G GHKGL++ + S H QL + LA LG +T +VA
Sbjct: 291 YRTNFGIGHSMREILEAHRAPSGRLGLGHKGLFDTVNNSLHFQLGLALASLGVITSLVAQ 350
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMYS+PPY ++A D+ T +L+THH +I GF++ GA AH AIF +RDYDP ++L R
Sbjct: 351 HMYSLPPYAFMAQDFTTMSALYTHHQYIAGFIMTGAFAHGAIFFIRDYDPIQNEGNVLAR 410
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
+L H++A+ISHL+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ
Sbjct: 411 MLEHKEALISHLSWVTLFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPVFAQWIQA 465
Query: 487 TH--------ALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHA 538
+H L +P AT+ + W G L A+ + L + +G DFLVHH A
Sbjct: 466 SHGKALYGFDVLLSSADSP-ATSASQSIWLPGWLDAINSSSNSLFLTIGPGDFLVHHAIA 524
Query: 539 FTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMY 598
+H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM
Sbjct: 525 LGLHTTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWML 584
Query: 599 NAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQ 658
N I F++ WK + +W QG F +SS + GWLRD+LW +SQ+I
Sbjct: 585 NTIGWTTFYWHWKHLA-LW-----QG-----NAAQFNESSTYLMGWLRDYLWLNSSQLIN 633
Query: 659 SYG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAT-- 712
Y +SLS + FL H VWA MFL S RGYWQELIE++ WAH + +A
Sbjct: 634 GYNPFGMNSLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLVRW 693
Query: 713 --QPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+ H+ +G + T AF +A
Sbjct: 694 KDKPVALSIVQARLVGLAHFSVGYVFTYAAFVIA 727
>gi|11467712 PSI P700 apoprotein A2 [Guillardia theta]
sp|O78507|PSAB_GUITH PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
gb|AAC35698.1| (AF041468) PSI P700 apoprotein A2 [Guillardia theta]
Length = 734
Score = 644 bits (1643), Expect = 0.0
Identities = 344/753 (45%), Positives = 451/753 (59%), Gaps = 73/753 (9%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P T IW A AHDF+SH E+ + +KIF++HFG L+IIFLW SG F
Sbjct: 8 FSQALAQDPATRR-IWYGLATAHDFESHDGMTEENLYQKIFASHFGHLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFR-GIQI-TSGFFQIWRA 147
H A N+E W+ +P ++P A +W P GQ + GG + I TSG + W
Sbjct: 67 HVAWQGNFEQWVLNPLKVKPIAHAIWDPHFGQPAVKAFTKGGVSYPVNIATSGVYHWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY +I L+ A++MLFAGW H K P LAWF++ ES LNHHL+GL G
Sbjct: 127 IGMRTNNDLYSGSIFLLILASVMLFAGWLHLQPKFRPGLAWFKNNESRLNHHLSGLFGFS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
S++W+GH +HV++P ++ L+ G D PH EG FFT NW
Sbjct: 187 SVAWSGHLIHVAIPESRGLHVGWDNFTKVYPHP----------------EGLKAFFTGNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
+ YA LTF GG P T LWLTDIAHHHLAI ++F+ AGHM
Sbjct: 231 SNYAANPDTADHIFGTTEGSGTAILTFLGGFHPQTQSLWLTDIAHHHLAIGVIFIFAGHM 290
Query: 309 YRTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
YRTNWGIGH +K+IL+AH+ P G GHKGL+E ++ S H QL + LA LG +T +VA
Sbjct: 291 YRTNWGIGHSLKEILDAHRAPSGRLGNGHKGLFETISNSLHFQLGLALASLGVITSLVAQ 350
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY++P Y ++A DY TQ +L+THH +I GFL+VGA AH AIF VRDYDP N++L R
Sbjct: 351 HMYALPSYAFIAKDYVTQAALYTHHQYIAGFLMVGAFAHGAIFFVRDYDPEQNKNNVLAR 410
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
+L H++AIISHL+WV +FLGFH+ GLY+HND + A G P+ I ++P+FAQWIQ
Sbjct: 411 MLEHKEAIISHLSWVSLFLGFHTLGLYVHNDVVVAFGTPEKQ-----ILVEPVFAQWIQA 465
Query: 487 THALAP-------GITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAF 539
+ A A +S W G L A+ + L +P+G DFLVHH A
Sbjct: 466 SSGKAMYGFDVLLSANTSIAKNASSNIWLPGWLEAINSGKNSLFLPIGPGDFLVHHAIAL 525
Query: 540 TIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN 599
+H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N
Sbjct: 526 ALHTTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLSMFWMLN 585
Query: 600 AISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQS 659
I V F++ WK +W QG G F +SS I GWLRD+LW +S +I
Sbjct: 586 TIGWVTFYWHWK-HVTIW-----QG-----NAGQFNESSTYIMGWLRDYLWLNSSPLING 634
Query: 660 YG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAT--- 712
Y +SLS + FL H +WA MFL S RGYWQELIE++VWAH + +A
Sbjct: 635 YNPFGMNSLSVWAWMFLFGHLIWATGFMFLISWRGYWQELIETLVWAHERTPLANLVRWR 694
Query: 713 -QPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+ H+ +G I T AF +A
Sbjct: 695 DKPVALSIVQARLVGLVHFTVGYIFTYAAFVIA 727
>gi|11466390 P700 apoprotein A2 of photosystem I [Mesostigma viride]
gb|AAF43834.1|AF166114_46 (AF166114) P700 apoprotein A2 of photosystem I [Mesostigma viride]
Length = 734
Score = 642 bits (1639), Expect = 0.0
Identities = 341/753 (45%), Positives = 455/753 (60%), Gaps = 73/753 (9%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + +KIF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAQDP-TTRRIWFGIATAHDFESHDGMTEENLYQKIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E W++DP H+RP A +W P GQ + GG G + I+ SG +Q W
Sbjct: 67 HVAWQGNFERWVADPLHVRPIAHAIWDPHFGQPAVEAFTRGGASGPVNISYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ S LY A+ L+ A++ LFAGW H + P L+WF++ ES LNHHL+GL G+
Sbjct: 127 IGMRSNTDLYIGALFLLITASMTLFAGWLHLQPQFKPSLSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+W GH +HV++P ++ + D LPH G +PFFT NW
Sbjct: 187 SLAWTGHLIHVAIPESRGQHVRWDNFLNVLPHP----------------AGLSPFFTGNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
A YA LTF GG P T LWLTDIAHHHLAIA+LF++AGHM
Sbjct: 231 AAYAQNPDSTSHIFSTSQGAGTAILTFLGGFHPQTQSLWLTDIAHHHLAIAVLFIVAGHM 290
Query: 309 YRTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
YRTN+GIGH +++ILEA + P G GH GLY+ + S H QL + LA LG +T +VA
Sbjct: 291 YRTNFGIGHSMREILEAQRPPGGRLGAGHSGLYDTVNNSLHFQLGLALASLGVITSVVAQ 350
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMYS+ PY +LA D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP +++L R
Sbjct: 351 HMYSLSPYAFLAQDFTTQAALYTHHQYIAGFIMTGAFAHGAIFFIRDYDPELNKDNVLAR 410
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
+L H++AIISHL+W +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ
Sbjct: 411 MLEHKEAIISHLSWASLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQA 465
Query: 487 THA-------LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAF 539
+H + ++ A + + W G L A+ + L + +G DFLVHH A
Sbjct: 466 SHGKSLYGFDVLLSSSSSFAASASDSIWLPGWLDAINSNSNSLFLTIGPGDFLVHHAIAL 525
Query: 540 TIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN 599
+H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N
Sbjct: 526 GLHTTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLN 585
Query: 600 AISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQS 659
I V F++ WK +W QG V F +SS + GWLRD+LW +SQ+I
Sbjct: 586 TIGWVTFYWHWK-HLTLW-----QGNV-----AQFDESSTYLMGWLRDYLWLNSSQLING 634
Query: 660 YG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAT--- 712
Y +SLS + FL H +WA MFL S RGYWQELIE++ WAH + +A
Sbjct: 635 YNPFGMNSLSVWAWMFLFGHLIWATGFMFLISWRGYWQELIETLAWAHERTPLANLVRWK 694
Query: 713 -QPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+ H+ +G + T AF +A
Sbjct: 695 DKPVALSIVQARVVGLAHFSVGYVFTYAAFLIA 727
>gi|11466702 psaB [Marchantia polymorpha]
sp|P06408|PSAB_MARPO PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||A2LVP7 photosystem I P700 apoprotein A2 - liverwort (Marchantia
polymorpha) chloroplast
emb|CAA28084.1| (X04465) psaB [Marchantia polymorpha]
Length = 734
Score = 641 bits (1636), Expect = 0.0
Identities = 346/754 (45%), Positives = 449/754 (58%), Gaps = 75/754 (9%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +++ P TT IW A AHDF+SH E+ + +KIF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLSQDP-TTRRIWFGIATAHDFESHDDMTEERLYQKIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+EAW DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFEAWGQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ ++ +++ L AGW H K PK++WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNQDLYNGALFLVILSSISLIAGWLHLQPKWKPKVSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+W GH VHV++P ++ + D LPH EG PFF W
Sbjct: 187 SLAWTGHLVHVAIPESRGEHVRWDNFLTKLPHP----------------EGLGPFFAGQW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
YA LTF GG P T LWLTDIAHHHLAIA++F+IAGHM
Sbjct: 231 NIYAQNVDSSNHAFGTSQGAGTAILTFIGGFHPQTQSLWLTDIAHHHLAIAVVFIIAGHM 290
Query: 309 YRTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
YRTN+GIGH IK+ILE H P G+GHKGLY+ + S H QL + LA LG +T +VA
Sbjct: 291 YRTNFGIGHSIKEILETHTPPGGRLGRGHKGLYDTINNSLHFQLGLALASLGVITSLVAQ 350
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMYS+PPY +LA D+ TQ +L+THH +I GF++ GA AH AIF +RDY+P +++L R
Sbjct: 351 HMYSLPPYAFLAQDFTTQAALYTHHQYIAGFIMTGAFAHGAIFFIRDYNPEQNKDNVLAR 410
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
+L H++AIISHL+W +FLGFH+ GLY+HND M A G P+ I ++PIFAQWIQ+
Sbjct: 411 MLEHKEAIISHLSWASLFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPIFAQWIQS 465
Query: 487 TH--------ALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHA 538
H L P G S+ W G L A+ N L + +G DFLVHH A
Sbjct: 466 AHGKALYGFDVLLSSTNNPAFNAGQSI-WLPGWLDAINNNSNSLFLTIGPGDFLVHHAIA 524
Query: 539 FTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMY 598
+H T LIL+KG L AR S+L+PDK G+ FPCDGPGRGGTC +SAWD +L +FWM
Sbjct: 525 LGLHTTTLILVKGALDARGSKLMPDKKEFGYSFPCDGPGRGGTCDISAWDAFYLAVFWML 584
Query: 599 NAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQ 658
N I V F++ WK +W QG F +SS + GWLRD+LW +SQ+I
Sbjct: 585 NTIGWVTFYWHWK-HITLW-----QG-----NAAQFNESSTYLMGWLRDYLWLNSSQLIN 633
Query: 659 SYG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAT-- 712
Y +SLS + FL H VWA MFL S RGYWQELIE++ WAH + +A
Sbjct: 634 GYNPFGMNSLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLVRW 693
Query: 713 --QPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+ H+ +G I T AF +A
Sbjct: 694 KDKPVALSIVQARLVGLAHFSVGYIFTYAAFLIA 727
>gi|11465751 Photosystem I p700 chlorophyll A apoprotein A2 [Porphyra purpurea]
sp|P51285|PSAB_PORPU PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||S73206 photosystem I protein A2 - red alga (Porphyra purpurea) chloroplast
gb|AAC08171.1| (U38804) Photosystem I p700 chlorophyll A apoprotein A2 [Porphyra
purpurea]
Length = 734
Score = 638 bits (1628), Expect = 0.0
Identities = 341/753 (45%), Positives = 452/753 (59%), Gaps = 73/753 (9%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +++ P TT IW A AHDF+SH E+ + +KIF++HFG L+IIFLW SG F
Sbjct: 8 FSQALSQDP-TTRRIWYGIATAHDFESHDGMTEENLYQKIFASHFGHLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNG-DVGGGFRGIQIT-SGFFQIWRA 147
H A N+E W+ +P ++P A +W P GQ L GG + I SG + W
Sbjct: 67 HVAWQGNFEQWILNPLKVKPIAHAIWDPHFGQPALKAFSKGGSAYPVNIAYSGVYHWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ LV +A++LF GW H K P L+WF++ ES LNHHL+GL G+
Sbjct: 127 IGMRTNQDLYTGALFLLVLSAILLFGGWLHLQPKFKPGLSWFKNNESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+W GH VHV++P ++ + G D LPH G PFF+ NW
Sbjct: 187 SLAWTGHLVHVAIPESRGQHVGWDNFTTVLPHP----------------AGLQPFFSGNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
+ YA LTF GG P + LWLTD+AHHHLAIA++F++AGHM
Sbjct: 231 SVYAQNPDTAQQLFGTNEGAGTAILTFLGGFHPQSQSLWLTDMAHHHLAIAVVFIVAGHM 290
Query: 309 YRTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
YRTNWGIGH +KDIL+AH+ P G GH+GL++ +T S H QL + LA LG +T +VA
Sbjct: 291 YRTNWGIGHNLKDILDAHRPPSGRLGAGHRGLFDTITNSLHMQLGLALAALGVITSLVAQ 350
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY+MPPY ++A D+ TQ SL+THH +I GFL+VGA AH AIF VRDYDP ++L R
Sbjct: 351 HMYAMPPYAFMAKDFTTQASLYTHHQYIAGFLMVGAFAHGAIFFVRDYDPEQNKGNVLAR 410
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
+L H++AIISHL+WV +FLGFH+ GLY+HNDTM A G P+ I ++P+FAQWIQ
Sbjct: 411 MLEHKEAIISHLSWVTLFLGFHTLGLYVHNDTMIAFGTPEKQ-----ILIEPVFAQWIQA 465
Query: 487 THALA-------PGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAF 539
+ A ++ AT S W G L A+ + L + +G DFLVHH A
Sbjct: 466 SSGKALYGFDVLLSSSSNIATQAGSNIWLPGWLEAINSGKNSLFLTIGPGDFLVHHAIAL 525
Query: 540 TIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN 599
+H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N
Sbjct: 526 GLHTTALILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLN 585
Query: 600 AISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQS 659
I V F++ WK +W QG T F +SS + GW RD+LW +S +I
Sbjct: 586 TIGWVTFYWHWK-HITIW-----QGNAT-----QFNESSTYLMGWFRDYLWLNSSPLING 634
Query: 660 YG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----A 711
Y ++LS + FL H VWA MFL S RGYWQELIE++ WAH + +A
Sbjct: 635 YNPYGMNNLSVWSWMFLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLIRWK 694
Query: 712 TQPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P A+SIVQ R VG+ H+ +G + T AF LA
Sbjct: 695 DKPVAMSIVQARLVGLAHFSVGYVLTYAAFVLA 727
>sp|P36492|PSAB_CHLMO PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||S41481 P700 chlorophyll a-apoprotein A2 - Chlamydomonas moewusii
gb|AAA84149.1| (M90641) P700 chlorophyll a-apoprotein A2 [Chlamydomonas moewusii]
Length = 735
Score = 637 bits (1626), Expect = 0.0
Identities = 341/754 (45%), Positives = 457/754 (60%), Gaps = 75/754 (9%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + +KIF++HFGQL+IIFLW SG F
Sbjct: 9 FSQGLAQDP-TTRRIWFGLAVAHDFESHDGMTEENLYQKIFASHFGQLAIIFLWTSGNLF 67
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRA 147
H A N+E W++DP H+RP A +W P G + GG G + I TSG +Q W
Sbjct: 68 HVAWQGNFEQWVTDPVHVRPIAHAIWDPHFGTSAVEAFTRGGASGPVNIATSGVYQWWYT 127
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + L+ ++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+
Sbjct: 128 IGMRTNQDLFTGSVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVS 187
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+W GH VHV++P ++ + G D LPH +G PF++ NW
Sbjct: 188 SLAWTGHLVHVAIPESRGRHVGWDNFLSVLPHP----------------QGLAPFWSGNW 231
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
A YA LTF GG P T LWL+D+AHHHLAIA++F++AGHM
Sbjct: 232 AAYAQNPDTASHAFGTSEGSGQAILTFLGGFHPQTQSLWLSDMAHHHLAIAVIFIVAGHM 291
Query: 309 YRTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
YRTN+GIGH ++ IL+AH P G GHKGL++ + S H QL + LA +G++T ++A
Sbjct: 292 YRTNFGIGHRLQAILDAHVPPSGNLGAGHKGLFDTVNNSLHFQLGLALASVGTITSMIAQ 351
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
H+YS+PPY YLA D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R
Sbjct: 352 HIYSLPPYAYLAIDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEANKGNVLAR 411
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
+L H++AIISHL+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ
Sbjct: 412 ILDHKEAIISHLSWVTLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQA 466
Query: 487 THA--------LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHA 538
L T+ +T G S+ W G L A+ N L + +G DFLVHH A
Sbjct: 467 AQGKTVYGFDLLLSSSTSVASTAGQSV-WLPGWLDAINNNQNTLFLTIGPGDFLVHHAIA 525
Query: 539 FTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMY 598
+H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D +L +FWM
Sbjct: 526 LGLHTTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYDAFYLAVFWML 585
Query: 599 NAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQ 658
N I V F+F WK + +W QG V F +SS + GWLRD+LW +SQ+I
Sbjct: 586 NTIGWVTFYFHWKHLA-LW-----QGNV-----AQFDESSTYLMGWLRDYLWLNSSQLIN 634
Query: 659 SYG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAT-- 712
Y +SLS + FL H ++A MFL S RGYWQELIE++VWAH K +A
Sbjct: 635 GYNPFGMNSLSVWAFCFLFGHLIYATGFMFLISWRGYWQELIETLVWAHEKTPLANIVYW 694
Query: 713 --QPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+ H+ +G I T AF +A
Sbjct: 695 KDKPVALSIVQARLVGLVHFSVGYIFTYAAFLIA 728
>gi|7525032 PSI P700 apoprotein A2 [Arabidopsis thaliana]
sp|P56767|PSAB_ARATH PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
dbj|BAA84384.1| (AP000423) PSI P700 apoprotein A2 [Arabidopsis thaliana]
Length = 734
Score = 636 bits (1623), Expect = 0.0
Identities = 344/746 (46%), Positives = 448/746 (59%), Gaps = 59/746 (7%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + + IF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAQDP-TTRRIWFGIATAHDFESHDDITEERLYQNIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E W+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFETWVQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGALGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L +AL L GW H K P+++WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNEDLYTGALFLLFLSALSLIGGWLHLQPKWKPRVSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLPI--------NQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAE 256
SL+W GH VHV++P N FLN LPH L Q LY +
Sbjct: 187 SLAWTGHLVHVAIPASRGEYVRWNNFLNV--------LPHPQGLGPLFTGQWNLYAQNPD 238
Query: 257 GATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIG 316
++ F + LT GG P T LWLTD+AHHHLAIAILFLIAGHMYRTN+GIG
Sbjct: 239 SSSHLFGTSQGSGTAILTLLGGFHPQTQSLWLTDMAHHHLAIAILFLIAGHMYRTNFGIG 298
Query: 317 HGIKDILEAH--KGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPY 374
H IKD+LEAH G G+GHKGLY+ + S H QL + LA LG +T +VA HMYS+P Y
Sbjct: 299 HSIKDLLEAHIPPGGRLGRGHKGLYDTINNSIHFQLGLALASLGVITSLVAQHMYSLPAY 358
Query: 375 PYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAI 434
++A D+ TQ +L+THH +I GF++ GA AH AIF +RDY+P +++L R+L H++AI
Sbjct: 359 AFIAQDFTTQAALYTHHQYIAGFIMTGAFAHGAIFFIRDYNPEQNEDNVLARMLDHKEAI 418
Query: 435 ISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTH------ 488
ISHL+W +FLGFH+ GLY+HND M A G P+ I ++PIFAQWIQ+ H
Sbjct: 419 ISHLSWASLFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPIFAQWIQSAHGKTSYG 473
Query: 489 --ALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
L + P G S+ W G L A+ L + +G DFLVHH A +H T L
Sbjct: 474 FDVLLSSTSGPAFNAGRSI-WLPGWLNAINENSNSLFLTIGPGDFLVHHAIALGLHTTTL 532
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
IL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N I V F
Sbjct: 533 ILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLNTIGWVTF 592
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYG----S 662
++ WK + G++S F +SS + GWLRD+LW +SQ+I Y +
Sbjct: 593 YWHWKHITLWQGNVS-----------QFNESSTYLMGWLRDYLWLNSSQLINGYNPFGMN 641
Query: 663 SLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----ATQPRALS 718
SLS + FL H VWA MFL S RGYWQELIE++ WAH + +A +P ALS
Sbjct: 642 SLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLIRWKDKPVALS 701
Query: 719 IVQGRAVGVTHYLLGGIATTWAFFLA 744
IVQ R VG+ H+ +G I T AF +A
Sbjct: 702 IVQARLVGLAHFSVGYIFTYAAFLIA 727
>sp|P09144|PSAB_CHLRE PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||B28341 photosystem I P700 apoprotein A2 - Chlamydomonas reinhardtii
chloroplast
emb|CAA29287.1| (X05848) P700 chlorophyll a-apoprotein A2 [Chlamydomonas
reinhardtii]
prf||1310243B gene ps1A2 [Chlamydomonas reinhardtii]
Length = 736
Score = 634 bits (1617), Expect = e-180
Identities = 345/755 (45%), Positives = 452/755 (59%), Gaps = 76/755 (10%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P T IW A AHDF+SH E+ + +KI S+HFGQLSIIFLW SG F
Sbjct: 9 FSQGLARDPATRI-IWYGLAMAHDFESHDVWTEENLYQKIISSHFGQLSIIFLWTSGNLF 67
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRA 147
H A N+E W++DP HIRP +W P GQ + GG G + I TSG +Q W
Sbjct: 68 HVAWQGNFEQWVTDPVHIRPICHAIWDPHFGQPAVEAFTRGGASGPVNISTSGVYQWWYT 127
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY ++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+
Sbjct: 128 IGMRTNQDLYVGSVFLALVSAIFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVS 187
Query: 207 SLSWAGHQVHVSLP-INQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLN 265
SL+W GH VHV++P + + + G D LPH +G TPFFT N
Sbjct: 188 SLAWTGHLVHVAIPELQRGQHVGWDNFLSVLPHP----------------QGLTPFFTGN 231
Query: 266 WAKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGH 307
WA YA LTF GG P T LWLTD+AHHHLAIA++F++AGH
Sbjct: 232 WAAYAQSPDTASHVFGTAQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGH 291
Query: 308 MYRTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVA 365
MYRTN+G GH ++ ILEAH P G GHKGL++ + S H Q+ + LA +G++T +VA
Sbjct: 292 MYRTNFGNGHRMQAILEAHTPPSGSLGAGHKGLFDTVNNSLHFQIGLALASVGTITSLVA 351
Query: 366 HHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLD 425
HMYS+PPY + A D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L
Sbjct: 352 QHMYSLPPYAFQAIDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLA 411
Query: 426 RVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQ 485
R+L H++A+ISHL+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ
Sbjct: 412 RMLDHKEALISHLSWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQ 466
Query: 486 NTHA--------LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIH 537
H L T+ G SL W G L A+ N L + +G DFLVHH
Sbjct: 467 AAHGKALYGFDFLLSSKTSAAFANGQSL-WLPGWLDAINNNQNSLFLTIGLGDFLVHHAI 525
Query: 538 AFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWM 597
A +H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D +L +FWM
Sbjct: 526 ALGLHTTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYDAFYLAVFWM 585
Query: 598 YNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVI 657
N I V F++ WK +W QG V F +SS + GWLRD+LW +SQ+I
Sbjct: 586 LNTIGWVTFYWHWK-HLTLW-----QGNV-----AQFDESSTYLMGWLRDYLWLNSSQLI 634
Query: 658 QSYG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAT- 712
Y +SLS + FL H ++A MFL S RGYWQELIE++VWAH K +A
Sbjct: 635 NGYNPFGMNSLSVWAWTFLFGHLIYATGFMFLISWRGYWQELIETLVWAHEKTPLANLVY 694
Query: 713 ---QPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+ H+ +G I T AF +A
Sbjct: 695 WKDKPVALSIVQARLVGLAHFSVGYIFTYAAFLIA 729
>dbj|BAB18343.1| (AB044417) photosystem I P700 chlorophyll a apoprotein A1
[Chlamydomonas debaryana]
Length = 346
Score = 633 bits (1615), Expect = e-180
Identities = 286/347 (82%), Positives = 322/347 (92%), Gaps = 1/347 (0%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESM+NHHL GL+GLGSL WAGHQ+HVSLPIN+ L+AGVDPKEIPLPH+FILNR +
Sbjct: 1 WFQNVESMMNHHLGGLIGLGSLGWAGHQIHVSLPINKLLDAGVDPKEIPLPHDFILNRAI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW+ Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSDYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
HMYRTNWGIGH ++++LEAH+GPFTG+GHKGLYEILTTSWHAQL+INLA+ GSL+IIVAH
Sbjct: 121 HMYRTNWGIGHSMREMLEAHRGPFTGEGHKGLYEILTTSWHAQLAINLALFGSLSIIVAH 180
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMYSMPPYPYLATDYGTQLSLFTHH WIGGF IVGAAAHAAIFMVRDYDPT YN+LLDR
Sbjct: 181 HMYSMPPYPYLATDYGTQLSLFTHHTWIGGFCIVGAAAHAAIFMVRDYDPTNNYNNLLDR 240
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
V+RHRDA+ISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQN
Sbjct: 241 VIRHRDAMISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQN 300
Query: 487 THALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLV 533
TH +AP +AP A TSLTW GGDLVAVG KVA++PI LGT+DFLV
Sbjct: 301 THFVAPQFSAPNALAATSLTW-GGDLVAVGGKVAMMPISLGTSDFLV 346
>gi|11467571 PSI, P700 apoprotein A2 [Odontella sinensis]
sp|P49480|PSAB_ODOSI PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||S78376 photosystem I P700 apoprotein A2 - Odontella sinensis chloroplast
emb|CAA91749.1| (Z67753) PSI, P700 apoprotein A2 [Odontella sinensis]
Length = 733
Score = 632 bits (1612), Expect = e-180
Identities = 337/753 (44%), Positives = 447/753 (58%), Gaps = 74/753 (9%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P T IW A AHD ++H E+ + +KIF++HFG L++IFLW +G F
Sbjct: 8 FSQALAQDPATRR-IWYGIATAHDLEAHDGMTEENLYQKIFASHFGHLAVIFLWTAGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRGIQIT-SGFFQIWRAS 148
H A N+E W++ P +RP A +W P G+ L G + I SG +Q W
Sbjct: 67 HVAWQGNFEQWVAKPLKVRPIAHSIWDPHFGESALKAFSKGNTYPVNIAFSGVYQWWYTI 126
Query: 149 GITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGS 207
G + +LY ++G L+ + +LFAGW H K P L+WF++ ES LNHHL+GL+G+ S
Sbjct: 127 GFRTNQELYLGSVGLLLLSCALLFAGWLHLQPKFRPSLSWFKNNESRLNHHLSGLMGVSS 186
Query: 208 LSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWA 267
L+W GH VHV+LP ++ ++ G D PH G TPFFT NW
Sbjct: 187 LAWTGHLVHVALPASRGVHVGWDNFLTTPPHP----------------AGLTPFFTGNWT 230
Query: 268 KYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMY 309
YA LTF GG P T LWL+DIAHH LAIA +F+IAGHMY
Sbjct: 231 VYAQNPDSPSHVYGTSEGAGTRILTFLGGFHPQTQSLWLSDIAHHQLAIAFVFIIAGHMY 290
Query: 310 RTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHH 367
RTN+GIGH +K+IL+AH+ P G GH GL+E +T S H QL + LA LG T + A H
Sbjct: 291 RTNFGIGHNMKEILDAHRPPGGRLGAGHVGLFETITNSLHIQLGLALACLGVATSLTAQH 350
Query: 368 MYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRV 427
MY++ PY YL+ D+ T+ +L+THH +I GFL+VGA AH AIF VRDYDP N++L R+
Sbjct: 351 MYALTPYAYLSKDFTTEAALYTHHQYIAGFLMVGAFAHGAIFFVRDYDPELNKNNVLARM 410
Query: 428 LRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQ-- 485
L H++AIISHL+W +FLGFH GLYIHNDT+ A G+P+ I +P+FA++IQ
Sbjct: 411 LEHKEAIISHLSWASLFLGFHVLGLYIHNDTVVAFGQPEKQ-----ILFEPLFAEYIQAA 465
Query: 486 ------NTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAF 539
N + L T P G + W G L A+ N L + +G DFLVHH A
Sbjct: 466 SGKAVYNFNVLLSSSTNPATIAGNQV-WLPGWLEAINNSKTDLFLKIGPGDFLVHHAIAL 524
Query: 540 TIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN 599
+HVT LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N
Sbjct: 525 GLHVTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAMFWMLN 584
Query: 600 AISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQS 659
I V F++ WK + +WG G F +SS I GWLRD+LW +S +I
Sbjct: 585 TIGWVTFYWHWKHMT-IWGG----------NPGQFDESSNYIMGWLRDYLWLNSSPLING 633
Query: 660 YG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----A 711
Y ++LS + FL H +WA MFL S RGYWQELIE++VWAH + +A
Sbjct: 634 YNPFGMNNLSVWAWMFLFGHLIWATGFMFLISWRGYWQELIETLVWAHERTPLANLIRWR 693
Query: 712 TQPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+ H+ +G I T AF +A
Sbjct: 694 DKPVALSIVQARLVGLVHFAVGYILTYAAFVIA 726
>gb|AAA84452.1| (M37526) alpha-apoprotein [Euglena gracilis]
Length = 733
Score = 631 bits (1611), Expect = e-180
Identities = 338/753 (44%), Positives = 447/753 (58%), Gaps = 74/753 (9%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A +HDF+SH E+ + +KIF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAQDP-TTRRIWFGIATSHDFESHDGMTENNLYQKIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E W+ DP HIRP A + P GQ + F G + I SG +Q W
Sbjct: 67 HVAWQGNFEQWIKDPLHIRPIAHAISDPHFGQPAIEAFTRSQFPGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + + LY ++ L A L LFAGW H K +PK++WF+D ES LNHHL+ L GL
Sbjct: 127 IGLRTNVDLYNGSMFLLFIATLALFAGWLHLEPKYSPKVSWFKDAESRLNHHLSALFGLS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+W+GH +HV++P ++ ++ D LPH PS G PFF NW
Sbjct: 187 SLAWSGHLIHVAIPESRGIHVRWDNFLSQLPH-------------PS---GLEPFFKGNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
+ Y++ LTF GG P T LWLTDIAHHHLAIA+LF++AGHM
Sbjct: 231 SLYSENPDSLTHVFGTASGAGTAVLTFLGGFHPETKSLWLTDIAHHHLAIAVLFIVAGHM 290
Query: 309 YRTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
YRTN+ IGH I DIL AHK P G GH GLYE + S H QL + LA LG +T +VA
Sbjct: 291 YRTNFAIGHRIDDILNAHKAPSGKLGLGHFGLYETINNSLHFQLGLALASLGVITSLVAQ 350
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMYS+ PY +L D T +L+THH +I GF++ GA AH AIF +RD+D ++L R
Sbjct: 351 HMYSLSPYAFLIQDRTTMAALYTHHQYIAGFIMTGAFAHGAIFFIRDFDEEKNKGNVLSR 410
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
+L H++AIISHL+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ+
Sbjct: 411 ILDHKEAIISHLSWVTLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQS 465
Query: 487 THA-------LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAF 539
H + + A + + W G L + +K L + +G DFLVHH A
Sbjct: 466 AHGKNIYELNILLSSDSSNAFSASQAIWLPGWLNGINDKSTSLFLQIGPGDFLVHHAIAL 525
Query: 540 TIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN 599
+H T LIL+KG L AR SRL+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N
Sbjct: 526 GLHTTTLILVKGALDARGSRLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLN 585
Query: 600 AISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQS 659
I F++ WK +W ++ G F +SS + GWLRD+LW +SQ+I
Sbjct: 586 TIGWTTFYWHWK-HITLWRNV-----------GQFNESSTYLMGWLRDYLWLNSSQLING 633
Query: 660 YG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----A 711
Y +SL+ +G FL H VWA MFL S RGYWQELIE++VWAH + +
Sbjct: 634 YNPFGMNSLAVWGWMFLFGHLVWATGFMFLISWRGYWQELIETLVWAHERTPITNVFKWT 693
Query: 712 TQPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+ H+ +G + T AF +A
Sbjct: 694 DKPVALSIVQARLVGLAHFSVGYVFTYAAFLIA 726
>gi|11467000 PSI P700 apoprotein, subunit 1b [Euglena gracilis]
sp|P19431|PSAB_EUGGR PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||S26072 photosystem I protein A2 - Euglena gracilis chloroplast
emb|CAA77911.1| (Z11874) PSII P700 chl. a apoprotein [Euglena gracilis]
emb|CAA50094.1| (X70810) PSI P700 apoprotein, subunit 1b [Euglena gracilis]
Length = 734
Score = 631 bits (1611), Expect = e-180
Identities = 341/753 (45%), Positives = 448/753 (59%), Gaps = 73/753 (9%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A +HDF+SH E+ + +KIF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAQDP-TTRRIWFGIATSHDFESHDGMTENNLYQKIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E W+ DP HIRP A + P GQ + F G + I SG +Q W
Sbjct: 67 HVAWQGNFEQWIKDPLHIRPIAHAISDPHFGQPAIEAFTRSQFPGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + + LY ++ L A L LFAGW H K +PK++WF+D ES LNHHL+ L GL
Sbjct: 127 IGLRTNVDLYNGSMFLLFIATLALFAGWLHLEPKYSPKVSWFKDAESRLNHHLSALFGLS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+W+GH +HV++P ++ ++ D LPH PS G PFF NW
Sbjct: 187 SLAWSGHLIHVAIPESRGIHVRWDNFLSQLPH-------------PS---GLEPFFKGNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
+ Y++ LTF GG P T LWLTDIAHHHLAIA+LF++AGHM
Sbjct: 231 SLYSENPDSLTHVFGTASGAGTAVLTFLGGFHPETKSLWLTDIAHHHLAIAVLFIVAGHM 290
Query: 309 YRTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
YRTN+ IGH I DIL AHK P G GH GLYE + S H QL + LA LG +T +VA
Sbjct: 291 YRTNFAIGHRIDDILNAHKAPSGKLGLGHFGLYETINNSLHFQLGLALASLGVITSLVAQ 350
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMYS+ PY +L D T +L+THH +I GF++ GA AH AIF +RD+D ++L R
Sbjct: 351 HMYSLSPYAFLIQDRTTMAALYTHHQYIAGFIMTGAFAHGAIFFIRDFDEEKNKGNVLSR 410
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
+L H++AIISHL+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ+
Sbjct: 411 ILDHKEAIISHLSWVTLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQS 465
Query: 487 THA-------LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAF 539
H + + A + + W G L + +K L + +G DFLVHH A
Sbjct: 466 AHGKNIYELNILLSSDSSNAFSASQAIWLPGWLNGINDKSTSLFLQIGPGDFLVHHAIAL 525
Query: 540 TIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN 599
+H T LIL+KG L AR SRL+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N
Sbjct: 526 GLHTTTLILVKGALDARGSRLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLN 585
Query: 600 AISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQS 659
I F++ WK +W QG V G F +SS + GWLRD+LW +SQ+I
Sbjct: 586 TIGWTTFYWHWK-HITLW-----QGNV-----GQFNESSTYLMGWLRDYLWLNSSQLING 634
Query: 660 YG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----A 711
Y +SL+ +G FL H VWA MFL S RGYWQELIE++VWAH + +
Sbjct: 635 YNPFGMNSLAVWGWMFLFGHLVWATGFMFLISWRGYWQELIETLVWAHERTPITNVFKWT 694
Query: 712 TQPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+ H+ +G + T AF +A
Sbjct: 695 DKPVALSIVQARLVGLAHFSVGYVFTYAAFLIA 727
>gi|11466786 PSI P700 apoprotein A2 [Oryza sativa]
sp|P12156|PSAB_ORYSA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||A2RZP7 photosystem I P700 apoprotein A2 - rice chloroplast
emb|CAA33995.1| (X15901) PSI P700 apoprotein A2 [Oryza sativa]
prf||1603356AA photosystem I P700 apoprotein A2 [Oryza sativa]
Length = 734
Score = 631 bits (1611), Expect = e-180
Identities = 343/746 (45%), Positives = 445/746 (58%), Gaps = 59/746 (7%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + + IF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAQDP-TTRRIWFGIATAHDFESHDDITEERLYQNIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E+W+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFESWIQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGAAGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L + L L GW H K P L+WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNEDLYTGALFLLFLSTLSLIGGWLHLQPKWKPSLSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLPI--------NQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAE 256
SL+W GH VHV++P N FL+ LP+ L L Q LY +
Sbjct: 187 SLAWTGHLVHVAIPASRGEYVRWNNFLDV--------LPYPQGLGPLLTGQWNLYAQNPD 238
Query: 257 GATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIG 316
+ F LT GG P T LWLTDIAHHHLAIA +FLIAGHMYRTN+GIG
Sbjct: 239 SSNHLFGTTQGAGTAILTLLGGFHPQTQSLWLTDIAHHHLAIAFIFLIAGHMYRTNFGIG 298
Query: 317 HGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPY 374
H IKD+LEAH P G+GHKGLY+ + S H QL + LA LG +T +VA HMYS+P Y
Sbjct: 299 HSIKDLLEAHTPPGGRLGRGHKGLYDTINNSIHFQLGLALASLGVITSLVAQHMYSLPSY 358
Query: 375 PYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAI 434
++A D+ TQ +L+THH +I GF++ GA AH AIF +RDY+P +++L R+L H++AI
Sbjct: 359 AFIAQDFTTQAALYTHHQYIAGFIMTGAFAHGAIFFIRDYNPEQNEDNVLARMLDHKEAI 418
Query: 435 ISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA----- 489
ISHL+W +FLGFH+ GLY+HND M A G P+ I ++PIFAQWIQ+ H
Sbjct: 419 ISHLSWASLFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPIFAQWIQSAHGKTTYG 473
Query: 490 ---LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
L + P G +L W G L AV L + +G DFLVHH A +H T L
Sbjct: 474 FDILLSSTSGPAFNAGRTL-WLPGWLNAVNENSNSLFLTIGPGDFLVHHAIALGLHTTTL 532
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
IL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N I V F
Sbjct: 533 ILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLNTIGWVTF 592
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYG----S 662
++ WK + G++S F +SS + GWLRD+LW +SQ+I Y +
Sbjct: 593 YWHWKHITLWQGNVS-----------QFNESSTYLMGWLRDYLWLNSSQLINGYNPFGMN 641
Query: 663 SLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----ATQPRALS 718
SLS + FL H VWA MFL S RGYWQELIE++ WAH + +A +P ALS
Sbjct: 642 SLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLIRWRDKPVALS 701
Query: 719 IVQGRAVGVTHYLLGGIATTWAFFLA 744
IVQ R VG+ H+ +G I T AF +A
Sbjct: 702 IVQARLVGLAHFSVGYIFTYAAFLIA 727
>gi|11465396 unknown; Photosystem I p700 chlorophyll A apoprotein A2 [Cyanidium
caldarium]
gb|AAF12881.1|AF022186_3 (AF022186) unknown; Photosystem I p700 chlorophyll A apoprotein A2
[Cyanidium caldarium]
Length = 734
Score = 631 bits (1610), Expect = e-180
Identities = 339/753 (45%), Positives = 453/753 (60%), Gaps = 73/753 (9%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS++++ P TT IW A +HDF++H + E+ + +KIF++HFG L+IIFLW SG F
Sbjct: 8 FSQSLSSDP-TTRRIWYGIATSHDFEAHDNVTEENLYQKIFASHFGHLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFR-GIQIT-SGFFQIWRA 147
H A N+EAW+++P ++P A +W P GQ L GG + I+ SG + W
Sbjct: 67 HVAWQGNFEAWIANPLKVKPVAHAIWDPHFGQAALKAFSRGGVNYPVDISYSGVYHWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ LV AAL +FAG H K P ++WF++ ES LNHHL+GL GL
Sbjct: 127 IGMRTSSDLYAGALFLLVLAALCMFAGRLHLQPKFKPSISWFKNNESRLNHHLSGLFGLS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+W GH +HV++P ++ + G N L +P+ G PFFT NW
Sbjct: 187 SLAWCGHLIHVAIPASRGQHIG-------------WNNFLTTPPHPA---GLKPFFTGNW 230
Query: 267 AKYA------------------DFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
+ Y+ LTF GG P T LWLTDIAHHHLAIA++F+IAGHM
Sbjct: 231 SDYSLNPDDPSHIFGTSSNSGKAILTFLGGFHPQTQSLWLTDIAHHHLAIAVIFIIAGHM 290
Query: 309 YRTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
YRTN+GIGH IKDILEAHK P G GHKGL+ ++ S H QL + LA LG L+ + A
Sbjct: 291 YRTNFGIGHNIKDILEAHKPPSGKMGLGHKGLFNTISNSLHFQLGLALACLGVLSSLTAQ 350
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
H+YS+PPY +++ D+ TQ +L+THH +I GFL+VGA AH AIF VRDYDP +++L R
Sbjct: 351 HLYSIPPYAFISRDFVTQAALYTHHQYIAGFLMVGAFAHGAIFFVRDYDPKQNKDNVLYR 410
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
+L H++AIISHL+WV +FLGFH+ GLY+HND + A G P+ I ++PIFAQWIQ
Sbjct: 411 MLEHKEAIISHLSWVSLFLGFHTLGLYVHNDVVVAFGNPEKQ-----ILIEPIFAQWIQA 465
Query: 487 THA-------LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAF 539
T + ++ AT W L A+ N L + +G DFLVHH A
Sbjct: 466 TSGKTLYGFNVLLASSSSSATQAAQSLWLPNWLEAINNNNNSLFLTIGPGDFLVHHAIAL 525
Query: 540 TIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN 599
+H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +LG+FWM N
Sbjct: 526 GLHTTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLGVFWMLN 585
Query: 600 AISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQS 659
I F++ WK +W + Q F +SS + GW RD+LW +S +I
Sbjct: 586 TIGWTTFYWHWK-HITIWQGNASQ----------FNESSTYLMGWFRDYLWLNSSPIING 634
Query: 660 YG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----A 711
Y ++LS + FL AH VWA MFL S RGYWQELIES+VWAH + +A
Sbjct: 635 YNPYGMNNLSVWAWMFLFAHLVWATGFMFLISWRGYWQELIESLVWAHERTPLANLITWK 694
Query: 712 TQPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+ H+ +G + T AF +A
Sbjct: 695 DKPVALSIVQARLVGLVHFSVGYVLTYAAFVIA 727
>gi|11465954 PSI P700 apoprotein A2 [Nicotiana tabacum]
sp|P06407|PSAB_TOBAC PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||A2NTP7 photosystem I P700 apoprotein A2 - common tobacco chloroplast
emb|CAA77351.1| (Z00044) PSI P700 apoprotein A2 [Nicotiana tabacum]
prf||1211235AB photosystem I P700 apoprotein A2 [Nicotiana tabacum]
Length = 734
Score = 631 bits (1609), Expect = e-179
Identities = 340/746 (45%), Positives = 447/746 (59%), Gaps = 59/746 (7%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + + IF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAQDP-TTRRIWFGIATAHDFESHDDITEERLYQNIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E+W+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFESWVQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGALGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L +A+ L AGW H K P ++WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNEDLYTGALFLLFLSAISLIAGWLHLQPKWKPSVSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLPI--------NQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAE 256
SL+W GH VHV++P N FL+ LPH L Q LY +
Sbjct: 187 SLAWTGHLVHVAIPASRGEYVRWNNFLDV--------LPHPQGLGPLFTGQWNLYAQNPD 238
Query: 257 GATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIG 316
++ F LT GG P T LWLTDIAHHHLAIA +FL+AGHMYRTN+GIG
Sbjct: 239 SSSHLFGTAQGAGTAILTLLGGFHPQTQSLWLTDIAHHHLAIAFIFLVAGHMYRTNFGIG 298
Query: 317 HGIKDILEAH--KGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPY 374
H +KD+L+AH G G+GHKGLY+ + S H QL + LA LG +T +VA HMYS+P Y
Sbjct: 299 HSMKDLLDAHIPPGGRLGRGHKGLYDTINNSLHFQLGLALASLGVITSLVAQHMYSLPAY 358
Query: 375 PYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAI 434
++A D+ TQ +L+THH +I GF++ GA AH AIF +RDY+P +++L R+L H++AI
Sbjct: 359 AFIAQDFTTQAALYTHHQYIAGFIMTGAFAHGAIFFIRDYNPEQNEDNVLARMLEHKEAI 418
Query: 435 ISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTH------ 488
ISHL+W +FLGFH+ GLY+HND M A G P+ I ++PIFAQWIQ+ H
Sbjct: 419 ISHLSWASLFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPIFAQWIQSAHGKTSYG 473
Query: 489 --ALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
L + P G S+ W G L AV L + +G DFLVHH A +H T L
Sbjct: 474 FDVLLSSTSGPAFNAGRSI-WLPGWLNAVNENSNSLFLTIGPGDFLVHHAIALGLHTTTL 532
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
IL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N I V F
Sbjct: 533 ILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLNTIGWVTF 592
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYG----S 662
++ WK + G++S F +SS + GWLRD+LW +SQ+I Y +
Sbjct: 593 YWHWKHITLWQGNVS-----------QFNESSTYLMGWLRDYLWLNSSQLINGYNPFGMN 641
Query: 663 SLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----ATQPRALS 718
SLS + FL H VWA MFL S RGYWQELIE++ WAH + +A +P ALS
Sbjct: 642 SLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLIRWRDKPVALS 701
Query: 719 IVQGRAVGVTHYLLGGIATTWAFFLA 744
IVQ R VG+ H+ +G I T AF +A
Sbjct: 702 IVQARLVGLAHFSVGYIFTYAAFLIA 727
>sp|Q33332|PSAB_ANTMA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
emb|CAA58958.1| (X84153) photosystem I subunit [Antirrhinum majus]
Length = 734
Score = 629 bits (1605), Expect = e-179
Identities = 338/746 (45%), Positives = 447/746 (59%), Gaps = 59/746 (7%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + + IF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAQDP-TTRRIWFGIATAHDFESHDDITEERLYQNIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E+W+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFESWVQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGALGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L +A+ L AGW H K P ++WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNEDLYSGALFLLFLSAISLIAGWLHLQPKWKPSVSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLP--------INQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAE 256
L+W GH VHV++P N FL+ LPH L Q LY +
Sbjct: 187 PLAWTGHLVHVAIPGSRGEYVRWNNFLDV--------LPHPQGLGPLFTGQWNLYAQNPD 238
Query: 257 GATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIG 316
++ F + LT GG P T LWLTD+AHHHLAIA +FL+AGHMYRTN+GIG
Sbjct: 239 SSSHLFGTSQGAGTAILTLLGGFHPQTQSLWLTDMAHHHLAIAFIFLVAGHMYRTNFGIG 298
Query: 317 HGIKDILEAH--KGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPY 374
H IKD+L+AH G G+GHKGLY+ + S H QL + LA LG +T +VA HMYS+P Y
Sbjct: 299 HSIKDLLDAHIPPGGRLGRGHKGLYDTINNSLHFQLGLALASLGVITSLVAQHMYSLPAY 358
Query: 375 PYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAI 434
++A D+ TQ +L+THH +I GF++ GA AH AIF +RDY+P +++L R+L H++AI
Sbjct: 359 AFIAQDFTTQAALYTHHQYIAGFIMTGAFAHGAIFFIRDYNPEQNQDNVLARMLDHKEAI 418
Query: 435 ISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTH------ 488
ISHL+W +FLGFH+ GLY+HND M A G P+ I ++PIFAQWIQ+ H
Sbjct: 419 ISHLSWASLFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPIFAQWIQSAHGKTSYG 473
Query: 489 --ALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
L + P G S+ W G L A+ L + +G DFLVHH A +H T L
Sbjct: 474 FDVLLSSTSGPAFNAGRSI-WLPGWLNAINENTNSLFLTIGPGDFLVHHAIALGLHTTTL 532
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
IL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N I V F
Sbjct: 533 ILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLNTIGWVTF 592
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYG----S 662
++ WK + G++S F +SS + GWLRD+LW +SQ+I Y +
Sbjct: 593 YWHWKHITLWQGNVS-----------QFNESSTYLMGWLRDYLWLNSSQLINGYNPFGMN 641
Query: 663 SLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----ATQPRALS 718
SLS + FL H VWA MFL S RGYWQELIE++ WAH + +A +P ALS
Sbjct: 642 SLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLIRWRDKPVALS 701
Query: 719 IVQGRAVGVTHYLLGGIATTWAFFLA 744
IVQ R VG+ H+ +G I T AF +A
Sbjct: 702 IVQARLVGLAHFSVGYIFTYAAFLIA 727
>sp|P25897|PSAB_SYNEN PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||S20923 photosystem I protein A2 - Synechococcus sp
emb|CAA45305.1| (X63768) photosystem I subunit Ib [Synechococcus sp.]
dbj|BAA01760.1| (D10986) photosystem I core protein B [Synechococcus vulcanus]
Length = 741
Score = 628 bits (1603), Expect = e-179
Identities = 343/765 (44%), Positives = 451/765 (58%), Gaps = 82/765 (10%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + +KIF++HFG L+IIFLW+SG F
Sbjct: 8 FSQDLAQDP-TTRRIWYAIAMAHDFESHDGMTEENLYQKIFASHFGHLAIIFLWVSGSLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDV-GGGFRGIQIT-SGFFQIWRA 147
H A N+E W+ DP + RP A +W P G+ ++ G + I SG + W
Sbjct: 67 HVAWQGNFEQWVQDPVNTRPIAHAIWDPQFGKAAVDAFTQAGASNPVDIAYSGVYHWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY AI L+ A+L LFAGW H K P L+WF++ ES LNHHLAGL G+
Sbjct: 127 IGMRTNGDLYQGAIFLLILASLALFAGWLHLQPKFRPSLSWFKNAESRLNHHLAGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+WAGH +HV++P ++ + G D +PH G PFFT NW
Sbjct: 187 SLAWAGHLIHVAIPESRGQHVGWDNFLSTMPHP----------------AGLAPFFTGNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
YA LTF GG P T LWLTD+AHHHLAIA+LF++AGHM
Sbjct: 231 GVYAQNPDTASHVFGTAQGAGTAILTFLGGFHPQTESLWLTDMAHHHLAIAVLFIVAGHM 290
Query: 309 YRTNWGIGHGIKDILEAH-------KGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLT 361
YRT +GIGH IK++++A +GPF H+G+YE S H QL +LA LG +T
Sbjct: 291 YRTQFGIGHSIKEMMDAKDFFGTKVEGPFN-MPHQGIYETYNNSLHFQLGWHLACLGVIT 349
Query: 362 IIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYN 421
+VA HMYS+PPY ++A D+ T +L+THH +I GFL+VGA AH AIF+VRDYDP
Sbjct: 350 SLVAQHMYSLPPYAFIAQDHTTMAALYTHHQYIAGFLMVGAFAHGAIFLVRDYDPAQNKG 409
Query: 422 DLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFA 481
++LDRVL+H++AIISHL+WV +FLGFH+ GLY+HND + A G P+ I ++P+FA
Sbjct: 410 NVLDRVLQHKEAIISHLSWVSLFLGFHTLGLYVHNDVVVAFGTPEKQ-----ILIEPVFA 464
Query: 482 QWIQNTHALA----------PGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADF 531
Q+IQ H P A A W G L A+ + L + +G DF
Sbjct: 465 QFIQAAHGKLLYGFDTLLSNPDSIASTAWPNYGNVWLPGWLDAINSGTNSLFLTIGPGDF 524
Query: 532 LVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVF 591
LVHH A +H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +
Sbjct: 525 LVHHAIALGLHTTTLILVKGALDARGSKLMPDKKDFGYAFPCDGPGRGGTCDISAWDAFY 584
Query: 592 LGLFWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWA 651
L +FWM N I V F++ WK VW +G V F +SS + GWLRD+LW
Sbjct: 585 LAMFWMLNTIGWVTFYWHWK-HLGVW-----EGNV-----AQFNESSTYLMGWLRDYLWL 633
Query: 652 QASQVIQSYG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLK 707
+SQ+I Y ++LS + FL H VWA MFL S RGYWQELIE++VWAH +
Sbjct: 634 NSSQLINGYNPFGTNNLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLVWAHERTP 693
Query: 708 VAPAT----QPRALSIVQGRAVGVTHYLLGGIATTWAFFLARIIA 748
+A +P ALSIVQ R VG+ H+ +G I T AF +A A
Sbjct: 694 LANLVRWKDKPVALSIVQARLVGLAHFSVGYILTYAAFLIASTAA 738
>sp|O52475|PSAB_MASLA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
gb|AAC12867.1| (AF038558) photosystem I PsaB [Fischerella PCC7605]
Length = 743
Score = 627 bits (1600), Expect = e-178
Identities = 337/762 (44%), Positives = 447/762 (58%), Gaps = 82/762 (10%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A +DF+SH E+ + +KIF+ HFG ++IIFLW S + F
Sbjct: 8 FSQDLAQDP-TTRRIWYAMATGNDFESHDGMTEENLYQKIFATHFGHVAIIFLWASSLLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFR-GIQIT-SGFFQIWRA 147
H A N+E W+ DP HIRP A +W P G+ + GG + I+ SG + W
Sbjct: 67 HVAWQGNFEQWIKDPIHIRPIAHAIWDPQFGKPAIEAFTQGGANYPVNISYSGIYHWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L+ A++ LFAGW H K P L+WF+ E LNHHLAGL G+
Sbjct: 127 IGMRTNNDLYMGAVFLLILASVFLFAGWLHLQPKFRPSLSWFKSAEPRLNHHLAGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+WAGH +HV++P ++ + G + LPH G PFFT NW
Sbjct: 187 SLAWAGHLIHVAIPESRGQHVGWNNFLSTLPHP----------------AGLQPFFTGNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
YA LTF GG P T LWLTD+AHHHLAIA++F+IAGHM
Sbjct: 231 GVYASDPDTANHVFGTSQGAGTAILTFLGGFHPQTESLWLTDMAHHHLAIAVIFIIAGHM 290
Query: 309 YRTNWGIGHGIKDILEAHKGPFTGQG--------HKGLYEILTTSWHAQLSINLAMLGSL 360
YRTN+GIGH IK++L + G G H+GLY+ S H QL I+LA LG++
Sbjct: 291 YRTNFGIGHSIKEMLNSKAGLVPGTNSEGQFNLPHQGLYDTYNNSLHFQLGIHLAALGTI 350
Query: 361 TIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
T +VA HMY+MPPY ++A DY TQ +L+THH +I FL++GA AH AIF VRDYDP
Sbjct: 351 TSLVAQHMYAMPPYAFIAKDYTTQAALYTHHQYIAIFLMLGAFAHGAIFWVRDYDPEQNK 410
Query: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
++LDRVL+H++AIISHL+WV +FLGFH+ GLY+HND + A G P+ I ++P+F
Sbjct: 411 GNVLDRVLKHKEAIISHLSWVSLFLGFHTLGLYVHNDVVVAFGTPEKQ-----ILIEPVF 465
Query: 481 AQWIQNTHALA----------PGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTAD 530
AQ+IQ +H P A A W G L A+ + L + +G D
Sbjct: 466 AQFIQASHGKVLYGLNVLLSNPDSVAYTAYPNYGNVWLQGWLDAINSGTNSLFLTIGPGD 525
Query: 531 FLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHV 590
FLVHH A +H TVLIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD
Sbjct: 526 FLVHHAFALALHTTVLILVKGALDARGSKLMPDKKDFGYAFPCDGPGRGGTCDISAWDAF 585
Query: 591 FLGLFWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLW 650
+L FW N + V F++ WK +W QG V F +SS + GW RD+LW
Sbjct: 586 YLATFWALNTVGWVTFYWHWK-HLGIW-----QGNV-----AQFNESSTYLMGWFRDYLW 634
Query: 651 AQASQVIQSYG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKL 706
A ++Q+I Y ++LS + FL H VWA MFL S RGYWQELIE++VWAH +
Sbjct: 635 ANSAQLINGYNPYGMNNLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLVWAHERT 694
Query: 707 KVAPAT----QPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+A +P ALSIVQ R VG+TH+ +G + T AF +A
Sbjct: 695 PLANLVRWKDKPVALSIVQARLVGLTHFTVGYVLTYAAFLIA 736
>pir||S60184 photosystem I protein A2 - garden snapdragon chloroplast
Length = 734
Score = 627 bits (1600), Expect = e-178
Identities = 337/746 (45%), Positives = 446/746 (59%), Gaps = 59/746 (7%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + + IF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAQDP-TTRRIWFGIATAHDFESHDDITEERLYQNIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E+W+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFESWVQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGALGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L +A+ L AGW H K P ++WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNEDLYSGALFLLFLSAISLIAGWLHLQPKWKPSVSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLP--------INQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAE 256
L+W GH VHV++P N FL+ LPH L Q LY +
Sbjct: 187 PLAWTGHLVHVAIPGSRGEYVRWNNFLDV--------LPHPQGLGPLFTGQWNLYAQNPD 238
Query: 257 GATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIG 316
++ F + LT GG P T LWLTD+AHHHLAIA +FL+AGHMYRTN+GIG
Sbjct: 239 SSSHLFGTSQGAGTAILTLLGGFHPQTQSLWLTDMAHHHLAIAFIFLVAGHMYRTNFGIG 298
Query: 317 HGIKDILEAH--KGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPY 374
H IKD+L+AH G G+GHKGLY+ + S H QL + LA LG +T +VA HMYS+P Y
Sbjct: 299 HSIKDLLDAHIPPGGRLGRGHKGLYDTINNSLHFQLGLALASLGVITSLVAQHMYSLPAY 358
Query: 375 PYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAI 434
++A D+ TQ +L+THH +I GF++ GA AH AIF +RDY+P +++L R+L H++AI
Sbjct: 359 AFIAQDFTTQAALYTHHQYIAGFIMTGAFAHGAIFFIRDYNPEQNQDNVLARMLDHKEAI 418
Query: 435 ISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTH------ 488
ISHL+W +FLGFH+ GLY+HND M A G P+ I ++PIFAQWIQ+ H
Sbjct: 419 ISHLSWASLFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPIFAQWIQSAHGKTSYG 473
Query: 489 --ALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
L + P G S+ W G L A+ L + +G DFLVHH A +H T L
Sbjct: 474 FDVLLSSTSGPAFNAGRSI-WLPGWLNAINENTNSLFLTIGPGDFLVHHAIALGLHTTTL 532
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
IL+KG L AR S+L+PDK + G+ FPCDGPGRGG C +SAWD +L +FWM N I V F
Sbjct: 533 ILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGACDISAWDAFYLAVFWMLNTIGWVTF 592
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYG----S 662
++ WK + G++S F +SS + GWLRD+LW +SQ+I Y +
Sbjct: 593 YWHWKHITLWQGNVS-----------QFNESSTYLMGWLRDYLWLNSSQLINGYNPFGMN 641
Query: 663 SLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----ATQPRALS 718
SLS + FL H VWA MFL S RGYWQELIE++ WAH + +A +P ALS
Sbjct: 642 SLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLIRWRDKPVALS 701
Query: 719 IVQGRAVGVTHYLLGGIATTWAFFLA 744
IVQ R VG+ H+ +G I T AF +A
Sbjct: 702 IVQARLVGLAHFSVGYIFTYAAFLIA 727
>gi|11497524 PSI P700 apoprotein A2 [Spinacia oleracea]
sp|P06512|PSAB_SPIOL PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||S00445 photosystem I protein A2 - spinach chloroplast
emb|CAB88725.1| (AJ400848) PSI P700 apoprotein A2 [Spinacia oleracea]
prf||1303218B gene psaB [Spinacia oleracea]
Length = 734
Score = 627 bits (1599), Expect = e-178
Identities = 338/746 (45%), Positives = 446/746 (59%), Gaps = 59/746 (7%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + + IF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAQDP-TTRRIWFGIATAHDFESHDDITEERLYQNIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E+W+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFESWVQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGALGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L + + L GW H K P ++WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNEDLYTGALFLLFLSVISLLGGWLHLQPKWKPSVSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLP--------INQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAE 256
SL+W GH VHV++P N FL+ LPH L Q LY +
Sbjct: 187 SLAWTGHLVHVAIPGSRGEYVRWNNFLDV--------LPHPQGLGPLFTGQWNLYAQNPD 238
Query: 257 GATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIG 316
++ F + LT GG P T LWLTD+AHHHLAIA +FL+AGHMYRTN+GIG
Sbjct: 239 SSSHLFGTSQGAGTAILTLLGGFHPQTQSLWLTDMAHHHLAIAFVFLVAGHMYRTNFGIG 298
Query: 317 HGIKDILEAH--KGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPY 374
H +KD+LEAH G G+GHKGLY+ + S H QL + LA LG +T +VA HMYS+P Y
Sbjct: 299 HSMKDLLEAHIPPGGRLGRGHKGLYDTINNSLHFQLGLALASLGVITSLVAQHMYSLPAY 358
Query: 375 PYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAI 434
++A D+ TQ +L+THH +I GF++ GA AH AIF +RDY+P +++L R+L H++AI
Sbjct: 359 AFIAQDFTTQAALYTHHQYIAGFIMTGAFAHGAIFFIRDYNPEQNEDNVLARMLDHKEAI 418
Query: 435 ISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTH------ 488
ISHL+W +FLGFH+ GLY+HND M A G P+ I ++PIFAQWIQ+ H
Sbjct: 419 ISHLSWASLFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPIFAQWIQSAHGKTSYG 473
Query: 489 --ALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
L + P G S+ W G L AV L + +G DFLVHH A +H T L
Sbjct: 474 FDVLLSSTSGPAFNAGRSI-WLPGWLNAVNENSNSLFLTIGPGDFLVHHAIALGLHTTTL 532
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
IL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N I V F
Sbjct: 533 ILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLNTIGWVTF 592
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYG----S 662
++ WK + G++S F +SS + GWLRD+LW +SQ+I Y +
Sbjct: 593 YWHWKHITLWQGNVS-----------QFNESSTYLMGWLRDYLWLNSSQLINGYNPFGMN 641
Query: 663 SLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----ATQPRALS 718
SLS + FL H VWA MFL S RGYWQELIE++ WAH + +A +P ALS
Sbjct: 642 SLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLIRWRDKPVALS 701
Query: 719 IVQGRAVGVTHYLLGGIATTWAFFLA 744
IVQ R VG+ H+ +G I T AF +A
Sbjct: 702 IVQARLVGLAHFSVGYIFTYAAFLIA 727
>gb|AAA84486.1| (M11203) P700 chlorophyll a-protein PSI-A2 [Zea mays]
Length = 735
Score = 626 bits (1596), Expect = e-178
Identities = 340/747 (45%), Positives = 443/747 (58%), Gaps = 60/747 (8%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + + IF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAQDP-TTRRIWFGIATAHDFESHDDITEERLYQNIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E+W+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFESWIQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGAAGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L + L L GW H K P L+WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNEDLYTGALFLLFLSTLSLIGGWLHLQPKWKPSLSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLP---------INQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFA 255
SL+W GH VHV++P N FL+ LP+ L L Q LY
Sbjct: 187 SLAWTGHLVHVAIPGSSRGEYVRWNNFLDV--------LPYPQGLGPLLTGQWNLYAQNP 238
Query: 256 EGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGI 315
+ + F LT GG P T LWLTDIAHHHLAIA +FLIAGHMYRTN+GI
Sbjct: 239 DSSNHLFGTTQGAGTAILTLLGGFHPQTQSLWLTDIAHHHLAIAFIFLIAGHMYRTNFGI 298
Query: 316 GHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPP 373
GH IKD+LEAH P G+GHKGLY+ + S H QL + LA LG +T +VA HMYS+P
Sbjct: 299 GHSIKDLLEAHTPPGGRLGRGHKGLYDTINNSIHFQLGLALASLGVITSLVAQHMYSLPA 358
Query: 374 YPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDA 433
Y ++A D+ TQ +L+THH +I GF++ GA AH AIF +RDY+P +++L R+L H++A
Sbjct: 359 YAFIAQDFTTQAALYTHHQYIAGFIMTGAFAHGAIFFIRDYNPEQNEDNVLARMLDHKEA 418
Query: 434 IISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA---- 489
IISHL+W +FLGFH+ G Y+HND M A G P+ I ++PIFAQWIQ+ H
Sbjct: 419 IISHLSWASLFLGFHTLGPYVHNDVMLAFGTPEKQ-----ILIEPIFAQWIQSAHGKTTY 473
Query: 490 ----LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTV 545
L P G ++ W G L AV L + +G DFLVHH A +H T
Sbjct: 474 GFDILLSSTNGPTFNAGRNI-WLPGWLNAVNENSNSLFLTIGPGDFLVHHAIALGLHTTT 532
Query: 546 LILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVI 605
LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N I V
Sbjct: 533 LILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLNTIGWVT 592
Query: 606 FHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYG---- 661
F++ WK + G++S F +SS + GWLRD+LW +SQ+I Y
Sbjct: 593 FYWHWKHITLWQGNVS-----------QFNESSTYLMGWLRDYLWLNSSQLINGYNPFGM 641
Query: 662 SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----ATQPRAL 717
+SLS + FL H VWA MFL S RGYWQELIE++ WAH + +A +P AL
Sbjct: 642 NSLSVWAWMFLFGHLVWATGFMFLISXRGYWQELIETLAWAHERTPLANLIRWRDKPVAL 701
Query: 718 SIVQGRAVGVTHYLLGGIATTWAFFLA 744
S+VQ R VG+ H+ +G I T AF +A
Sbjct: 702 SVVQARLVGLAHFSVGYIFTYAAFLIA 728
>sp|P17155|PSAB_SYNP2 PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||S06902 photosystem I protein A2 - Synechococcus sp. (PCC 7002)
gb|AAA88634.1| (M18165) chlorophyll a/b apoprotein [Synechococcus sp.]
prf||1410329C P700 chlorophyll a 81.4kD protein [Synechococcus sp.]
Length = 733
Score = 626 bits (1596), Expect = e-178
Identities = 339/752 (45%), Positives = 447/752 (59%), Gaps = 72/752 (9%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF++H E+ + +KIF++HFG L+IIFLW SG F
Sbjct: 8 FSQDLAQDP-TTRRIWYGIATAHDFETHDGMTEENLYQKIFASHFGHLAIIFLWTSGTLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDV-GGGFRGIQIT-SGFFQIWRA 147
H A N+E W+ DP ++RP A +W P GQ ++ G + I SG + +
Sbjct: 67 HVAWQGNFEQWIKDPLNVRPIAHAIWDPHFGQGAIDAFTQAGASNPVNIAYSGVYHWFYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+T+ LY A+ L+ ++L LFAGW H K P LAWF+D ES LNHHLAGL G+
Sbjct: 127 IGMTTNADLYQGAVFLLILSSLFLFAGWLHLQPKFRPSLAWFKDAESRLNHHLAGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+W GH VHV++P ++ ++ G D PH G PFFT NW
Sbjct: 187 SLAWTGHLVHVAIPESRGVHVGWDNFLSVKPHP----------------AGLMPFFTGNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
YA LTF GG P T LWLTDIAHHHLAIA+LF+IAGHM
Sbjct: 231 GVYAQNPDTAGHVFGTSQGAGTAILTFLGGFHPQTESLWLTDIAHHHLAIAVLFIIAGHM 290
Query: 309 YRTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAH 366
YRTN IGH IK+IL AH+ P G GHKGLY+ + S H QL + LA LG +T +VA
Sbjct: 291 YRTNVRIGHSIKEILNAHQPPSGKLGAGHKGLYDTVNNSLHFQLGLALASLGVITSLVAQ 350
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMYS+P Y ++A DY TQ +L+THH +I GFL+VGA AH AIF VRDYDP +++L R
Sbjct: 351 HMYSLPSYAFIAKDYTTQAALYTHHQYIAGFLMVGAFAHGAIFFVRDYDPEANKDNVLYR 410
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
+L H++A+ISHL+WV +FLGFH+ GLY+HND + A G P+ I ++P+FAQ++Q
Sbjct: 411 MLEHKEALISHLSWVSLFLGFHTLGLYVHNDVVVAFGTPEKQ-----ILIEPVFAQFVQA 465
Query: 487 THALAPG------ITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 540
A A + S + G L A+ + L + +G DFLVHH A
Sbjct: 466 ASGKALYGFDVLLSNADSIASNASAAYLPGWLDAINSGSNSLFLNIGPGDFLVHHAIALG 525
Query: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
+H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N
Sbjct: 526 LHTTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAMFWMLNT 585
Query: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
+ + F++ WK +W QG V F + S + GW RD+LWA ++Q+I Y
Sbjct: 586 LGWLTFYWHWK-HLGIW-----QGNV-----AQFNEKSTYLMGWFRDYLWANSAQLINGY 634
Query: 661 G----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAT---- 712
++LS + FL H VWA MFL S RGYWQELIE+IVWAH + +A
Sbjct: 635 NPYGVNNLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETIVWAHERTPLANIVRWKD 694
Query: 713 QPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+ H+ +G + T AF +A
Sbjct: 695 KPVALSIVQARLVGLAHFTVGYVLTYAAFLIA 726
>gi|7524711 PSI P700 apoprotein A2 [Pinus thunbergii]
sp|P41640|PSAB_PINTH PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||T07544 photosystem I protein A2 - Japanese black pine chloroplast
dbj|BAA04420.1| (D17510) PSI P700 apoprotein A2 [Pinus thunbergii]
Length = 734
Score = 625 bits (1595), Expect = e-178
Identities = 339/738 (45%), Positives = 446/738 (59%), Gaps = 43/738 (5%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AH F+SH E+ + KIF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAQDP-TTRRIWFGIATAHHFESHDDITEERLYHKIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+EAW+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFEAWVRDPLHVRPIAHAIWDPHFGQPAIEAFTRGGAPGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L + + L AG H K P ++WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNEDLYAGALFLLFLSVIFLIAGRLHLQPKWRPSVSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAEGATPFFTL 264
SL+W GH VHV++P ++ ++ D LPH L Q LY + ++ F
Sbjct: 187 SLAWTGHLVHVAIPESRGVHVRWDNFLDVLPHPEGLEPLFTGQWNLYAQNPDSSSHLFGT 246
Query: 265 NWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIKDILE 324
+ LTF GG P T LWLTD+AHHHLAIA++F IAGHMYRTN+GIGH ++DILE
Sbjct: 247 SQGAGTAILTFLGGFHPQTQSLWLTDMAHHHLAIALVFSIAGHMYRTNFGIGHSMEDILE 306
Query: 325 AHKGP--FTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLATDYG 382
AH P G+GHKGLY + S H QL + LA LG +T +VA HMYS+PPY ++A D+
Sbjct: 307 AHVPPGGLLGRGHKGLYNTINNSLHFQLGLALASLGVITSLVAQHMYSLPPYAFIAQDFT 366
Query: 383 TQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWVC 442
TQ +L+THH +I GF++ GA AH AIF++RDY+P +++L R+L ++AIISHL+WV
Sbjct: 367 TQAALYTHHQYIAGFIMTGAFAHGAIFLIRDYNPEQNKDNVLARMLEQKEAIISHLSWVS 426
Query: 443 IFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------LAPGI 494
+ LGFH+ GLY+HND M A G P+ I ++PIFAQWIQ+ H L
Sbjct: 427 LLLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPIFAQWIQSAHGKTLYGFDILLSST 481
Query: 495 TAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLF 554
+ P G S+ W G L A+ + L +G DFLVHH A +H T LIL+KG L
Sbjct: 482 SGPSFDAGKSI-WLPGWLNAINDNNNSLFSTIGPGDFLVHHAIALGLHTTTLILVKGALD 540
Query: 555 ARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKMQS 614
AR SRL+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N I V F++ WK
Sbjct: 541 ARGSRLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLNTIGWVTFYWHWK-HI 599
Query: 615 DVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYG----SSLSAYGLF 670
+W QG V F +SS + GW RD+LW +SQ+I Y +SLS +
Sbjct: 600 TLW-----QGNV-----AQFNESSTYLMGWSRDYLWLNSSQLINGYNPFGMNSLSVWAWM 649
Query: 671 FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAT----QPRALSIVQGRAVG 726
FL H VWA MFL S RGYWQELIE++ WAH + +A +P ALSIVQ R VG
Sbjct: 650 FLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLVRWRDKPVALSIVQARLVG 709
Query: 727 VTHYLLGGIATTWAFFLA 744
+ H+ +G I T AF +A
Sbjct: 710 LAHFSVGYIFTYAAFLIA 727
>gi|11467191 PSI P700 apoprotein A2 [Zea mays]
sp|P04967|PSAB_MAIZE PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||S58551 photosystem I protein A2 - maize chloroplast
emb|CAA60285.1| (X86563) PSI P700 apoprotein A2 [Zea mays]
Length = 735
Score = 625 bits (1595), Expect = e-178
Identities = 340/747 (45%), Positives = 443/747 (58%), Gaps = 60/747 (8%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + + IF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAQDP-TTRRIWFGIATAHDFESHDDITEERLYQNIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E+W+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFESWIQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGAAGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L + L L GW H K P L+WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNEDLYTGALFLLFLSTLSLIGGWLHLQPKWKPSLSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLP---------INQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFA 255
SL+W GH VHV++P N FL+ LP+ L L Q LY
Sbjct: 187 SLAWTGHLVHVAIPGSSRGEYVRWNNFLDV--------LPYPQGLGPLLTGQWNLYAQNP 238
Query: 256 EGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGI 315
+ + F LT GG P T LWLTDIAHHHLAIA +FLIAGHMYRTN+GI
Sbjct: 239 DSSNHLFGTTQGAGTAILTLLGGFHPQTQSLWLTDIAHHHLAIAFIFLIAGHMYRTNFGI 298
Query: 316 GHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPP 373
GH IKD+LEAH P G+GHKGLY+ + S H QL + LA LG +T +VA HMYS+P
Sbjct: 299 GHSIKDLLEAHTPPGGRLGRGHKGLYDTINNSIHFQLGLALASLGVITSLVAQHMYSLPA 358
Query: 374 YPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDA 433
Y ++A D+ TQ +L+THH +I GF++ GA AH AIF +RDY+P +++L R+L H++A
Sbjct: 359 YAFIAQDFTTQAALYTHHQYIAGFIMTGAFAHGAIFFIRDYNPEQNEDNVLARMLDHKEA 418
Query: 434 IISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA---- 489
IISHL+W +FLGFH+ G Y+HND M A G P+ I ++PIFAQWIQ+ H
Sbjct: 419 IISHLSWASLFLGFHTLGPYVHNDVMLAFGTPEKQ-----ILIEPIFAQWIQSAHGKTTY 473
Query: 490 ----LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTV 545
L P G ++ W G L AV L + +G DFLVHH A +H T
Sbjct: 474 GFDILLSSTNGPTFNAGRNI-WLPGWLNAVNENSNSLFLTIGPGDFLVHHAIALGLHTTT 532
Query: 546 LILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVI 605
LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N I V
Sbjct: 533 LILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLNTIGWVT 592
Query: 606 FHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYG---- 661
F++ WK + G++S F +SS + GWLRD+LW +SQ+I Y
Sbjct: 593 FYWHWKHITLWQGNVS-----------QFNESSTYLMGWLRDYLWLNSSQLINGYNPFGM 641
Query: 662 SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----ATQPRAL 717
+SLS + FL H VWA MFL S RGYWQELIE++ WAH + +A +P AL
Sbjct: 642 NSLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLIRWRDKPVAL 701
Query: 718 SIVQGRAVGVTHYLLGGIATTWAFFLA 744
S+VQ R VG+ H+ +G I T AF +A
Sbjct: 702 SVVQARLVGLAHFSVGYIFTYAAFLIA 728
>sp|P31088|PSAB_ANAVA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||I39616 photosystem I protein A2 - Anabaena variabilis
gb|AAA18489.1| (L26326) PsaB [Anabaena variabilis]
Length = 741
Score = 624 bits (1591), Expect = e-177
Identities = 337/761 (44%), Positives = 446/761 (58%), Gaps = 82/761 (10%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A +DF+SH E+ + +KIF+ HFG L+IIFLW S + F
Sbjct: 8 FSQDLAQDP-TTRRIWYAMAMGNDFESHDGMTEENLYQKIFATHFGHLAIIFLWASSLLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E W+ DP H+RP A +W P G+ + G G + I SG + W
Sbjct: 67 HVAWQGNFEQWIKDPLHVRPIAHAIWDPHFGKPAIEAFTQAGANGPVNIAYSGVYHWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + +LY ++ L+FA+L LFAGW H K P LAWF+ ES LNHHLAGL G+
Sbjct: 127 IGMRTNTELYTGSVFLLLFASLFLFAGWLHLQPKFRPSLAWFKSAESRLNHHLAGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+WAGH +HV++P ++ + G D PH G PFFT NW
Sbjct: 187 SLAWAGHLIHVAIPESRGQHVGWDNFLTTAPHP----------------AGLQPFFTGNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
YA LTF GG P T LWLTDIAHHHLAIA+LF++AGHM
Sbjct: 231 GVYAQNPDTAGHIFSTSQGSGTAILTFLGGFHPQTESLWLTDIAHHHLAIAVLFIVAGHM 290
Query: 309 YRTNWGIGHGIKDILEAH-------KGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLT 361
YRTN+GIGH IK+++ A +GPF H+G+Y+ S H QL +LA LG +T
Sbjct: 291 YRTNFGIGHSIKEMMNAKTFFGKPVEGPFN-MPHQGIYDTYNNSLHFQLGWHLACLGVVT 349
Query: 362 IIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYN 421
VA HMYS+P Y ++A DY TQ +L+THH +I FL+VGA AH AIF+VRDYDP
Sbjct: 350 SWVAQHMYSLPSYAFIAKDYTTQAALYTHHQYIAIFLMVGAFAHGAIFLVRDYDPEQNKG 409
Query: 422 DLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFA 481
++L+RVL+H++AIISHL+WV +FLGFH+ GLY+HND + A G P+ I ++P+FA
Sbjct: 410 NVLERVLQHKEAIISHLSWVSLFLGFHTLGLYVHNDVVVAFGTPEKQ-----ILIEPVFA 464
Query: 482 QWIQNTHALA----------PGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADF 531
Q+IQ H P A A + W G L A+ + L + +G DF
Sbjct: 465 QFIQAAHGKVLYGLDTLLSNPDSVAYTAYPNYANVWLPGWLDAINSGTNSLFLTIGPGDF 524
Query: 532 LVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVF 591
LVHH A +H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +
Sbjct: 525 LVHHAIALGLHTTTLILVKGALDARGSKLMPDKKDFGYAFPCDGPGRGGTCDISAWDSFY 584
Query: 592 LGLFWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWA 651
L LFW N + V F++ WK +W QG V F ++S + GW RD+LWA
Sbjct: 585 LSLFWALNTVGWVTFYWHWK-HLGIW-----QGNV-----AQFNENSTYLMGWFRDYLWA 633
Query: 652 QASQVIQSYG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLK 707
++Q+I Y ++LS + FL H VWA MFL S RGYWQELIE++VWAH +
Sbjct: 634 NSAQLINGYNPYGVNNLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLVWAHERTP 693
Query: 708 VAP----ATQPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+A +P ALSIVQ R VG+ H+ +G + T AF +A
Sbjct: 694 IANLIRWKDKPVALSIVQARVVGLAHFTVGYVLTYAAFLIA 734
>gi|11467294 PsaB subunit of photosystem I reaction center [Cyanophora paradoxa]
sp|P48113|PSAB_CYAPA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||T06839 probable photosystem I protein A2 - Cyanophora paradoxa cyanelle
gb|AAA81182.1| (U30821) PsaB subunit of photosystem I reaction center [Cyanophora
paradoxa]
Length = 737
Score = 623 bits (1589), Expect = e-177
Identities = 343/758 (45%), Positives = 447/758 (58%), Gaps = 82/758 (10%)
Query: 33 SRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYFH 91
S+ +A+ P TT IW A A+DF+++ E+ + +KIF++HFG L+IIFLW SG FH
Sbjct: 9 SQALAQDP-TTRRIWYGIATANDFETNDGITEENLYQKIFASHFGHLAIIFLWTSGNLFH 67
Query: 92 GARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNG-DVGGGFRGIQIT-SGFFQIWRAS 148
A N+E W+ DP + RP A + P GQ + G + I+ SG +Q W
Sbjct: 68 VAWQGNFEQWVKDPLNTRPIAHAISDPHFGQRAIEAFSQAGASSPVNISYSGVYQWWYTQ 127
Query: 149 GITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGS 207
G+ + +LY AI L+ +AL LFAGW H K P L+WF++ ES LNHHL GL G S
Sbjct: 128 GMRTNEELYNGAIFLLILSALSLFAGWLHLQPKFRPNLSWFKNAESRLNHHLGGLFGTSS 187
Query: 208 LSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWA 267
L+W GH VHV++P ++ + G D PH G PFFT NW
Sbjct: 188 LAWTGHIVHVAIPESRGQHVGWDNFLQVAPHP----------------AGLQPFFTGNWG 231
Query: 268 KYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMY 309
Y + LTF GG P T LWLTDIAHHHLAIA+LF++AGHMY
Sbjct: 232 VYTENPDTANHVFGSSDGAGTAILTFLGGFHPQTQSLWLTDIAHHHLAIAVLFIVAGHMY 291
Query: 310 RTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHH 367
RTN+GIGH IK+IL H+ P G GH GLY+ + S H QL + LA LG +T +VA H
Sbjct: 292 RTNFGIGHSIKEILNGHRPPGGRLGAGHVGLYDTVNNSLHFQLGLALAALGVITSLVAQH 351
Query: 368 MYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRV 427
MYS+PPY YLA D+ TQ +L+THH +I GFL+VGA AH AIF+VRDYD N++L R+
Sbjct: 352 MYSIPPYAYLARDFTTQAALYTHHQYIAGFLMVGAFAHGAIFLVRDYDAEQNKNNVLARI 411
Query: 428 LRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNT 487
+ H++AIISHL+WV +FLGFH+ GLY+HND + A G P+ I ++P+FAQWIQ+
Sbjct: 412 IDHKEAIISHLSWVSLFLGFHTLGLYVHNDVVQAFGTPEKQ-----ILIEPVFAQWIQSV 466
Query: 488 HAL-----------APGIT--APGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVH 534
H A IT APG+ W G L A+ + L + +G DFLVH
Sbjct: 467 HGKSLYGFEVLLNNADSITRVAPGS---AQPIWLPGWLDAINSGNNSLFLTIGPGDFLVH 523
Query: 535 HIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGL 594
H A +H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +
Sbjct: 524 HAIALGLHTTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAV 583
Query: 595 FWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQAS 654
FWM N I F++ WK VW QG V F +SS + GW RD+LW +S
Sbjct: 584 FWMLNTIGWTTFYWHWK-HLGVW-----QGNV-----AQFNESSTYLMGWFRDYLWLNSS 632
Query: 655 QVIQSYG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP 710
Q+I Y ++LS + FL H +WA MFL S RGYWQELIE++VWAH + +A
Sbjct: 633 QLINGYNPFGMNNLSVWAWMFLFGHLIWATGFMFLISWRGYWQELIETLVWAHERTPLAN 692
Query: 711 AT----QPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+ H+ +G I T AF +A
Sbjct: 693 LVRWKDKPVALSIVQARLVGLAHFAVGYIVTYAAFLIA 730
>emb|CAB67138.1| (AJ271079) PSI P700 apoprotein A2 [Oenothera elata subsp. hookeri]
Length = 734
Score = 621 bits (1583), Expect = e-176
Identities = 338/738 (45%), Positives = 443/738 (59%), Gaps = 43/738 (5%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + + IF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAQDP-TTRRIWFGIATAHDFESHDDITEERLHQNIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+EAW+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFEAWVQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGALGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L +A+ L AGW H K P ++WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNEDLYTGALFLLFLSAISLIAGWLHLQPKWKPSVSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAEGATPFFTL 264
SL+W GH VHV++P ++ + + LPH L Q LY + ++ F
Sbjct: 187 SLAWTGHLVHVAIPGSRGESVRWNNFLDVLPHPEGLGPLFTGQWNLYAQNPDSSSHLFGT 246
Query: 265 NWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIKDILE 324
+ LT GG P T LWLTD+AHHHLAIA LFLIAGHMYRTN+GIGH IKD+LE
Sbjct: 247 SQGSGTAILTLLGGFHPQTQSLWLTDMAHHHLAIAFLFLIAGHMYRTNFGIGHSIKDLLE 306
Query: 325 AHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLATDYG 382
AH P G+GHKGLY+ + S H QL + LA LG +T +VA HMYS+P Y ++A D+
Sbjct: 307 AHTPPGGRLGRGHKGLYDTINNSIHFQLGLALASLGVITSLVAQHMYSLPAYAFIAQDFT 366
Query: 383 TQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWVC 442
TQ +L+THH +I GF++ GA AH AIF +RDY+P +++L R+L H +AIISHL+W
Sbjct: 367 TQAALYTHHQYIAGFIMTGAFAHGAIFFIRDYNPEQNEDNVLARMLDHIEAIISHLSWAS 426
Query: 443 IFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTH--------ALAPGI 494
+FLGFH+ GLY+HND M A G P+ I ++PIFAQW Q+ H L
Sbjct: 427 LFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPIFAQWRQSAHGKTSYGFDVLLSST 481
Query: 495 TAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLF 554
+P S+ W G L A+ L + +G DFLVHH A H T LIL+KG L
Sbjct: 482 NSPAFNAVRSI-WLPGWLNAINENSNSLFLTIGPGDFLVHHAIALGSHTTTLILVKGALD 540
Query: 555 ARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKMQS 614
AR S+L+PDK +LG+ FPCDGPGRGGTC +SAWD +L +FWM N I ++ WK
Sbjct: 541 ARGSKLMPDKKDLGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLNTIGWATLYWHWK-HI 599
Query: 615 DVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYG----SSLSAYGLF 670
+W QG V F +SS + GWLRD+LW +SQ+I + +SLS +
Sbjct: 600 TLW-----QGFV-----AQFNESSTYLMGWLRDYLWLNSSQLINGFNPFGLNSLSVWAWM 649
Query: 671 FLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----ATQPRALSIVQGRAVG 726
FL H VWA MFL S RGYWQELIE++ WAH + +A +P ALSIVQ R VG
Sbjct: 650 FLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLIRWRDKPVALSIVQARLVG 709
Query: 727 VTHYLLGGIATTWAFFLA 744
+ H+ +G I T AF +A
Sbjct: 710 LAHFSVGYIFTYAAFLIA 727
>sp|P05311|PSAB_PEA PHOTOSYSTEM I P700 CHLOROPHYLL A APOPROTEIN A2
pir||S00704 photosystem I protein A2 - garden pea chloroplast
emb|CAA29004.1| (X05423) P700 chlorophyll a-apoproteins 82 KD protein [Pisum
sativum]
Length = 734
Score = 615 bits (1569), Expect = e-175
Identities = 341/745 (45%), Positives = 439/745 (58%), Gaps = 57/745 (7%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ IA+ P TT IW A AHDF+SH E + + IF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGIAQDP-TTRRIWFGIATAHDFESHDDITEGRLYQNIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRGI--QITSGFFQIWRA 147
H A N+EAW+ DP H+RP A +W P GQ + GG G SG +Q W
Sbjct: 67 HVAWQGNFEAWVQDPFHVRPIAHAIWDPHFGQPAVEAFTRGGALGPVNNAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY AI L + + L AGW H K P ++WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNEDLYTGAIFLLFLSFISLLAGWLHLQPKWKPSVSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLP--------INQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAE 256
SL+WAGH VHV++P N FL+ LP+ L L Q LY
Sbjct: 187 SLAWAGHLVHVAIPGSRGEYVRWNNFLDV--------LPYPQGLGPLLTGQWNLYAQNPS 238
Query: 257 GATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIG 316
+ F LT GG P T LWLTD+AHHHLAIA LFLI G MYRTN+GIG
Sbjct: 239 SSNHLFGTTQGAGTAILTILGGFHPQTQSLWLTDVAHHHLAIAFLFLIGGLMYRTNFGIG 298
Query: 317 HGIKDILEAH--KGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPY 374
H IK ILEAH G G+GHKGLY+ + S H QL + LA LG +T +VA HMYS+P Y
Sbjct: 299 HSIKYILEAHIPPGGRLGRGHKGLYDTINNSIHFQLGLALASLGVITSLVAQHMYSLPAY 358
Query: 375 PYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAI 434
++A D+ TQ +L+THH +I GF++ GA AH IF +RDY+P +++L R+L H++AI
Sbjct: 359 AFIAQDFTTQAALYTHHQYIAGFIMTGAFAHGPIFFIRDYNPEQNADNVLARMLEHKEAI 418
Query: 435 ISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALAP-G 493
ISHL+W +FLGFH+ GLY+HND M A G P+ I ++PIFAQWIQ+ H G
Sbjct: 419 ISHLSWASLFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPIFAQWIQSAHGKTTYG 473
Query: 494 ITAP-GATTGTSLT-----WGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLI 547
P +T G +L W G L A+ L + +G DFLVHH A +H T LI
Sbjct: 474 FDIPLSSTNGPALNAGRNIWLPGWLNAINENSNSLFLTIGPGDFLVHHAIALGLHTTTLI 533
Query: 548 LLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFH 607
L+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N I V F+
Sbjct: 534 LVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDDFYLAVFWMLNTIGWVTFY 593
Query: 608 FSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYG----SS 663
+ WK + G++S F +SS + GWLRD+LW +SQ+I +S
Sbjct: 594 WHWKHITLWRGNVS-----------QFNESSTYLMGWLRDYLWLNSSQLINGITPLVCNS 642
Query: 664 LSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----ATQPRALSI 719
LS + FL H VWA MFL S RGYWQELIE++ WAH + +A +P ALSI
Sbjct: 643 LSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLIRWRDKPVALSI 702
Query: 720 VQGRAVGVTHYLLGGIATTWAFFLA 744
VQ R VG+ H+ +G I T AF +A
Sbjct: 703 VQARLVGLVHFSVGYIFTYAAFLIA 727
>prf||1303353B gene psaA2 [Pisum sativum]
Length = 734
Score = 614 bits (1565), Expect = e-174
Identities = 341/745 (45%), Positives = 440/745 (58%), Gaps = 57/745 (7%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ IA+ P TT IW A AHDF+SH E + + IF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGIAQDP-TTRRIWFGIATAHDFESHDDITEGRLYQNIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+EAW+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFEAWVQDPFHVRPIAHAIWDPHFGQPAVEAFTRGGALGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY AI L + + L AGW H K P ++WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNEDLYTGAIFLLFLSFISLLAGWLHLQPKWKPSVSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLP--------INQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAE 256
SL+WA H VHV++P N FL+ LP+ L L Q LY
Sbjct: 187 SLAWASHLVHVAIPGSRGEYVRWNNFLDV--------LPYPQGLGPLLTGQWNLYAQNPS 238
Query: 257 GATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIG 316
+ F LT GG P T LWLTD+AHHHLAIA LFLI G MYRTN+GIG
Sbjct: 239 SSNHLFGTTQGAGTAILTILGGFHPQTQSLWLTDVAHHHLAIAFLFLIGGLMYRTNFGIG 298
Query: 317 HGIKDILEAH--KGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPY 374
H IK ILEAH G G+GHKGLY+ + S H QL + LA LG +T +VA HMYS+P Y
Sbjct: 299 HSIKYILEAHIPPGGRLGRGHKGLYDTINNSIHFQLGLALASLGVITSLVAQHMYSLPAY 358
Query: 375 PYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAI 434
++A D+ TQ +L+THH +I GF++ GA AH IF +RDY+P +++L R+L H++AI
Sbjct: 359 AFIAQDFTTQAALYTHHQYIAGFIMTGAFAHGPIFFIRDYNPEQNADNVLARMLEHKEAI 418
Query: 435 ISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALAP-G 493
ISHL+W +FLGFH+ GLY+HND M A G P+ I ++PIFAQWIQ+ H G
Sbjct: 419 ISHLSWASLFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPIFAQWIQSAHGKTTYG 473
Query: 494 ITAP-GATTGTSLT-----WGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLI 547
P +T G +L W G L A+ L + +G DFLVHH A +H T LI
Sbjct: 474 FDIPLSSTNGPALNAGRNIWLPGWLNAINENSNSLFLTIGPGDFLVHHAIALGLHTTTLI 533
Query: 548 LLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFH 607
L+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N I V F+
Sbjct: 534 LVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDDFYLAVFWMLNTIGWVTFY 593
Query: 608 FSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYG----SS 663
+ WK + G++S F +SS + GWLRD+LW +SQ+I +S
Sbjct: 594 WHWKHITLWRGNVS-----------QFNESSTYLMGWLRDYLWLNSSQLINGITPLVCNS 642
Query: 664 LSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----ATQPRALSI 719
LS + FL H VWA MFL S RGYWQELIE++ WAH + +A +P ALSI
Sbjct: 643 LSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLIRWRDKPVALSI 702
Query: 720 VQGRAVGVTHYLLGGIATTWAFFLA 744
VQ R VG+ H+ +G I T AF +A
Sbjct: 703 VQARLVGLVHFSVGYIFTYAAFLIA 727
>emb|CAB64209.1| (AJ133190) photosystem I subunit PsaB [Synechococcus sp. WH7803]
Length = 738
Score = 613 bits (1564), Expect = e-174
Identities = 337/757 (44%), Positives = 447/757 (58%), Gaps = 77/757 (10%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + +K+FS HFG L+II LW+SG F
Sbjct: 8 FSQGLAQDP-TTRRIWYGIATAHDFESHDGMTEEKLYQKLFSTHFGHLAIIGLWVSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E W++DP H+RP A +W P GQ ++ G + I SG + +
Sbjct: 67 HIAWQGNFEQWVADPLHVRPIAHAIWDPHFGQGAIDAFTQAGASSPVNIAYSGLYHWFYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+T+ +LY +I ++ +A LFAGW H K P LAWF++ ES LNHHLA L G
Sbjct: 127 IGMTTNAELYQGSIFMMILSAWALFAGWLHLQPKFRPSLAWFKNAESRLNHHLAVLFGFS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
S++W GH VHV++P ++ + G D +PH G PFFT NW
Sbjct: 187 SIAWTGHLVHVAIPESRGQHVGWDNFLNVMPHP----------------AGLGPFFTGNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
YA LTF GG P T LWLTDIAHHHLAI ++F+IAGHM
Sbjct: 231 GVYAQNPDTMGQVFGSAEGSGTAILTFLGGFHPQTEALWLTDIAHHHLAIGVIFVIAGHM 290
Query: 309 YRTNWGIGHGIKDILEAHKGPF-----TGQGHKGLYEILTTSWHAQLSINLAMLGSLTII 363
YRTN+GIGH I++ILEAH P G GHKGLY+ + S H QL + LA LG +T +
Sbjct: 291 YRTNFGIGHSIREILEAHNPPQGTPGDLGAGHKGLYDTINNSLHFQLGLALASLGVVTSL 350
Query: 364 VAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDL 423
VA HMY+MP Y ++A DY TQ +L+THH +I FL+ GA AH AIF +RDYDP +++
Sbjct: 351 VAQHMYAMPSYAFIAKDYTTQAALYTHHQYIAIFLMCGAFAHGAIFFIRDYDPEANKDNV 410
Query: 424 LDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQW 483
L R+L H++AIISHL+WV +FLGFH+ GLY+HND + A G P+ I ++P+FAQ+
Sbjct: 411 LARMLEHKEAIISHLSWVSLFLGFHTLGLYVHNDVVVAFGTPEKQ-----ILVEPVFAQF 465
Query: 484 IQNTHALA-PGITAPGATTG-----TSLTWGGGDLVAVGNKVAL--LPIPLGTADFLVHH 535
+Q A G A TG + + GG + A+ L +P+G DFLVHH
Sbjct: 466 VQAASGKAIYGFDVLLANTGGVAANANAAYMGGWMDAINGVRGSNDLFLPIGPGDFLVHH 525
Query: 536 IHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLF 595
A +H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +F
Sbjct: 526 AIALGLHTTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVF 585
Query: 596 WMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQ 655
W N + V F++ WK + +W QG V F +SS + GW RD+LW +SQ
Sbjct: 586 WALNTVGWVTFYWHWKHLA-IW-----QGNV-----AQFNESSTYLMGWFRDYLWLNSSQ 634
Query: 656 VIQSY----GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPA 711
+I Y ++L+ + FL H VWA MFL S RGYWQELIE+IVWAH + +A
Sbjct: 635 LINGYNPFGSNNLAVWAWMFLFGHLVWATGFMFLISWRGYWQELIETIVWAHQRTPLANL 694
Query: 712 T----QPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+P ALSIVQ R VG+ H+ +G I T AF +A
Sbjct: 695 VGWRDKPVALSIVQARVVGLAHFTIGYILTYAAFLIA 731
>emb|CAB64201.1| (AJ133191) photosystem I subunit PsaB [Prochlorococcus sp.
CCMP1378]
Length = 742
Score = 593 bits (1512), Expect = e-168
Identities = 330/762 (43%), Positives = 441/762 (57%), Gaps = 83/762 (10%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
F++ +A+ P TT IW A AHDF+SH E+ + +K+FS HFG L+II LW++G F
Sbjct: 8 FNQGLAQDP-TTRRIWYGIATAHDFESHDGMTEEKLYQKLFSTHFGHLAIIALWVAGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E ++ DPTH+RP A +W P G I G G + I SG + W
Sbjct: 67 HIAWQGNFEQFVLDPTHVRPIAHAIWDPHFGSGITEAMTQAGASGPVNIAYSGLYHWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + QL+ +I + A LFAGW H K P LAWF++ ES LNHHLA L G
Sbjct: 127 IGMRTNEQLFQASIFMSILACWTLFAGWLHLQPKFRPSLAWFKNNESRLNHHLAVLFGFS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
S++W GH VHV++P ++ ++ G D LPH G PFFTLNW
Sbjct: 187 SIAWTGHLVHVAIPESRGIHVGWDNWLTVLPHP----------------AGLAPFFTLNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
YA TF GGL P + LWLTDIAHHH+AI +F+IAGHM
Sbjct: 231 GAYAQNPDTLEQVFGTSEGAGTAIFTFLGGLHPQSEALWLTDIAHHHIAIGTVFIIAGHM 290
Query: 309 YRTNWGIGHGIKDILEAH---------KGPFTGQGHKGLYEILTTSWHAQLSINLAMLGS 359
YR +GIGH +K+I EAH KG F G H G+YE +T S H QL + LA LG
Sbjct: 291 YRNTFGIGHSLKEITEAHNTRHPLDPHKGSF-GINHDGIYETVTNSLHFQLGLALAALGV 349
Query: 360 LTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTR 419
T +VA HM ++P Y ++A DY TQ +L+THH +I FL+VGA AH AIF VRDYDP
Sbjct: 350 ATSLVAQHMGALPSYAFIARDYTTQSALYTHHQYIAMFLMVGAFAHGAIFFVRDYDPELN 409
Query: 420 YNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPI 479
+++L RVL ++A+ISHL+WV + LGFH+ G+Y+HND + A G P+ I ++P+
Sbjct: 410 KDNVLARVLGTKEALISHLSWVTMILGFHTLGIYVHNDVVVAFGNPEKQ-----ILIEPV 464
Query: 480 FAQWIQ--------NTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLP-IPLGTAD 530
FAQ++Q +AL T+ + SL + + + AL +P+G AD
Sbjct: 465 FAQFVQAAQGKMMYGFNALLSDPTSSASLAANSLPGNHYWMDLINRQDALSAFLPIGPAD 524
Query: 531 FLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHV 590
FLVHH A +H T LIL+KG L AR ++LIPDK +LG+ FPCDGPGRGGTC S+WD +
Sbjct: 525 FLVHHAIALGLHTTALILIKGALDARGTKLIPDKKDLGYAFPCDGPGRGGTCDSSSWDAM 584
Query: 591 FLGLFWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLW 650
+L +FW N I+ V F++ WK + +W QG V F +S + GW RD+LW
Sbjct: 585 YLAMFWALNLIAWVTFYWHWKHLA-IW-----QGNV-----AQFNESGTYLMGWFRDYLW 633
Query: 651 AQASQVIQSYG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKL 706
++Q+I Y +SLS + FL H VWA MFL S RGYWQELIE++VWAH +
Sbjct: 634 LNSAQLINGYNPFGVNSLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLVWAHQRT 693
Query: 707 KVAPAT----QPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+A +P ALSIVQ R VG+ H+ +G I T AF +A
Sbjct: 694 PIANLVGWRDKPVALSIVQARLVGLAHFTIGNILTFGAFVIA 735
>emb|CAB64199.1| (AJ133192) photosystem I subunit PsaB [Prochlorococcus marinus]
Length = 747
Score = 585 bits (1493), Expect = e-166
Identities = 324/766 (42%), Positives = 437/766 (56%), Gaps = 86/766 (11%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
F++ +A+ P TT IW A AHDF+SH E+ + +K+F+ HFG L+II LW++G F
Sbjct: 8 FNQGLAQDP-TTRRIWYGIATAHDFESHDGMTEEQLYQKLFATHFGHLAIIGLWVAGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNG-DVGGGFRGIQIT-SGFFQIWRA 147
H A N+E W++DP HIRP A +W P GQ I + G + I SG + W
Sbjct: 67 HIAWQGNFEQWVTDPLHIRPIAHAIWDPHFGQGITDALTQAGATSPVNIAYSGLYHWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + QL+ AI + +LFAGW H K P LAWF++ E+ LNHHLA L G
Sbjct: 127 IGMRTNAQLFQGAIFLNILVCWLLFAGWLHLQPKFRPSLAWFKNAEAQLNHHLAVLFGFS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
S++W GH VHV++P ++ + G D LPH +G PFF+LNW
Sbjct: 187 SIAWTGHLVHVAIPESRGQHVGWDNWLSVLPHP----------------QGLAPFFSLNW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
YA TF GGL P + LWLTDIAHHHLAI ++F+IAGHM
Sbjct: 231 GAYAQNPDSLDAVFGTSQGAGTAIFTFLGGLHPQSESLWLTDIAHHHLAIGVMFIIAGHM 290
Query: 309 YRTNWGIGHGIKDILEAHK-------------GPFTGQGHKGLYEILTTSWHAQLSINLA 355
YR +GIGH +K+I EAH G G H G++E + S H QL + LA
Sbjct: 291 YRNTFGIGHTLKEITEAHNTDNPNDPHTTAKDGRHFGVNHNGIFETVNNSLHFQLGLALA 350
Query: 356 MLGSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYD 415
LG+ +VA HM ++P Y ++A DY TQ +L+THH +I FL+VGA +H AIF VRDYD
Sbjct: 351 SLGAACSLVAQHMGALPSYAFIARDYTTQSALYTHHQYIAMFLMVGAFSHGAIFFVRDYD 410
Query: 416 PTTRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQ 475
P ++L RVL ++A+ISHL+WV + LGFH+ G+Y+HND + A G P+ I
Sbjct: 411 PELNKGNVLARVLETKEALISHLSWVTMLLGFHTLGIYVHNDVVVAFGNPEKQ-----IL 465
Query: 476 LQPIFAQWIQN-THALAPGITAPGATTGTSLTWGGGDLVA-------VGNKVALLP-IPL 526
++P+FAQ IQ + + GI A A +S T + + + AL +P+
Sbjct: 466 VEPVFAQAIQAFSGKVMYGINALLANANSSATLAANSMPGNHYWMDMINRQDALTNFLPI 525
Query: 527 GTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSA 586
G ADFLVHH A +H T LIL+KG L AR S+LIPDK + G+ FPCDGPGRGGTC SA
Sbjct: 526 GPADFLVHHAIALGLHTTALILIKGALDARGSKLIPDKKDFGYAFPCDGPGRGGTCDSSA 585
Query: 587 WDHVFLGLFWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLR 646
WD +L +FW N I+ + F++ WK + +W QG V F +S + GW R
Sbjct: 586 WDATYLAMFWALNTIAWITFYWHWKHLA-IW-----QGNV-----AQFNESGTYLMGWFR 634
Query: 647 DFLWAQASQVIQSYG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWA 702
D+LW +SQ+I Y ++LS + FL H +WA MFL S RGYWQELIE++VWA
Sbjct: 635 DYLWLNSSQLINGYNPFGVNALSPWAWMFLFGHLIWATGFMFLISWRGYWQELIETLVWA 694
Query: 703 HNKLKVAPAT----QPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
H + +A +P ALSIVQ R VG+TH+ +G T AF +A
Sbjct: 695 HQRTPIANLVGWRDKPVALSIVQARLVGLTHFTVGNFVTFGAFVIA 740
>dbj|BAA83447.1| (AB013671) photosystem I P700 apoprotein A2 [Sphagnum fallax]
Length = 591
Score = 540 bits (1377), Expect = e-152
Identities = 281/590 (47%), Positives = 366/590 (61%), Gaps = 24/590 (4%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS +++ P TT IW A AHDF+SH E+ + +KIF++HFGQL+IIFLW SG F
Sbjct: 8 FSXGLSQDP-TTRRIWFGIATAHDFESHDDMTEERLYQKIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+EAW+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFEAWVQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L +A+ L AGW H K P ++WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRNNQDLYTGALFLLFLSAIFLIAGWLHLQPKWKPSISWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAEGATPFFTL 264
SL+W GH VHV++P ++ + LPH L Q +Y + + F
Sbjct: 187 SLAWTGHLVHVAIPESRGEHVRWGNLLTALPHPQGLGPFFAGQWNIYAQNPDSNSHLFGT 246
Query: 265 NWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIKDILE 324
+ LTF GG P T LWLTDIAHHHLAIA++F+IAGHMYRTN+GIGH IK+ILE
Sbjct: 247 SQGSGTAILTFLGGFHPQTQSLWLTDIAHHHLAIAVVFIIAGHMYRTNFGIGHSIKEILE 306
Query: 325 AHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLATDYG 382
AH P G+GHKGLY+ + S H QL + LA LG +T +VA HMYS+PPY +LA D+
Sbjct: 307 AHTPPGGRLGRGHKGLYDTINNSLHFQLGLALASLGVITSLVAQHMYSLPPYAFLAQDFT 366
Query: 383 TQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWVC 442
TQ +L+THH +I GF++ GA AH AIF +RDY+P N++L R+L H++AIISHL+W
Sbjct: 367 TQAALYTHHQYIAGFIMTGAFAHGAIFFIRDYNPEQNKNNVLARMLEHKEAIISHLSWAS 426
Query: 443 IFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTH--------ALAPGI 494
+FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ+ H L
Sbjct: 427 LFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPVFAQWIQSAHGKALYGFDVLLSSA 481
Query: 495 TAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLF 554
+P G SL W G L A+ N L + +G DFLVHH A +H T LIL+KG L
Sbjct: 482 DSPAFNAGQSL-WLPGWLDAINNNSNSLFLTIGPGDFLVHHAIALGLHTTTLILVKGALD 540
Query: 555 ARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVV 604
AR S+L+PDK G+ FPCDGPGRGGTC +SAWD +L +FWM N I V
Sbjct: 541 ARGSKLMPDKKEFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLNTIGWV 590
>dbj|BAA83457.1| (AB013681) photosystem I P700 apoprotein A2 [Adiantum
capillus-veneris]
Length = 586
Score = 527 bits (1343), Expect = e-148
Identities = 274/585 (46%), Positives = 361/585 (60%), Gaps = 24/585 (4%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +++ P TT IW A AHDF+SH E+ + +KIF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLSQDP-TTRRIWFGIATAHDFESHDDTTEERLYQKIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E W+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFETWVQDPLHVRPIAHAIWDPHFGQPAVEAYTRGGSAGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY AI L AAL L A W H K P ++WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRNNEDLYTGAIFLLAVAALFLLASWLHLQPKWKPSISWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAEGATPFFTL 264
SL+W GH +HV++P + + + PH L Q +Y + T F
Sbjct: 187 SLAWTGHLIHVAIPEARGGHVRWNDLLTTAPHPQGLGPFFSGQWSVYSQNPDSNTHLFNS 246
Query: 265 NWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIKDILE 324
N TF GG P T LWLTD+AHHHLAIA++F+IAGHMYRTN+GIGH +K+IL+
Sbjct: 247 NQGAGTAIPTFLGGFHPQTQSLWLTDMAHHHLAIAVVFIIAGHMYRTNFGIGHSMKEILD 306
Query: 325 AH--KGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLATDYG 382
AH G G+GH+GLY+ + S H QL + LA LG +T +VA HMYS+P Y +LA DY
Sbjct: 307 AHIPPGGKLGRGHRGLYDTVNNSLHFQLGLALAALGVITSLVAQHMYSLPAYAFLAQDYT 366
Query: 383 TQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWVC 442
TQ +L+THH +I GF++ GA AH AIF +RDYDP +++L R+L H++AIISHL+W
Sbjct: 367 TQAALYTHHQYIAGFIMAGAFAHGAIFFLRDYDPEQNRDNVLARMLEHKEAIISHLSWAS 426
Query: 443 IFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTH--------ALAPGI 494
+FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ+ H L +
Sbjct: 427 LFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPVFAQWIQSAHGKVSYGFDVLLSSV 481
Query: 495 TAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLF 554
+P + G L W G L AV + L + +G DFLVHH A +H T LIL+KG L
Sbjct: 482 DSPASNAGRGL-WLPGWLDAVNSSNNSLFLTIGPGDFLVHHAIALGLHTTTLILVKGALD 540
Query: 555 ARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN 599
AR S+L+PDK G+ FPCDGPGRGGTC +SAWD +L +FWM N
Sbjct: 541 ARGSKLMPDKKEFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLN 585
>dbj|BAA83452.1| (AB013676) photosystem I P700 apoprotein A2 [Haplomitrium mnioides]
Length = 604
Score = 524 bits (1334), Expect = e-147
Identities = 272/582 (46%), Positives = 360/582 (61%), Gaps = 24/582 (4%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ + + P TT IW A AH F+SH E+ + ++IF++HFGQL+IIFLW SG F
Sbjct: 30 FSQGLXQXP-TTRRIWFGXATAHXFESHDDITEERLYQRIFASHFGQLAIIFLWTSGNLF 88
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+EAW DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 89 HVAWQGNFEAWGQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIAYSGVYQWWYT 148
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ ++ +++ L AGW H K P ++WF++ ES LNHHL+GL G+
Sbjct: 149 IGLRTNQDLYLGALFLVICSSIFLIAGWLHLQPKWKPSVSWFKNAESRLNHHLSGLFGVS 208
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAEGATPFFTL 264
SL+W GH VHV++P ++ + D PH L Q +Y + + F
Sbjct: 209 SLAWTGHLVHVAIPESRGEHVRWDTLLSTSPHPQGLGPFFAGQWNVYSQNVDSSDHLFGT 268
Query: 265 NWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIKDILE 324
+ LTF GG P T GLWLTDIAHHHLAIA++F+IAGHMYRTN+GIGH IK+ILE
Sbjct: 269 SRGAGTAILTFLGGFHPQTQGLWLTDIAHHHLAIAVVFIIAGHMYRTNFGIGHSIKEILE 328
Query: 325 AHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLATDYG 382
AH P G+GHKGLY+ L S H QL + LA LG +T +VA HMYS+PPY +LA D+
Sbjct: 329 AHTPPGGRLGRGHKGLYDTLNNSLHLQLGLALASLGVITSLVAQHMYSLPPYAFLAQDFT 388
Query: 383 TQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWVC 442
TQ + +THH +I GF++ GA AH AIF +RDY+P +++L R+L H++AIISHL+W
Sbjct: 389 TQAASYTHHQYIAGFIMTGAFAHGAIFFIRDYNPDQNKDNVLARMLEHKEAIISHLSWAS 448
Query: 443 IFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTH--------ALAPGI 494
+FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ+ H L P
Sbjct: 449 LFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPVFAQWIQSAHGKALYGFDVLLPSP 503
Query: 495 TAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLF 554
P G SL W G L A+ + L + +G DFLVHH A +H T LIL+KG L
Sbjct: 504 DGPAFNAGRSL-WLPGWLDAINSGSNSLFLTIGPGDFLVHHAIALGLHTTTLILVKGALD 562
Query: 555 ARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFW 596
AR S+L+PDK G+ FPCDGPGRGGTC +SAWD +L + W
Sbjct: 563 ARGSKLMPDKKEFGYSFPCDGPGRGGTCDISAWDAFYLAVXW 604
>gb|AAK08971.1| (AY026898) photosystem I P700 protein subunit B [Prochlorothrix
hollandica]
Length = 661
Score = 516 bits (1314), Expect = e-145
Identities = 289/690 (41%), Positives = 389/690 (55%), Gaps = 80/690 (11%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+ H E+ + +KIF++HFG L+IIFLW SG F
Sbjct: 8 FSQDLAQDP-TTRRIWYGIATAHDFEMHDGMTEENLYQKIFASHFGHLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFR-GIQIT-SGFFQIWRA 147
H A N+ W++DP + RP A +W P G+ + + I SG + W
Sbjct: 67 HVAWQGNFAQWVADPLNTRPIAHAIWDPHFGEAAIEAFTQSDASCAVNIAYSGVYHWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY +I +V AALMLFAGW H K P LAWF++ ES LNHHLAGL G+
Sbjct: 127 IGMRTAADLYQGSIFLMVLAALMLFAGWLHLQPKFRPSLAWFKNAESRLNHHLAGLFGVS 186
Query: 207 SLSWAGHQVHVSLPIN--QFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTL 264
SL+WAGH +HV++P + + +N G P P G PFF+
Sbjct: 187 SLAWAGHLIHVAIPASRGETVNWGNFMSTPPHP------------------AGLAPFFSG 228
Query: 265 NWAKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
NW+ YA LTF GG P T +WLTDIAHHHLAIA++F+IAG
Sbjct: 229 NWSVYAANPDTTGHVFNTSEGAGTAILTFLGGFHPQTQAMWLTDIAHHHLAIAVIFIIAG 288
Query: 307 HMYRTNWGIGHGIKDILEAHKGP-------FTGQGHKGLYEILTTSWHAQLSINLAMLGS 359
HMY+TN+GIGH +K+IL+ H+ P G GH GLY+ + S H QL + LA LG
Sbjct: 289 HMYKTNFGIGHSMKEILKTHRSPEGTPFGGMLGAGHDGLYDTINNSLHFQLGLALAALGV 348
Query: 360 LTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTR 419
+T +VA HMYSMP Y ++A D+ TQ +L+ HH +I FL+ GA AH AIF VRDYD
Sbjct: 349 VTSLVAQHMYSMPSYAFIAQDFTTQAALYVHHQYIAIFLMCGAFAHGAIFFVRDYDHEAN 408
Query: 420 YNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPI 479
N++L RVL H++AIISHL+WV +FLGFH+ L +HND + A P+ I L+P+
Sbjct: 409 KNNVLGRVLEHKEAIISHLSWVSLFLGFHTLAL-VHNDVVVAFATPEKQ-----ILLEPV 462
Query: 480 FAQWIQNTHALA-----PGITAPGATTGTSLTWGGGD---LVAVGNKVALLPIPLGTADF 531
FAQ++Q A + P + + + G L A+ + V L + +G DF
Sbjct: 463 FAQFVQAASGKALYGFNVLLVNPDSAASIASDYIAGPHFWLDAINSGVNSLFLTIGPGDF 522
Query: 532 LVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVF 591
LVHH A +H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +
Sbjct: 523 LVHHAIALGLHTTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDSFY 582
Query: 592 LGLFWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWA 651
L +FW N + F++ WK S +W Q F +SS + GW RD+LW
Sbjct: 583 LAVFWALNTAGWLTFYWHWKHLS-IWQDNVAQ----------FNESSTYLMGWFRDYLWL 631
Query: 652 QASQVIQSY----GSSLSAYGLFFLGAHFV 677
++Q+I Y L+ + FL H V
Sbjct: 632 NSAQLINGYNPFGSQQLAVWAWMFLFGHLV 661
>gb|AAD49556.1| (AF107783) PsaA [Trichodesmium sp. IMS101]
Length = 274
Score = 510 bits (1300), Expect = e-143
Identities = 223/274 (81%), Positives = 257/274 (93%)
Query: 43 TTWIWNLHADAHDFDSHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWL 102
TTWIWNLHADAHDFDSHTSDLED+SRKIFSAHFG+L+IIF+WLSG YFHGA+FSNYEAWL
Sbjct: 1 TTWIWNLHADAHDFDSHTSDLEDVSRKIFSAHFGRLAIIFVWLSGAYFHGAKFSNYEAWL 60
Query: 103 SDPTHIRPSAQVVWPIVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIG 162
+DPT I+PSAQVVWP++GQ ILNGD+GGGF GIQITSG FQ+WRASGIT+E QLYCTAIG
Sbjct: 61 ADPTGIKPSAQVVWPVLGQGILNGDMGGGFHGIQITSGLFQLWRASGITNEYQLYCTAIG 120
Query: 163 GLVFAALMLFAGWFHYHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPIN 222
GLV AALM+FAGWFHYHK APKL WFQ+VESM+NHHLAGLLGLG LSWAGHQ+HVSLPIN
Sbjct: 121 GLVMAALMMFAGWFHYHKKAPKLEWFQNVESMMNHHLAGLLGLGCLSWAGHQIHVSLPIN 180
Query: 223 QFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPV 282
+ L++GV P++IPLPHEF+ N+ L+A+LYPSFA+G TPFFTLNW +YADFLTF+GGL+PV
Sbjct: 181 KLLDSGVAPEDIPLPHEFLFNKSLMAELYPSFAKGLTPFFTLNWGEYADFLTFKGGLNPV 240
Query: 283 TGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIG 316
TGGLWL+D AHHHLA+A+LF++AGHMYRTNWGIG
Sbjct: 241 TGGLWLSDTAHHHLALAVLFIVAGHMYRTNWGIG 274
>gb|AAD44699.1| (AF130032) PSI P700 apoprotein A2 [Heterocapsa triquetra]
Length = 776
Score = 469 bits (1195), Expect = e-131
Identities = 290/781 (37%), Positives = 401/781 (51%), Gaps = 127/781 (16%)
Query: 46 IWNLHADAHDFDSHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDP 105
I N+H + H S ++ +IF +HFG L+IIFLW +G FH NYE W+ +P
Sbjct: 36 IGNIHDIESYYGIHGSQ---VNLQIFLSHFGHLAIIFLWAAGNLFHIGWNGNYELWILNP 92
Query: 106 THIRPSAQVVWP----IVGQEILNG--DVGGGFRGIQIT-SGFFQIWRASGITSELQLYC 158
P A +W G+ + G GG + ++ SG + A G TS ++Y
Sbjct: 93 ISTMPIAHGIWDPHFGAQGEAVWGGVSATSGGSEAVVVSYSGIYNWLYAVGFTSVYEIYN 152
Query: 159 TAIGGLVFAALMLFAGWFH------------------------------YHKAA------ 182
I + A L G H Y K A
Sbjct: 153 FVIVLELLAVAALLLGKTHLIYNEELIQWLGTNKPYYRVGSDNEEVVSLYKKLALDMKVY 212
Query: 183 PKLAW--------FQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEI 234
P W F LN H+ L+G SL+WAGH +HV++P ++ + + P
Sbjct: 213 PMFIWPFRIFLAAFDASGLRLNFHIGALIGFTSLAWAGHLIHVAIPASRGIYHTISPVG- 271
Query: 235 PLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------------------FLTFR 276
+ L LYP F++ NWA YA LTF
Sbjct: 272 ----DSTFAEPSLKALYP--------FYSGNWAYYAQDIDKDNHIFGSTVGAGKAILTFL 319
Query: 277 GGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNW-GIGHGIKDILEAHKGPFTGQGH 335
GG+ T L+LTDIAHHHLAI +LF+ A H+Y + + G GH I+DIL + +
Sbjct: 320 GGVKSDTASLYLTDIAHHHLAIGVLFIWAAHLYSSLYKGFGHRIRDILVS--------AN 371
Query: 336 KGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIG 395
G+ S+H QL++ A + +T +V H+YS+ PYPYLA DY T ++L+ HH WI
Sbjct: 372 SGMMIRSMNSYHLQLALACAGVSVITSVVGQHIYSLAPYPYLAYDYVTTVALYLHHSWIA 431
Query: 396 GFLIVGAAAHAAIFMVRDYD---PTTRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGL 452
L++ A AHA IF+VRDY TT D++ RVL H+ AIISHL+WV ++LGFH+ G+
Sbjct: 432 SLLMMAAFAHAGIFLVRDYTVNPKTTTGEDIIGRVLAHKAAIISHLSWVSLWLGFHTLGV 491
Query: 453 YIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALAPGITAPGATTGTSLTWGGGDL 512
YIHNDT++A G PQ+ +I ++PIFAQ IQ+ A G T G T + + G +
Sbjct: 492 YIHNDTVTAFGEPQN-----SILIEPIFAQIIQS----ASGKTLYGTTLFSVVNPSSGWV 542
Query: 513 VAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFP 572
+V L +P+G D L HH A +HVTVLIL+KG L AR S+L+PDK + G+ F
Sbjct: 543 QSVNKSFGSLLLPIGPGDLLAHHAIALGLHVTVLILMKGALDARGSKLMPDKIHFGYGFA 602
Query: 573 CDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGG 632
CDGPGRGGTC +SAWD +L +FWM N + IF+F WK + +W +I+ Q
Sbjct: 603 CDGPGRGGTCDISAWDSFYLAMFWMLNTNAWTIFYFHWK-ELTLWQNITFQ--------- 652
Query: 633 NFAQSSITINGWLRDFLWAQASQVIQSY----GSSLSAYGLFFLGAHFVWAFSLMFLFSG 688
F +SS +NGW RD+LW + +I+ Y + LS + FL AH WA MFL S
Sbjct: 653 -FDESSNYLNGWFRDYLWFNSGSLIRGYDALGANDLSVWAWIFLAAHLCWATGFMFLISW 711
Query: 689 RGYWQELIESIVWAHNKLKV------APATQPRALSIVQGRAVGVTHYLLGGIATTWAFF 742
RGYWQELI+ I++ H K + A P ALSIVQ R +G+ H+ +G I T AF
Sbjct: 712 RGYWQELIDIILYMHLKTPILYDIWNAGVYTPVALSIVQARFIGLVHFAVGFIITYAAFI 771
Query: 743 L 743
+
Sbjct: 772 V 772
>dbj|BAB18384.1| (AB044458) photosystem I P700 chlorophyll a apoprotein A2
[Astrephomene gubernaculifera]
Length = 498
Score = 462 bits (1176), Expect = e-129
Identities = 241/518 (46%), Positives = 314/518 (60%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LYC
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYCG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +AL LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSALFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTSSH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH I+
Sbjct: 165 IFGTAQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRIQ 224
Query: 321 DILEAHKGPFT--GQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVPPSKNLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++AIISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAIISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18396.1| (AB044470) photosystem I P700 chlorophyll a apoprotein A2
[Chlamydomonas reinhardtii]
Length = 498
Score = 462 bits (1175), Expect = e-129
Identities = 240/518 (46%), Positives = 314/518 (60%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNISTSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAIFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQSPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTAQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
ILEAH P G GHKGL++ + S H QL + LA +G++T +VA HMYS+PPY + A
Sbjct: 225 AILEAHTPPSGSLGAGHKGLFDTVNNSLHFQLGLALASVGTITSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEALISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFANGQSL-WLPGWLDAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18378.1| (AB044452) photosystem I P700 chlorophyll a apoprotein A2
[Pandorina morum]
Length = 498
Score = 461 bits (1173), Expect = e-128
Identities = 240/518 (46%), Positives = 315/518 (60%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + L LY
Sbjct: 1 TDPVHIRPIAHKIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNLDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTANH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F+IAGHMYRTN+GIGH ++
Sbjct: 165 VFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIIAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVAPSGGLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKTLYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLDAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18371.1| (AB044445) photosystem I P700 chlorophyll a apoprotein A2
[Yamagishiella unicocca]
Length = 498
Score = 460 bits (1170), Expect = e-128
Identities = 238/518 (45%), Positives = 314/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALLSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 AFGTAQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVAPSGNLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18397.1| (AB044471) photosystem I P700 chlorophyll a apoprotein A2
[Vitreochlamys ordinata]
Length = 498
Score = 459 bits (1169), Expect = e-128
Identities = 238/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNISTSGVYQWWYTQGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +AL LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSALFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLTVLPHP----------------QGLTPFFTGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTSQGSGQAILTFLGGFHPQTQSLWLTDNAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
ILE+H P G GHKG+++ + S H QL + LA LG++T +VA HMYS+PPY + A
Sbjct: 225 AILESHVAPSGSMGAGHKGIFDTVNNSLHFQLGLALASLGTITSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEALISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+W +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWASLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
++ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSSSSAAYSNGQSL-WLPGWLDAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18395.1| (AB044469) photosystem I P700 chlorophyll a apoprotein A2
[Chlamydomonas debaryana]
Length = 498
Score = 459 bits (1169), Expect = e-128
Identities = 244/518 (47%), Positives = 310/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNISTSGVYQWWYTQGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +AL LFAGW H P L+WF+D ES LNHHLAGL G SL+W GH VHV+
Sbjct: 61 SVFLALASALFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLAGLFGTSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHPL----------------GLTPFFTGNWAAYAKNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P + LWLTD AHHHLAIA++F++AGHMYRTN+GIGH +K
Sbjct: 165 VFGTSEGSGQAILTFLGGFHPQSQSLWLTDNAHHHLAIAVIFIVAGHMYRTNFGIGHRMK 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GH GL++ + S H QL + LA +G++T +VA HMYSMPPY + A
Sbjct: 225 AILDAHVAPQGKMGAGHTGLFDTVNNSLHFQLGLALASVGTITSLVAQHMYSMPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++AIISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAIISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTH--------AL 490
+WV +FLGFH+ GLY HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWVSLFLGFHTLGLYTHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKALYGFDVL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
+ G SL W G L AV N L + +G DFLVHH A +HVTVLIL+K
Sbjct: 400 LSSSNSAAFANGQSL-WLPGWLDAVNNNQNSLFLTIGPGDFLVHHAIALGLHVTVLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18391.1| (AB044465) photosystem I P700 chlorophyll a apoprotein A2 [Gonium
viridistellatum]
Length = 498
Score = 459 bits (1169), Expect = e-128
Identities = 238/518 (45%), Positives = 314/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LYC
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYCG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +AL LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSALFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTALH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL++H P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDSHVPPSGGLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18380.1| (AB044454) photosystem I P700 chlorophyll a apoprotein A2
[Pandorina morum]
Length = 498
Score = 459 bits (1169), Expect = e-128
Identities = 238/518 (45%), Positives = 314/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPALSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 IFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHTAPSGRMGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKTLYGFDLL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18370.1| (AB044444) photosystem I P700 chlorophyll a apoprotein A2
[Yamagishiella unicocca]
Length = 498
Score = 459 bits (1169), Expect = e-128
Identities = 238/518 (45%), Positives = 314/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVAPSGNMGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18383.1| (AB044457) photosystem I P700 chlorophyll a apoprotein A2
[Pandorina colemaniae]
Length = 498
Score = 459 bits (1168), Expect = e-128
Identities = 238/518 (45%), Positives = 314/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPALSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 IFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVAPSGNMGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKTLYGFDLL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18361.1| (AB044435) photosystem I P700 chlorophyll a apoprotein A2 [Eudorina
elegans]
Length = 498
Score = 459 bits (1168), Expect = e-128
Identities = 238/518 (45%), Positives = 314/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 IFGTTQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHTPPSGGLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNRGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKTLYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18382.1| (AB044456) photosystem I P700 chlorophyll a apoprotein A2
[Pandorina morum]
Length = 498
Score = 458 bits (1167), Expect = e-128
Identities = 238/518 (45%), Positives = 314/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHKIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAIFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTANH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVAPSGGLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLDAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18385.1| (AB044459) photosystem I P700 chlorophyll a apoprotein A2
[Astrephomene gubernaculifera]
Length = 498
Score = 458 bits (1166), Expect = e-127
Identities = 240/518 (46%), Positives = 313/518 (60%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYTG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +AL LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSALFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTSSH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH I+
Sbjct: 165 IFGTAQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRIQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVPPAGNLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++AIISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAIISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18377.1| (AB044451) photosystem I P700 chlorophyll a apoprotein A2
[Volvulina boldii]
Length = 498
Score = 458 bits (1166), Expect = e-127
Identities = 238/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPALSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTQSH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVAPSGNMGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFANGQSL-WLPGWLDAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18368.1| (AB044442) photosystem I P700 chlorophyll a apoprotein A2
[Platydorina caudata]
Length = 498
Score = 458 bits (1166), Expect = e-127
Identities = 238/518 (45%), Positives = 314/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTGSH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTAQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHIAPSGNMGTGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPVFAQWIQAAHGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18379.1| (AB044453) photosystem I P700 chlorophyll a apoprotein A2
[Pandorina morum]
Length = 498
Score = 457 bits (1164), Expect = e-127
Identities = 238/518 (45%), Positives = 314/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHKIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTANH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F+IAGHMYRTN+GIGH ++
Sbjct: 165 VFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIIAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMY++PPY + A
Sbjct: 225 AILDAHVAPSGGLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYALPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLDAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18374.1| (AB044448) photosystem I P700 chlorophyll a apoprotein A2
[Volvulina steinii]
Length = 498
Score = 457 bits (1164), Expect = e-127
Identities = 238/518 (45%), Positives = 312/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + + LY
Sbjct: 1 TDPVHIRPIAHKIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNIDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTANH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH I
Sbjct: 165 VFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRIS 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
ILEAH P G GHKGL+ + S H QL + LA +G++ +VA H Y++PPY +LA
Sbjct: 225 AILEAHTAPSGKMGVGHKGLFATVNNSLHFQLGLALASVGTICSLVAQHTYALPPYAFLA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEALISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+W +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWASLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLDAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18351.1| (AB044425) photosystem I P700 chlorophyll a apoprotein A2 [Volvox
carteri f. kawasakiensis]
Length = 498
Score = 457 bits (1163), Expect = e-127
Identities = 238/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQTPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTAQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
ILEAH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILEAHTPPSGGLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18389.1| (AB044463) photosystem I P700 chlorophyll a apoprotein A2 [Gonium
pectorale]
Length = 498
Score = 457 bits (1162), Expect = e-127
Identities = 238/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +AL LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSALFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 IFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVPPTGSLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18387.1| (AB044461) photosystem I P700 chlorophyll a apoprotein A2 [Gonium
multicoccum]
Length = 498
Score = 457 bits (1162), Expect = e-127
Identities = 237/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAIFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTAAH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVAPSGSLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18372.1| (AB044446) photosystem I P700 chlorophyll a apoprotein A2
[Volvulina compacta]
Length = 498
Score = 457 bits (1162), Expect = e-127
Identities = 237/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTAQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHTAPAGKMGTGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18369.1| (AB044443) photosystem I P700 chlorophyll a apoprotein A2
[Yamagishiella unicocca]
Length = 498
Score = 457 bits (1162), Expect = e-127
Identities = 237/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALLSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQSPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVAPSGNMGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18381.1| (AB044455) photosystem I P700 chlorophyll a apoprotein A2
[Pandorina morum]
Length = 498
Score = 456 bits (1161), Expect = e-127
Identities = 237/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALLSAIFLFAGWLHLQPNFQPALSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 LFGTAQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVAPSGNMGLGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18353.1| (AB044427) photosystem I P700 chlorophyll a apoprotein A2 [Volvox
barberi]
Length = 498
Score = 456 bits (1161), Expect = e-127
Identities = 237/518 (45%), Positives = 314/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPIHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAIFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTSIH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTAEGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVAPSGNMGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP +++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKSNVLARMLDHKEALISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18352.1| (AB044426) photosystem I P700 chlorophyll a apoprotein A2 [Volvox
dissipatrix]
Length = 498
Score = 456 bits (1161), Expect = e-127
Identities = 237/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAIFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHTPPSGGLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18350.1| (AB044424) photosystem I P700 chlorophyll a apoprotein A2 [Volvox
aureus]
Length = 498
Score = 456 bits (1161), Expect = e-127
Identities = 237/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALLSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQSSDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTAQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHTPPSGGLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18365.1| (AB044439) photosystem I P700 chlorophyll a apoprotein A2 [Eudorina
unicocca]
dbj|BAB18366.1| (AB044440) photosystem I P700 chlorophyll a apoprotein A2 [Eudorina
unicocca]
Length = 498
Score = 456 bits (1160), Expect = e-127
Identities = 237/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQTPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 IFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHTPPSGGLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18360.1| (AB044434) photosystem I P700 chlorophyll a apoprotein A2 [Eudorina
illinoisensis]
dbj|BAB18364.1| (AB044438) photosystem I P700 chlorophyll a apoprotein A2 [Eudorina
elegans]
dbj|BAB18367.1| (AB044441) photosystem I P700 chlorophyll a apoprotein A2 [Eudorina
cylindrica]
Length = 498
Score = 456 bits (1160), Expect = e-127
Identities = 237/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTAQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHTPPSGGLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18356.1| (AB044430) photosystem I P700 chlorophyll a apoprotein A2
[Pleodorina californica]
dbj|BAB18357.1| (AB044431) photosystem I P700 chlorophyll a apoprotein A2
[Pleodorina japonica]
Length = 498
Score = 456 bits (1160), Expect = e-127
Identities = 237/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQSPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTAQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHTPPSGGLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18373.1| (AB044447) photosystem I P700 chlorophyll a apoprotein A2
[Volvulina pringsheimii]
Length = 498
Score = 455 bits (1159), Expect = e-127
Identities = 237/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPIHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTAQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVAPGGKMGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18355.1| (AB044429) photosystem I P700 chlorophyll a apoprotein A2 [Volvox
rousseletii]
Length = 498
Score = 455 bits (1159), Expect = e-127
Identities = 236/518 (45%), Positives = 314/518 (60%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPIHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAIFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTSLH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
+TF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTAEGSGQAIITFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVAPSGNMGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP +++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKSNVLARMLDHKEALISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18392.1| (AB044466) photosystem I P700 chlorophyll a apoprotein A2
[Tetrabaena socialis]
Length = 498
Score = 455 bits (1158), Expect = e-127
Identities = 235/517 (45%), Positives = 311/517 (59%), Gaps = 52/517 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTVGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +AL LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSALFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAVYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFSTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMY++PPY + A
Sbjct: 225 AILDAHTPPGGGLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYALPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALA------- 491
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H A
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKALYGFDFL 399
Query: 492 PGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKG 551
+ A + W G L A+ N L + +G DFLVHH A +H T LIL+KG
Sbjct: 400 LSSSNSSAFANSQSLWLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVKG 459
Query: 552 VLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 460 ALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18390.1| (AB044464) photosystem I P700 chlorophyll a apoprotein A2 [Gonium
quadratum]
Length = 498
Score = 455 bits (1158), Expect = e-127
Identities = 237/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYSG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPALSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSILPHP----------------QGLTPFFTGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVAPSGSLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKTLYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLDAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18375.1| (AB044449) photosystem I P700 chlorophyll a apoprotein A2
[Volvulina steinii]
dbj|BAB18376.1| (AB044450) photosystem I P700 chlorophyll a apoprotein A2
[Volvulina steinii]
Length = 498
Score = 455 bits (1158), Expect = e-127
Identities = 236/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAVYAQNPDTANH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFGTAQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMY++PPY + A
Sbjct: 225 AILDAHIAPSGKMGVGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYALPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+W +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWASLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLDAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18388.1| (AB044462) photosystem I P700 chlorophyll a apoprotein A2 [Gonium
octonarium]
Length = 498
Score = 455 bits (1157), Expect = e-126
Identities = 236/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 IFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMY++PPY + A
Sbjct: 225 AILDAHVAPSGKLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYALPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLNIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18354.1| (AB044428) photosystem I P700 chlorophyll a apoprotein A2 [Volvox
globator]
Length = 498
Score = 453 bits (1153), Expect = e-126
Identities = 236/518 (45%), Positives = 313/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + L+
Sbjct: 1 TDPIHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNEDLFVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAVYAQNPDTSLH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 VFSTAEGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVAPSGNLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEALISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKVLYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18398.1| (AB044472) photosystem I P700 chlorophyll a apoprotein A2
[Lobomonas monstruosa]
Length = 498
Score = 451 bits (1148), Expect = e-125
Identities = 235/518 (45%), Positives = 311/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +A+ LFAGW H AP L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSAVFLFAGWLHLQPNFAPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G PFF+ NWA YA
Sbjct: 121 IPESRGQHVGWDNFMSVLPHP----------------QGLAPFFSGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 IFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDAHVPPQGGLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
+ G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSSNSAAFANGQSL-WLPGWLEAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18386.1| (AB044460) photosystem I P700 chlorophyll a apoprotein A2
[Astrephomene perforata]
Length = 498
Score = 450 bits (1144), Expect = e-125
Identities = 234/518 (45%), Positives = 311/518 (59%), Gaps = 54/518 (10%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P G+ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGEPAVEAFTRGGASGPVNIATSGVYQWWYTIGMRTNQDLYSG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +AL LFA W H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSALFLFASWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTSSH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 IFGTAQGSGKAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL++H P G GHKGL+ + S H QL + LA +G++ +VA HMYS+PPY + A
Sbjct: 225 AILDSHVPPSGNLGTGHKGLFNTVNNSLHFQLGLALASVGTICSLVAQHMYSLPPYAFQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ L
Sbjct: 345 SWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAQGKALYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
T+ + G +L W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSKTSAAFSNGQNL-WLPGWLEAINNNQNSLFLNIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
G L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 459 GALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 496
>dbj|BAB18363.1| (AB044437) photosystem I P700 chlorophyll a apoprotein A2 [Eudorina
elegans]
Length = 479
Score = 441 bits (1122), Expect = e-122
Identities = 227/490 (46%), Positives = 298/490 (60%), Gaps = 52/490 (10%)
Query: 129 GGGFRGIQI-TSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLA 186
GG + I TSG +Q W G+ + LY ++ + +A+ LFAGW H P L+
Sbjct: 10 GGASGPVNIATSGVYQWWYTIGMRTNQDLYVGSVFLALVSAVFLFAGWLHLQPNFQPSLS 69
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WF+D ES LNHHL+GL G+ SL+W GH VHV++P ++ + G D LPH
Sbjct: 70 WFKDAESRLNHHLSGLFGVSSLAWTGHLVHVAIPESRGQHVGWDNFLSVLPHP------- 122
Query: 247 LAQLYPSFAEGATPFFTLNWAKYAD------------------FLTFRGGLDPVTGGLWL 288
+G TPFFT NWA YA LTF GG P T LWL
Sbjct: 123 ---------QGLTPFFTGNWAAYAQNPDTASHIFGTTQGSGQAILTFLGGFHPQTQSLWL 173
Query: 289 TDIAHHHLAIAILFLIAGHMYRTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSW 346
TD+AHHHLAIA++F++AGHMYRTN+GIGH ++ ILEAH P G GHKGL++ + S
Sbjct: 174 TDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQAILEAHTPPSGNLGAGHKGLFDTVNNSL 233
Query: 347 HAQLSINLAMLGSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHA 406
H QL + LA +G++ +VA HMYS+PPY + A D+ TQ +L+THH +I GF++ GA AH
Sbjct: 234 HFQLGLALASVGTICSLVAQHMYSLPPYAFQAIDFTTQAALYTHHQYIAGFIMCGAFAHG 293
Query: 407 AIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQ 466
AIF +RDYDP ++L R+L H++A+ISHL+WV +FLGFH+ GLY+HND M A G P+
Sbjct: 294 AIFFIRDYDPEQNKGNVLARMLDHKEAVISHLSWVSLFLGFHTLGLYVHNDVMQAFGTPE 353
Query: 467 DMFSDTAIQLQPIFAQWIQNTHA--------LAPGITAPGATTGTSLTWGGGDLVAVGNK 518
I ++P+FAQWIQ H L T+ G SL W G L A+ N
Sbjct: 354 KQ-----ILIEPVFAQWIQAAHGKTLYGFDFLLSSKTSAAFANGQSL-WLPGWLDAINNN 407
Query: 519 VALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGR 578
L + +G DFLVHH A +H T LIL+KG L AR S+L+PDK + G+ FPCDGPGR
Sbjct: 408 QNSLFLTIGPGDFLVHHAIALGLHTTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGR 467
Query: 579 GGTCQVSAWD 588
GGTC +SA+D
Sbjct: 468 GGTCDISAYD 477
>dbj|BAB18359.1| (AB044433) photosystem I P700 chlorophyll a apoprotein A2
[Pleodorina indica]
Length = 479
Score = 436 bits (1108), Expect = e-121
Identities = 224/490 (45%), Positives = 297/490 (59%), Gaps = 52/490 (10%)
Query: 129 GGGFRGIQI-TSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLA 186
GG + I TSG +Q W G+ + LY ++ + +A+ LFAGW H P L+
Sbjct: 10 GGASGPVNIATSGVYQWWYTIGMRTNQDLYVGSVFLALVSAIFLFAGWLHLQPNFQPSLS 69
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WF+D ES LNHHL+GL G+ SL+W GH VHV++P ++ + G D LPH
Sbjct: 70 WFKDAESRLNHHLSGLFGVSSLAWTGHLVHVAIPESRGQHVGWDNFLSVLPHP------- 122
Query: 247 LAQLYPSFAEGATPFFTLNWAKYAD------------------FLTFRGGLDPVTGGLWL 288
+G TPFFT NWA YA LTF GG P T LWL
Sbjct: 123 ---------QGLTPFFTGNWAAYAQNPDTASHVFGTAQGSGQAILTFLGGFHPQTQSLWL 173
Query: 289 TDIAHHHLAIAILFLIAGHMYRTNWGIGHGIKDILEAHKGPF--TGQGHKGLYEILTTSW 346
TD+AHHHLAIA++F++AGHMYRTN+GIGH ++ IL+AH P G GHKGL++ + S
Sbjct: 174 TDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQAILDAHTPPGGGLGAGHKGLFDTVNNSL 233
Query: 347 HAQLSINLAMLGSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHA 406
H QL + LA +G++ +VA HMYS+PPY + A D+ TQ +L+THH +I GF++ GA AH
Sbjct: 234 HFQLGLALASVGTICSLVAQHMYSLPPYAFQAIDFTTQAALYTHHQYIAGFIMCGAFAHG 293
Query: 407 AIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQ 466
AIF +RDYDP ++L R+L H++A+ISHL+WV +FLGFH+ GLY+HND M A G P+
Sbjct: 294 AIFFIRDYDPEQNKGNVLARMLDHKEAVISHLSWVSLFLGFHTLGLYVHNDVMQAFGTPE 353
Query: 467 DMFSDTAIQLQPIFAQWIQNTHA--------LAPGITAPGATTGTSLTWGGGDLVAVGNK 518
I ++P+FAQWIQ L + + G SL W G L A+ N
Sbjct: 354 KQ-----ILIEPVFAQWIQAAQGKALYGFDFLLSSKNSAAFSNGQSL-WLPGWLEAINNN 407
Query: 519 VALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGR 578
L + +G DFLVHH A +H T LIL+KG L AR S+L+PDK + G+ FPCDGPGR
Sbjct: 408 QNSLFLTIGPGDFLVHHAIALGLHTTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGR 467
Query: 579 GGTCQVSAWD 588
GGTC +SA+D
Sbjct: 468 GGTCDISAYD 477
>dbj|BAB18399.1| (AB044473) photosystem I P700 chlorophyll a apoprotein A2
[Paulschulzia pseudovolvox]
Length = 468
Score = 413 bits (1051), Expect = e-114
Identities = 221/490 (45%), Positives = 289/490 (58%), Gaps = 54/490 (11%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNISTSGVYQWWYTVGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +AL LFAGW H AP L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSALFLFAGWLHLQPNFAPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQSPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F++AGHMYRTN+GIGH ++
Sbjct: 165 LFGTAEGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIVAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLA 378
IL+AH P G GHKGL++ + S H QL + LA +G++T +VA HMYS+PPY Y A
Sbjct: 225 AILDAHVPPSGSLGAGHKGLFDTVNNSLHFQLGLALASVGTITSLVAQHMYSLPPYAYQA 284
Query: 379 TDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHL 438
D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R+L H++A+ISHL
Sbjct: 285 IDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLARMLDHKEAVISHL 344
Query: 439 NWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------L 490
+W +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ H L
Sbjct: 345 SWASLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQAAHGKTLYGFDFL 399
Query: 491 APGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLK 550
+ G SL W G L A+ N L + +G DFLVHH A +H T LIL+K
Sbjct: 400 LSSSNSAAFANGQSL-WLPGWLGAINNNQNSLFLTIGPGDFLVHHAIALGLHTTTLILVK 458
Query: 551 GVLFARSSRL 560
G L AR S+L
Sbjct: 459 GALDARGSKL 468
>dbj|BAA83440.1| (AB013664) photosystem I P700 apoprotein A1 [Anthoceros punctatus]
Length = 194
Score = 346 bits (878), Expect = 6e-94
Identities = 159/179 (88%), Positives = 168/179 (93%), Gaps = 1/179 (0%)
Query: 29 PGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKIFSAHFGQLSIIFLWLSGM 88
PG I KGP TTTWIWNLHADAHDFDSHT+DLE++SRK+FSAHFGQL+IIF+WLSGM
Sbjct: 17 PGRHGAGI-KGPSTTTWIWNLHADAHDFDSHTNDLEEVSRKVFSAHFGQLAIIFIWLSGM 75
Query: 89 YFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGGFRGIQITSGFFQIWRAS 148
YFHGARF NYEAWL DPTHI+PSAQVVWPIVGQEILNGDVGGGF+GIQITSGFFQIWRAS
Sbjct: 76 YFHGARFPNYEAWLGDPTHIKPSAQVVWPIVGQEILNGDVGGGFQGIQITSGFFQIWRAS 135
Query: 149 GITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDVESMLNHHLAGLLGLGS 207
GITSELQLY TAIGGL+FAALMLFAGWFHYHKAAPKL WFQDVESMLNHHLAGLLGLGS
Sbjct: 136 GITSELQLYSTAIGGLIFAALMLFAGWFHYHKAAPKLVWFQDVESMLNHHLAGLLGLGS 194
>gb|AAD10229.1| (U72056) photosystem I protein A1 [Anabaena sp. CA]
Length = 194
Score = 328 bits (833), Expect = 1e-88
Identities = 148/192 (77%), Positives = 166/192 (86%)
Query: 21 TSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISRKIFSAHFGQLSI 80
TSFE+WA+PGHF RT+A+GP TTTWIWNLHA AHDFD+HTSDLEDISRKIF+A FG L++
Sbjct: 2 TSFEKWAQPGHFDRTLARGPKTTTWIWNLHALAHDFDTHTSDLEDISRKIFAARFGHLAV 61
Query: 81 IFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGGFRGIQITSG 140
+ +WLSGM FHGA+FSNYEA LSDP +++PSAQVVW IVGQ+ILNGDVGGGF GIQITSG
Sbjct: 62 VTIWLSGMIFHGAKFSNYEARLSDPMNVKPSAQVVWSIVGQDILNGDVGGGFHGIQITSG 121
Query: 141 FFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDVESMLNHHLA 200
FQ+WR GIT+ QLY TAIGGLV A L LFAGWFHYHK APKL WFQ+VESMLNHHLA
Sbjct: 122 LFQVWRGWGITNSFQLYVTAIGGLVLAGLFLFAGWFHYHKRAPKLEWFQNVESMLNHHLA 181
Query: 201 GLLGLGSLSWAG 212
LLG GSL W G
Sbjct: 182 VLLGCGSLGWTG 193
>dbj|BAA83428.1| (AB013652) photosystem I P700 apoprotein A1 [Physcomitrella patens]
Length = 149
Score = 298 bits (754), Expect = 2e-79
Identities = 135/149 (90%), Positives = 146/149 (97%)
Query: 53 AHDFDSHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSA 112
AHDFDSHT+DLE+ISRK+FSAHFGQL++IF+WLSGMYFHGARFSNYEAWLSDPTHI+PSA
Sbjct: 1 AHDFDSHTNDLEEISRKVFSAHFGQLAVIFIWLSGMYFHGARFSNYEAWLSDPTHIKPSA 60
Query: 113 QVVWPIVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLF 172
QVVWPIVGQEILNGDVGGGF+GIQITSGFFQ+WRASGITSELQ Y TAIGGL+FAALMLF
Sbjct: 61 QVVWPIVGQEILNGDVGGGFQGIQITSGFFQLWRASGITSELQRYTTAIGGLIFAALMLF 120
Query: 173 AGWFHYHKAAPKLAWFQDVESMLNHHLAG 201
AGWFHYHKAAPKLAW Q+VESMLNHHLAG
Sbjct: 121 AGWFHYHKAAPKLAWCQNVESMLNHHLAG 149
>dbj|BAB18344.1| (AB044418) photosystem I P700 chlorophyll a apoprotein A1
[Chlamydomonas debaryana]
Length = 151
Score = 294 bits (745), Expect = 2e-78
Identities = 137/152 (90%), Positives = 145/152 (95%), Gaps = 1/152 (0%)
Query: 534 HHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLG 593
HHIHAFTIHVT LILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLG
Sbjct: 1 HHIHAFTIHVTTLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLG 60
Query: 594 LFWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQA 653
LFWMYNAISV IFHFSWKMQSDVWG+++ G V+HITGGNFAQS+ TINGWLRDFLWA++
Sbjct: 61 LFWMYNAISVDIFHFSWKMQSDVWGTVTASG-VSHITGGNFAQSANTINGWLRDFLWAES 119
Query: 654 SQVIQSYGSSLSAYGLFFLGAHFVWAFSLMFL 685
SQVIQSYGS+LSAYGL FLGAHFVWAFSLMFL
Sbjct: 120 SQVIQSYGSALSAYGLIFLGAHFVWAFSLMFL 151
>dbj|BAA83446.1| (AB013670) photosystem I P700 apoprotein A1 [Sphagnum fallax]
Length = 142
Score = 287 bits (726), Expect = 4e-76
Identities = 131/142 (92%), Positives = 141/142 (99%)
Query: 61 SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVG 120
+DLE+ISRK+FSAHFGQL+IIF+WLSGMYFHGARFSNYEAWLSDPTHI+PSAQVVWPIVG
Sbjct: 1 NDLEEISRKVFSAHFGQLAIIFIWLSGMYFHGARFSNYEAWLSDPTHIKPSAQVVWPIVG 60
Query: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHK 180
QEILNGDVGGGF+GIQITSGFFQ+WRASGITSELQLY TAIGGL+FAALMLFAGWFHYHK
Sbjct: 61 QEILNGDVGGGFQGIQITSGFFQLWRASGITSELQLYSTAIGGLIFAALMLFAGWFHYHK 120
Query: 181 AAPKLAWFQDVESMLNHHLAGL 202
AAPKLAWFQ+VESMLNHHLAGL
Sbjct: 121 AAPKLAWFQNVESMLNHHLAGL 142
>dbj|BAA83456.1| (AB013680) photosystem I P700 apoprotein A1 [Adiantum
capillus-veneris]
Length = 145
Score = 285 bits (722), Expect = 1e-75
Identities = 129/145 (88%), Positives = 141/145 (96%)
Query: 51 ADAHDFDSHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRP 110
ADAHDFDSHT+DLEDISRKIFSAHFGQL+IIF+ LSGMYFHGARFSNYE+WL+DPTH++P
Sbjct: 1 ADAHDFDSHTNDLEDISRKIFSAHFGQLAIIFIRLSGMYFHGARFSNYESWLNDPTHVKP 60
Query: 111 SAQVVWPIVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALM 170
SAQVVWPIVGQEILNGDVGGGF+G+QITSGFFQ+WRASGITSELQLYCTA+G LVFA LM
Sbjct: 61 SAQVVWPIVGQEILNGDVGGGFQGVQITSGFFQLWRASGITSELQLYCTAVGALVFAGLM 120
Query: 171 LFAGWFHYHKAAPKLAWFQDVESML 195
FAGWFHYHKAAPKLAWFQ+VESML
Sbjct: 121 FFAGWFHYHKAAPKLAWFQNVESML 145
>dbj|BAA83451.1| (AB013675) photosystem I P700 apoprotein A1 [Haplomitrium mnioides]
Length = 143
Score = 279 bits (706), Expect = 9e-74
Identities = 128/143 (89%), Positives = 137/143 (95%)
Query: 58 SHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWP 117
SHT+DLE+ISRK+F AHFGQL+II +WLSGMYFHGARFSNYEAWLSDPTHI+PSAQVVWP
Sbjct: 1 SHTNDLEEISRKVFGAHFGQLAIILIWLSGMYFHGARFSNYEAWLSDPTHIKPSAQVVWP 60
Query: 118 IVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFH 177
IVGQEILNGDVGGGF+GIQITSGFFQ+WRASGITSELQL+ TAIGGLV AALMLFAGWFH
Sbjct: 61 IVGQEILNGDVGGGFQGIQITSGFFQLWRASGITSELQLHSTAIGGLVLAALMLFAGWFH 120
Query: 178 YHKAAPKLAWFQDVESMLNHHLA 200
YHKAAPKL WFQDVESMLNH LA
Sbjct: 121 YHKAAPKLVWFQDVESMLNHRLA 143
>dbj|BAA83434.1| (AB013658) photosystem I P700 apoprotein A1 [Coleochaete
nitellarum]
Length = 153
Score = 278 bits (704), Expect = 2e-73
Identities = 127/149 (85%), Positives = 138/149 (92%)
Query: 48 NLHADAHDFDSHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTH 107
NLHADAH F HT+DLE+ISRK+FSAHFGQL++IF+WLSGMYFHGARFSNYEAWLSDP +
Sbjct: 1 NLHADAHXFXRHTTDLEEISRKVFSAHFGQLAVIFIWLSGMYFHGARFSNYEAWLSDPIN 60
Query: 108 IRPSAQVVWPIVGQEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFA 167
I+PSAQVVWPIVGQEILNGDVGGGF+G+QITSGFFQIWRASGIT+ELQLY TAIGGLV A
Sbjct: 61 IKPSAQVVWPIVGQEILNGDVGGGFQGVQITSGFFQIWRASGITTELQLYSTAIGGLVAA 120
Query: 168 ALMLFAGWFHYHKAAPKLAWFQDVESMLN 196
LM AGWFHYHKAAP LAWFQDVES LN
Sbjct: 121 TLMFIAGWFHYHKAAPTLAWFQDVESTLN 149
>dbj|BAB18348.1| (AB044422) photosystem I P700 chlorophyll a apoprotein A1
[Paulschulzia pseudovolvox]
Length = 141
Score = 249 bits (629), Expect = 1e-64
Identities = 105/138 (76%), Positives = 127/138 (91%)
Query: 187 WFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDL 246
WFQ+VESMLNHHL GLLGLGSL WAGHQ+HVSLP+N+ L+AGVDPKEIPLPH+ + NR +
Sbjct: 1 WFQNVESMLNHHLGGLLGLGSLGWAGHQIHVSLPVNKLLDAGVDPKEIPLPHDLLFNRGI 60
Query: 247 LAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAG 306
+A LYPSFA+G PFFTLNW++Y+DFLTF+GGL+PVTGGLWL+D AHHH+AIA+LFL+AG
Sbjct: 61 MADLYPSFAKGIAPFFTLNWSEYSDFLTFKGGLNPVTGGLWLSDTAHHHVAIAVLFLVAG 120
Query: 307 HMYRTNWGIGHGIKDILE 324
HMYRTNWGIGH +K+IL+
Sbjct: 121 HMYRTNWGIGHSMKEILD 138
>dbj|BAA83435.1| (AB013659) photosystem I P700 apoprotein A2 [Coleochaete
nitellarum]
Length = 301
Score = 243 bits (615), Expect = 4e-63
Identities = 135/311 (43%), Positives = 179/311 (57%), Gaps = 40/311 (12%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +++ P TT IW A AHDF+SH E+ + +KIF++HFGQL++IFLW SG F
Sbjct: 8 FSQGLSQDP-TTRRIWFGIATAHDFESHDDITEEKLYQKIFASHFGQLAVIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+EAW DP H+RP A +W P GQ + GG G + I+ SG +Q W
Sbjct: 67 HVAWQGNFEAWTKDPLHVRPIAHAIWDPHFGQPAVEAFTRGGASGPVNISYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + L LY A+ L AA+ L AGW H K P ++WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNLDLYRGALFLLALAAISLVAGWLHLQPKWKPSVSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+W+GH VHV++P ++ + LPH +G PFF+ W
Sbjct: 187 SLAWSGHLVHVAIPESRGETVRWENFLTTLPHP----------------QGLAPFFSGQW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
+ YA LTF GG P T LWLTDIAHHH+AIA+LF++AGHM
Sbjct: 231 SVYAQNADSSNHLFGTSEGAGTAILTFLGGFHPQTQSLWLTDIAHHHIAIAVLFIVAGHM 290
Query: 309 YRTNWGIGHGI 319
YRTN+GIGH I
Sbjct: 291 YRTNFGIGHSI 301
>dbj|BAB18394.1| (AB044468) photosystem I P700 chlorophyll a apoprotein A2
[Basichlamys sacculifera]
Length = 227
Score = 236 bits (597), Expect = 6e-61
Identities = 111/229 (48%), Positives = 149/229 (64%), Gaps = 12/229 (5%)
Query: 367 HMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR 426
HMY++PPY + A D+ TQ +L+THH +I GF++ GA AH AIF +RDYDP ++L R
Sbjct: 2 HMYALPPYAFQAIDFTTQAALYTHHQYIAGFIMCGAFAHGAIFFIRDYDPEQNKGNVLAR 61
Query: 427 VLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQN 486
+L H++A+ISHL+WV +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ
Sbjct: 62 MLDHKEAVISHLSWVSLFLGFHTLGLYVHNDVMQAFGTPEKQ-----ILIEPVFAQWIQA 116
Query: 487 THALA-------PGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAF 539
H A + A + + W G L A+ N L + +G DFLVHH A
Sbjct: 117 AHGKALYGFDFLLSSSNSSAFSNSQSLWLPGWLEAINNNQNSLFLTIGPGDFLVHHAIAL 176
Query: 540 TIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
+H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SA+D
Sbjct: 177 GLHTTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAYD 225
>dbj|BAB18393.1| (AB044467) photosystem I P700 chlorophyll a apoprotein A2
[Basichlamys sacculifera]
Length = 271
Score = 221 bits (557), Expect = 3e-56
Identities = 124/287 (43%), Positives = 162/287 (56%), Gaps = 40/287 (13%)
Query: 103 SDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQI-TSGFFQIWRASGITSELQLYCT 159
+DP HIRP A +W P GQ + GG G + I TSG +Q W G+ + LY
Sbjct: 1 TDPVHIRPIAHAIWDPHFGQPAVEAFTRGGASGPVNIATSGVYQWWYTVGMRTNQDLYVG 60
Query: 160 AIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVS 218
++ + +AL LFAGW H P L+WF+D ES LNHHL+GL G+ SL+W GH VHV+
Sbjct: 61 SVFLALVSALFLFAGWLHLQPNFQPSLSWFKDAESRLNHHLSGLFGVSSLAWTGHLVHVA 120
Query: 219 LPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYAD------- 271
+P ++ + G D LPH +G TPFFT NWA YA
Sbjct: 121 IPESRGQHVGWDNFLSVLPHP----------------QGLTPFFTGNWAAYAQNPDTASH 164
Query: 272 -----------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIK 320
LTF GG P T LWLTD+AHHHLAIA++F+ AGHMYRTN+GIGH ++
Sbjct: 165 LFGTSQGSGQAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIGAGHMYRTNFGIGHRMQ 224
Query: 321 DILEAHKGPF--TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVA 365
IL+AH P G GHKGL++ + S H QL + LA +G++ +VA
Sbjct: 225 AILDAHTPPAGGLGAGHKGLFDTVNNSLHFQLGLALASVGTICSLVA 271
>dbj|BAA83429.1| (AB013653) photosystem I P700 apoprotein A2 [Physcomitrella patens]
Length = 291
Score = 215 bits (542), Expect = 2e-54
Identities = 126/301 (41%), Positives = 164/301 (53%), Gaps = 40/301 (13%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS +++ P TT IW A A DF+SH E+ + +KIF++HFGQL+IIFLW SG F
Sbjct: 8 FSXGLSQDP-TTRRIWFGXATAXDFESHDDMTEERLYQKIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+EAW DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFEAWGQDPLHVRPIAHAIWDPQFGQPAVEAFTRGGASGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY +I L +AL L AGW H K P ++WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNQDLYGGSIFLLFVSALFLIAGWLHLQPKWKPSVSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNW 266
SL+W GH VHV++P ++ + + LPH +G PFF W
Sbjct: 187 SLAWTGHLVHVAIPESRGEHVRWNNLLTALPH----------------PQGLGPFFAGQW 230
Query: 267 AKYAD------------------FLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHM 308
YA LTF GG P T LWLTD+AHHHLAIA++F+IAGHM
Sbjct: 231 NVYAQNPDSNSHLFGTSEGAGTAILTFLGGFHPQTQSLWLTDMAHHHLAIAVIFIIAGHM 290
Query: 309 Y 309
Y
Sbjct: 291 Y 291
>dbj|BAA83441.1| (AB013665) photosystem I P700 apoprotein A2 [Anthoceros punctatus]
Length = 282
Score = 198 bits (498), Expect = 2e-49
Identities = 115/276 (41%), Positives = 160/276 (57%), Gaps = 8/276 (2%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +++ P TT IW A AHDF+SH + E+ + +KIF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLSQDP-TTRRIWFGIATAHDFESHDNITEERLYQKIFASHFGQLAIIFLWTSGNSF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+EAW+ DP H+RP A +W P G+ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFEAWVQDPLHVRPIAHAIWDPHFGEPAVEAFTRGGALGPVNIAYSGVYQWWYI 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L AA+ L AGW H K P ++WF++ ES NHHL+GL G+
Sbjct: 127 IGLRTIQDLYIGALFLLFLAAVFLIAGWLHLQPKWKPSVSWFKNAESRPNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAEGATPFFTL 264
SL+WAGH +HV++ ++ + D LPH L Q +Y A+ ++ F+
Sbjct: 187 SLAWAGHLIHVAISESRGEHVRWDNLLTALPHPQGLKPFFEGQWGIYAENADSSSHLFST 246
Query: 265 NWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
+ TF GG P T LWLTDIAHH+LAIA+
Sbjct: 247 SEGAGTAIPTFIGGFHPQTQSLWLTDIAHHYLAIAV 282
>dbj|BAA83430.1| (AB013654) photosystem I P700 apoprotein A2 [Physcomitrella patens]
Length = 169
Score = 168 bits (421), Expect = 2e-40
Identities = 82/167 (49%), Positives = 104/167 (62%), Gaps = 14/167 (8%)
Query: 430 HRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA 489
H++AIISHL+W +FLGFH+ GLY+HND M A G P+ I ++P+FAQWIQ+ H
Sbjct: 1 HKEAIISHLSWASLFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPVFAQWIQSAHG 55
Query: 490 --------LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTI 541
L +P G +L W G L A+ N L + +G DFLVHH A +
Sbjct: 56 KALYGFDVLLSSADSPAFNAGQTL-WLPGWLDAINNNSNSLFLTIGPGDFLVHHAIALGL 114
Query: 542 HVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
H T LIL+KG L AR S+L+PDK G+ FPCDGPGRGGTC +SAWD
Sbjct: 115 HTTTLILVKGALDARGSKLMPDKKEFGYSFPCDGPGRGGTCDISAWD 161
>dbj|BAA83436.1| (AB013660) photosystem I P700 apoprotein A2 [Coleochaete
nitellarum]
Length = 166
Score = 166 bits (416), Expect = 9e-40
Identities = 81/167 (48%), Positives = 104/167 (61%), Gaps = 14/167 (8%)
Query: 430 HRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA 489
H++AIISHL+W +FLGFH+ GLY+HND + A G P+ I ++P+FAQWIQ+ H
Sbjct: 1 HKEAIISHLSWASLFLGFHTLGLYVHNDVVQAFGNPEKQ-----ILIEPVFAQWIQSAHG 55
Query: 490 --------LAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTI 541
L +P G +L W G L +V N L + +G DFLVHH A +
Sbjct: 56 KSLYGFDVLLSSTGSPAFNAGQTL-WLPGWLDSVNNASNSLFLTIGPGDFLVHHAIALGL 114
Query: 542 HVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWD 588
H T LIL+KG L AR S+L+PDK G+ FPCDGPGRGGTC +SAWD
Sbjct: 115 HTTTLILVKGALDARGSKLMPDKKEFGYSFPCDGPGRGGTCDISAWD 161
>dbj|BAA83442.1| (AB013666) photosystem I P700 apoprotein A2 [Anthoceros punctatus]
Length = 155
Score = 140 bits (350), Expect = 5e-32
Identities = 72/160 (45%), Positives = 89/160 (55%), Gaps = 14/160 (8%)
Query: 448 HSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHA--------LAPGITAPGA 499
H+ GLY+HND M A G P+ I ++P+FAQWIQ+ H L P
Sbjct: 1 HTLGLYVHNDVMLAFGTPEKQ-----ILIEPVFAQWIQSAHGKTLYGFDLLLSSADRPAF 55
Query: 500 TTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLFARSSR 559
G S+ W G L A N L + +G DFLVHH A +H T LIL+KG L AR +
Sbjct: 56 NAGQSI-WLPGWLDATNNNSNSLFLTIGPGDFLVHHAIALGLHTTTLILVKGALDARGFK 114
Query: 560 LIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN 599
L+PDK G+ FPCDGPGRGGTC +SAWD L + W N
Sbjct: 115 LMPDKKEFGYSFPCDGPGRGGTCDISAWDAFXLAVSWXLN 154
>emb|CAA25804.1| (X01647) open reading frame (ORF 704) [Marchantia polymorpha]
Length = 144
Score = 109 bits (271), Expect = 9e-23
Identities = 60/130 (46%), Positives = 76/130 (58%), Gaps = 13/130 (10%)
Query: 628 HIT-----GGNFAQSSITINGWLRDFLWAQASQVIQSYG----SSLSAYGLFFLGAHFVW 678
HIT F +SS + GWLRD+LW +SQ+I Y +SLS + FL H VW
Sbjct: 8 HITLWQGNAAQFNESSTYLMGWLRDYLWLNSSQLINGYNPFGMNSLSVWAWMFLFGHLVW 67
Query: 679 AFSLMFLFSGRGYWQELIESIVWAHNKLKVAPAT----QPRALSIVQGRAVGVTHYLLGG 734
A MFL S RGYWQELIE++ WAH + +A +P ALSIVQ R VG+ H+ +G
Sbjct: 68 ATGFMFLISWRGYWQELIETLAWAHERTPLANLVRWKDKPVALSIVQARLVGLAHFSVGY 127
Query: 735 IATTWAFFLA 744
I T AF +A
Sbjct: 128 IFTYAAFLIA 137
>emb|CAA58957.1| (X84152) photosystem I subunit [Antirrhinum majus]
Length = 135
Score = 97.9 bits (240), Expect = 4e-19
Identities = 56/129 (43%), Positives = 76/129 (58%), Gaps = 5/129 (3%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + + IF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAQDP-TTRRIWFGIATAHDFESHDDITEERLYQNIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E+W+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFESWVQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGALGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQL 156
G+ + L
Sbjct: 127 IGLRTNEDL 135
>pir||S18319 photosystem I protein A2 - Chlamydomonas reinhardtii chloroplast
(fragment)
Length = 69
Score = 93.2 bits (228), Expect = 1e-17
Identities = 40/67 (59%), Positives = 50/67 (73%)
Query: 526 LGTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVS 585
+G DFLVHH A +H T LIL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +S
Sbjct: 2 IGPGDFLVHHAIALGLHTTTLILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDIS 61
Query: 586 AWDHVFL 592
A+D +L
Sbjct: 62 AYDAFYL 68
>pir||T31454 reaction center core polypeptide pshA - Heliobacillus mobilis
gb|AAC84025.1| (AF080002) reaction center core polypeptide PshA [Heliobacillus
mobilis]
Length = 609
Score = 84.3 bits (205), Expect = 5e-15
Identities = 63/223 (28%), Positives = 101/223 (45%), Gaps = 16/223 (7%)
Query: 529 ADFLVHHIHAFTIHVTVLILLKGVLFARSS---RLIPDKANLGFRFPCDGPGRGGTCQVS 585
+DF+ H A +H T++ L + V F++ S + KA FPC GP GGTC +S
Sbjct: 386 SDFVAAHAIAGGLHFTMVPLWRMVFFSKVSPWTTKVGMKAKRDGEFPCLGPAYGGTCSIS 445
Query: 586 AWDHVFLGLFWMYNAISVVIFHFS--W-----KMQSDVWGSISDQGVVTHITGGNFAQSS 638
D +L +F+ I+ F+ W S+V+ ++ A S+
Sbjct: 446 LVDQFYLAIFFSLQVIAPAWFYLDGCWMGSFVATSSEVYKQAAELFKANPTWFSLHAVSN 505
Query: 639 ITINGWLRDFLWAQASQVIQSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIES 698
T G+ ++ +Q +++ + Y L LGAHF+WAF+ LF RG E
Sbjct: 506 FTSEGYF-SYIISQTTRMFKYYDGHLVQ---ALLGAHFIWAFTFSMLFQYRGSRDEGAMV 561
Query: 699 IVWAHNK--LKVAPATQPRALSIVQGRAVGVTHYLLGGIATTW 739
+ WAH + L A RALS+ +G+A+G + + W
Sbjct: 562 LKWAHEQVGLGFAGKVYNRALSLKEGKAIGTFLFFKMTVLCMW 604
>pir||A48290 reaction center core polypeptide - Heliobacillus mobilis
gb|AAA02821.1| (L19604) core polypeptide [Heliobacillus mobilis]
Length = 609
Score = 71.4 bits (172), Expect = 4e-11
Identities = 60/220 (27%), Positives = 98/220 (44%), Gaps = 10/220 (4%)
Query: 529 ADFLVHHIHAFTIHVTVLILLKGVLFARSS---RLIPDKANLGFRFPCDGPGRGGTCQVS 585
+DF+ H A +H T++ L + V F++ S + KA FPC GP GGTC +S
Sbjct: 386 SDFVAAHAIAGGLHFTMVPLWRMVFFSKVSPWTTKVGMKAKRDGEFPCLGPAYGGTCSIS 445
Query: 586 AWDHVFLGLFWMYNAISVVIFHFS--WKMQSDVWGSISDQGVVTHITGGNFAQSSI-TIN 642
D +L +F+ I+ F+ W M S V S + N S+ ++
Sbjct: 446 LVDQFYLAIFFSLQVIAPAWFYLDGCW-MGSFVATSSEVYKQAAELFKANPTWFSLHAVS 504
Query: 643 GWLRDFLWAQAS-QVIQSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVW 701
+ + A +S + + +++ + AHF+WAF+ LF RG E + W
Sbjct: 505 NFTSEVTSATSSLKPLVCSNTTMVTWFKPCWAAHFIWAFTFSMLFQYRGSRDEGAMVLKW 564
Query: 702 AHNK--LKVAPATQPRALSIVQGRAVGVTHYLLGGIATTW 739
AH + L A RALS+ +G+A+G + + W
Sbjct: 565 AHEQVGLGFAGKVYNRALSLKEGKAIGTFLFFKMTVLCMW 604
>gb|AAA92988.1| (L27262) P700 chlorophyll a-apoprotein A1 [Chlamydomonas zebra]
Length = 41
Score = 56.6 bits (134), Expect = 1e-06
Identities = 26/41 (63%), Positives = 34/41 (82%), Gaps = 3/41 (7%)
Query: 1 MIIRSPEPE---VKILVDRDPIKTSFEEWAKPGHFSRTIAK 38
M I +PE E VKI VDR+P++TSFE+WAKPGHFSR+++K
Sbjct: 1 MTISTPEREAKKVKIAVDRNPVETSFEKWAKPGHFSRSLSK 41
>gb|AAA92987.1| (L27259) P700 chlorophyll a-apoprotein A1 [Chlamydomonas
gelatinosa]
Length = 26
Score = 54.7 bits (129), Expect = 4e-06
Identities = 21/26 (80%), Positives = 26/26 (99%)
Query: 15 DRDPIKTSFEEWAKPGHFSRTIAKGP 40
DR+P++TSFE+WAKPGHFSRT+AKGP
Sbjct: 1 DRNPVETSFEKWAKPGHFSRTLAKGP 26
>gb|AAB20423.2| (S67792) polypeptide of the photosystem I reaction center
[Chlamydomonas reinhardtii]
Length = 67
Score = 51.9 bits (122), Expect = 3e-05
Identities = 24/42 (57%), Positives = 29/42 (68%)
Query: 526 LGTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANL 567
+G DFLVHH A +H T LIL+KG L AR S+L+PDK L
Sbjct: 2 IGPGDFLVHHAIALGLHTTTLILVKGALDARGSKLMPDKKTL 43
>pir||A46265 P840 (photosystem 1-like) reaction center large chain - Chlorobium
limicola f.sp. thiosulfatophilum
gb|AAA74735.1| (M94675) P840 reaction center large subunit [Chlorobium limicola f.
thiosulfatophilum]
Length = 730
Score = 49.2 bits (115), Expect = 2e-04
Identities = 46/175 (26%), Positives = 72/175 (40%), Gaps = 34/175 (19%)
Query: 530 DFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPD---KANLGFRFPCDGPGRGGTCQVSA 586
D+++ H+ +++ L++ FA +S L D K N + FPC GP GGTC VS
Sbjct: 481 DWVMAHVITAGSLFSLIALVRIAFFAHTSPLWDDLGLKKN-SYSFPCLGPVYGGTCGVSI 539
Query: 587 WDHVFLGLFWMYNAISVVIFHF--SW---------KMQSDVWGSISDQGVVTHITGGNFA 635
D ++ + W +S V ++ +W + W SI+ + H T G F
Sbjct: 540 QDQLWFAMLWGIKGLSAVCWYIDGAWIASMMYGVPAADAKAWDSIAH--LQHHYTSGIFY 597
Query: 636 QSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRG 690
+ W + + S S LS + +G H VW S F RG
Sbjct: 598 ------------YFWTETVTIFSS--SHLST--ILMIG-HLVWFISFAVWFEDRG 635
>sp|P10977|CARV_CANAL VACUOLAR ASPARTIC PROTEASE PRECURSOR (ASPARTATE PROTEASE) (ACP)
gb|AAA79879.1| (U36754) vacuolar aspartic proteinase precursor [Candida albicans]
Length = 419
Score = 35.2 bits (79), Expect = 3.1
Identities = 24/79 (30%), Positives = 38/79 (47%), Gaps = 6/79 (7%)
Query: 343 TTSWHAQLSINLAM---LGSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMW---IGG 396
T SW Q ++ A L LT+ A + +++ PY Y+ G+ +S+FT + IG
Sbjct: 330 TKSWSGQYQVDCAKRDSLPDLTLTFAGYNFTLTPYDYILEVSGSCISVFTPMDFPQPIGD 389
Query: 397 FLIVGAAAHAAIFMVRDYD 415
IVG A + + D D
Sbjct: 390 LAIVGDAFLRKYYSIYDLD 408
>dbj|BAB08016.1| (AB026513) cytochrome c oxidase subunit I [Campages sp. Cam]
Length = 406
Score = 34.8 bits (78), Expect = 4.0
Identities = 66/292 (22%), Positives = 107/292 (36%), Gaps = 36/292 (12%)
Query: 174 GWFHYHKAAPKLAWFQDVESM--LNHHLAGLLG-LGSLSWAGHQVHVSLPINQFLNAGVD 230
GW Y + +A F + + HLAG LG++++ V + + GV+
Sbjct: 101 GWTLYPPLSGSVAHFGGSVDLAIFSLHLAGASSILGAINFISTVVRLKVD-------GVN 153
Query: 231 PKEIPLPHEFILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTD 290
+ IPL ++ +L L GA + F GG DPV
Sbjct: 154 HEHIPLFVWSVVITVVLLLLSLPVLAGAITMLLTDRNFNTSFFDPSGGGDPVLFQHLFWF 213
Query: 291 IAHHHLAIAIL--FLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHA 348
H + I IL F I H+ + HG K+ H G GL + + H
Sbjct: 214 FGHPEVYILILPGFGIISHI-----AMLHGRKEEAFGHLGMIYAMLSIGLLGFIVWAHH- 267
Query: 349 QLSINL-----AMLGSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAA 403
++ + A S T+I+A + + +LAT +G+ + W GF+
Sbjct: 268 MFTVGMDVDTRAYFTSATMIIAVPT-GIKVFSWLATIFGSSSKFYPQVYWALGFVF---- 322
Query: 404 AHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWV----CIFLGFHSFG 451
+F + N LD VL + +H ++V +F F FG
Sbjct: 323 ----LFTIGGLTGVVLANSSLDTVLHDTYYVTAHFHYVLSMGAVFALFAGFG 370
>emb|CAB45538.1| (AJ242660) photosystem I P700 chlorophyll [Sinapis alba]
Length = 17
Score = 34.8 bits (78), Expect = 4.0
Identities = 16/16 (100%), Positives = 16/16 (100%)
Query: 1 MIIRSPEPEVKILVDR 16
MIIRSPEPEVKILVDR
Sbjct: 1 MIIRSPEPEVKILVDR 16
>pir||S14661 photosystem I protein psaA - maize (fragment)
emb|CAA41109.1| (X58080) psaA product, photosystem I protein [Zea mays]
Length = 18
Score = 34.8 bits (78), Expect = 4.0
Identities = 16/18 (88%), Positives = 16/18 (88%)
Query: 1 MIIRSPEPEVKILVDRDP 18
MIIRS EPEVKI VDRDP
Sbjct: 1 MIIRSSEPEVKIAVDRDP 18
>gb|AAG19813.1| (AE005065) Vng1524c [Halobacterium sp. NRC-1]
Length = 458
Score = 34.0 bits (76), Expect = 6.9
Identities = 27/91 (29%), Positives = 41/91 (44%), Gaps = 11/91 (12%)
Query: 454 IHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALAPGITA------PGATTGTSLTW 507
I ++ + LGR +++ +++P I + L G T P T S TW
Sbjct: 271 ISDEIQAGLGRTGELWGIDHTEIEP---DVITSAKGLRVGATVANEDLFPSETGRLSSTW 327
Query: 508 GGGDLVAVGNKVALLPIPLGTADFLVHHIHA 538
G GD+VA VA I T+D L+ H+ A
Sbjct: 328 GAGDIVAAAQGVA--TIDAITSDGLLDHVVA 356
>gi|11499293 hypothetical protein [Archaeoglobus fulgidus]
pir||F69462 hypothetical protein AF1703 - Archaeoglobus fulgidus
gb|AAB89558.1| (AE000986) A. fulgidus predicted coding region AF1703
[Archaeoglobus fulgidus]
Length = 208
Score = 34.0 bits (76), Expect = 6.9
Identities = 28/82 (34%), Positives = 40/82 (48%), Gaps = 7/82 (8%)
Query: 484 IQNTHALAPG-ITAPGATTGTSLTWGGGDLVAVGNKVALLP----IPLGTADFLVHH--I 536
I ++ ALAPG +TA A G W GG V+VG+ LP I G A L H +
Sbjct: 12 ISSSGALAPGPLTAATAAIGVKRGWKGGFWVSVGHAAVELPLVILIATGVAVVLTHSSAL 71
Query: 537 HAFTIHVTVLILLKGVLFARSS 558
++ V++L L A+S+
Sbjct: 72 SLLSVAGGVMLLSFAFLTAKSA 93
Database: nr
Posted date: Mar 14, 2001 1:25 PM
Number of letters in database: 201,912,487
Number of sequences in database: 640,428
Lambda K H
0.326 0.141 0.464
Gapped
Lambda K H
0.270 0.0470 0.230
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 332955997
Number of Sequences: 640428
Number of extensions: 14779056
Number of successful extensions: 39312
Number of sequences better than 10.0: 206
Number of HSP's better than 10.0 without gapping: 202
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 38199
Number of HSP's gapped (non-prelim): 224
length of query: 750
length of database: 201,912,487
effective HSP length: 49
effective length of query: 701
effective length of database: 170,531,515
effective search space: 119542592015
effective search space used: 119542592015
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.8 bits)
X3: 64 (24.9 bits)
S1: 40 (21.6 bits)
S2: 75 (33.6 bits)
Lotus japonicus(Cp)