Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
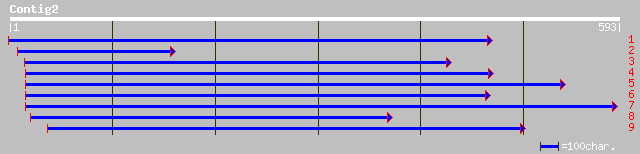
Query= KMC000038A_C02 KMC000038A_c02
(593 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T04011 hypothetical protein T5L19.200 - Arabidopsis thalian... 74 1e-12
ref|NP_192745.2| KH domain protein; protein id: At4g10070.1 [Ara... 74 1e-12
gb|AAN71223.1| GM32356p [Drosophila melanogaster] 63 2e-09
ref|NP_695271.1| narrowly conserved hypothetical protein [Bifido... 60 2e-08
gb|AAC47010.1| fibroin-3 60 2e-08
>pir||T04011 hypothetical protein T5L19.200 - Arabidopsis thaliana
gi|4539010|emb|CAB39631.1| putative DNA-directed RNA
polymerase [Arabidopsis thaliana]
gi|7267703|emb|CAB78130.1| putative DNA-directed RNA
polymerase [Arabidopsis thaliana]
Length = 748
Score = 73.9 bits (180), Expect = 1e-12
Identities = 59/146 (40%), Positives = 69/146 (46%), Gaps = 15/146 (10%)
Frame = -3
Query: 591 PAADGYNH--PPAASGPVYAQPGGQP---TYSQPSAQPAASYAQGYGSYPSQQTYPEQSA 427
P +D YN+ A+GP Y Q QP TY Q AQ AA A GYG Q A
Sbjct: 628 PPSDAYNNGTQTPATGPAYQQQSVQPASSTYDQTGAQQAA--AAGYGG---------QVA 676
Query: 426 PNNAVYGYQAPQDPSYGSAAAPAYSAAQSGQPGYAQPTPTQTGYE-----QSAGYAAVPA 262
P Y Y Q P+YGS AA Y+Q PTQTGYE Q+A YA P
Sbjct: 677 PTGG-YTYPTSQ-PAYGSQAA------------YSQAAPTQTGYEQQPATQAAVYATAPG 722
Query: 261 AS-----APAAAYPQYDTAQVYGAPR 199
+ +P +AY QYD +QVY PR
Sbjct: 723 TAPVKTQSPQSAYAQYDASQVYATPR 748
Score = 39.3 bits (90), Expect = 0.038
Identities = 41/134 (30%), Positives = 50/134 (36%), Gaps = 17/134 (12%)
Frame = -3
Query: 564 PAASGPVYAQPGGQPTYSQPSAQPAASYAQGYG-SYPSQQTYPEQS---APNNAVYGYQA 397
P SGPV + G P SQ S S+ YG + P QT +Q+ Y
Sbjct: 468 PPPSGPVPSPAFGGPPLSQVSYGYGQSHGPEYGHAAPYSQTGYQQTYGQTYEQPKYDSNP 527
Query: 396 PQDPSYGSAAAPAYSAAQSG-----QPGY----AQPTPTQTGYEQSAGYAAVPAASAP-- 250
P P YG + PA QSG QPG Q P Q GY AA + P
Sbjct: 528 PMQPPYGGSYPPA-GGGQSGYYQMQQPGVRPYGMQQGPVQQGYGPPQPAAAASSGDVPYQ 586
Query: 249 --AAAYPQYDTAQV 214
A P Y + +
Sbjct: 587 GATPAAPSYGSTNM 600
Score = 36.6 bits (83), Expect = 0.25
Identities = 39/137 (28%), Positives = 43/137 (30%), Gaps = 18/137 (13%)
Frame = -3
Query: 576 YNHPPAASGPVYAQPGGQPTYSQPSAQPAASYAQGYG-SYP----SQQTYPEQSAPNNAV 412
Y H S Y Q GQ TY QP YG SYP Q Y + P
Sbjct: 499 YGHAAPYSQTGYQQTYGQ-TYEQPKYDSNPPMQPPYGGSYPPAGGGQSGYYQMQQPGVRP 557
Query: 411 YG---------YQAPQDPSYGSAAAPAYSAAQSGQPGYAQ----PTPTQTGYEQSAGYAA 271
YG Y PQ + S+ Y A P Y P Q GY S G
Sbjct: 558 YGMQQGPVQQGYGPPQPAAAASSGDVPYQGATPAAPSYGSTNMAPQQQQYGYTSSDG--- 614
Query: 270 VPAASAPAAAYPQYDTA 220
YP Y +A
Sbjct: 615 ----PVQQQTYPSYSSA 627
Score = 32.3 bits (72), Expect = 4.7
Identities = 42/145 (28%), Positives = 49/145 (32%), Gaps = 45/145 (31%)
Frame = -3
Query: 579 GYNHPPAASGPVYAQPGGQPTYSQPSAQPAASYAQGY---GSYPSQQTY---------PE 436
GYN P A P PGG P + S P A + Y G Y SQ +Y P
Sbjct: 377 GYNQP--AYRP--QGPGGPPQWG--SRGPHAPHPYDYHPRGPYSSQGSYYNSPGFGGYPP 430
Query: 435 QSAPNNAVYGYQAPQDPSY----------------------GSAAAPA----------YS 352
Q P YG Q P Y G +PA Y
Sbjct: 431 QHMPPRGGYGTDWDQRPPYSGPYNYYGRQGAQSAGPVPPPSGPVPSPAFGGPPLSQVSYG 490
Query: 351 AAQSGQPGYAQPTP-TQTGYEQSAG 280
QS P Y P +QTGY+Q+ G
Sbjct: 491 YGQSHGPEYGHAAPYSQTGYQQTYG 515
>ref|NP_192745.2| KH domain protein; protein id: At4g10070.1 [Arabidopsis thaliana]
Length = 725
Score = 73.9 bits (180), Expect = 1e-12
Identities = 59/146 (40%), Positives = 69/146 (46%), Gaps = 15/146 (10%)
Frame = -3
Query: 591 PAADGYNH--PPAASGPVYAQPGGQP---TYSQPSAQPAASYAQGYGSYPSQQTYPEQSA 427
P +D YN+ A+GP Y Q QP TY Q AQ AA A GYG Q A
Sbjct: 605 PPSDAYNNGTQTPATGPAYQQQSVQPASSTYDQTGAQQAA--AAGYGG---------QVA 653
Query: 426 PNNAVYGYQAPQDPSYGSAAAPAYSAAQSGQPGYAQPTPTQTGYE-----QSAGYAAVPA 262
P Y Y Q P+YGS AA Y+Q PTQTGYE Q+A YA P
Sbjct: 654 PTGG-YTYPTSQ-PAYGSQAA------------YSQAAPTQTGYEQQPATQAAVYATAPG 699
Query: 261 AS-----APAAAYPQYDTAQVYGAPR 199
+ +P +AY QYD +QVY PR
Sbjct: 700 TAPVKTQSPQSAYAQYDASQVYATPR 725
Score = 39.3 bits (90), Expect = 0.038
Identities = 41/134 (30%), Positives = 50/134 (36%), Gaps = 17/134 (12%)
Frame = -3
Query: 564 PAASGPVYAQPGGQPTYSQPSAQPAASYAQGYG-SYPSQQTYPEQS---APNNAVYGYQA 397
P SGPV + G P SQ S S+ YG + P QT +Q+ Y
Sbjct: 445 PPPSGPVPSPAFGGPPLSQVSYGYGQSHGPEYGHAAPYSQTGYQQTYGQTYEQPKYDSNP 504
Query: 396 PQDPSYGSAAAPAYSAAQSG-----QPGY----AQPTPTQTGYEQSAGYAAVPAASAP-- 250
P P YG + PA QSG QPG Q P Q GY AA + P
Sbjct: 505 PMQPPYGGSYPPA-GGGQSGYYQMQQPGVRPYGMQQGPVQQGYGPPQPAAAASSGDVPYQ 563
Query: 249 --AAAYPQYDTAQV 214
A P Y + +
Sbjct: 564 GATPAAPSYGSTNM 577
Score = 36.6 bits (83), Expect = 0.25
Identities = 39/137 (28%), Positives = 43/137 (30%), Gaps = 18/137 (13%)
Frame = -3
Query: 576 YNHPPAASGPVYAQPGGQPTYSQPSAQPAASYAQGYG-SYP----SQQTYPEQSAPNNAV 412
Y H S Y Q GQ TY QP YG SYP Q Y + P
Sbjct: 476 YGHAAPYSQTGYQQTYGQ-TYEQPKYDSNPPMQPPYGGSYPPAGGGQSGYYQMQQPGVRP 534
Query: 411 YG---------YQAPQDPSYGSAAAPAYSAAQSGQPGYAQ----PTPTQTGYEQSAGYAA 271
YG Y PQ + S+ Y A P Y P Q GY S G
Sbjct: 535 YGMQQGPVQQGYGPPQPAAAASSGDVPYQGATPAAPSYGSTNMAPQQQQYGYTSSDG--- 591
Query: 270 VPAASAPAAAYPQYDTA 220
YP Y +A
Sbjct: 592 ----PVQQQTYPSYSSA 604
Score = 32.3 bits (72), Expect = 4.7
Identities = 42/145 (28%), Positives = 49/145 (32%), Gaps = 45/145 (31%)
Frame = -3
Query: 579 GYNHPPAASGPVYAQPGGQPTYSQPSAQPAASYAQGY---GSYPSQQTY---------PE 436
GYN P A P PGG P + S P A + Y G Y SQ +Y P
Sbjct: 354 GYNQP--AYRP--QGPGGPPQWG--SRGPHAPHPYDYHPRGPYSSQGSYYNSPGFGGYPP 407
Query: 435 QSAPNNAVYGYQAPQDPSY----------------------GSAAAPA----------YS 352
Q P YG Q P Y G +PA Y
Sbjct: 408 QHMPPRGGYGTDWDQRPPYSGPYNYYGRQGAQSAGPVPPPSGPVPSPAFGGPPLSQVSYG 467
Query: 351 AAQSGQPGYAQPTP-TQTGYEQSAG 280
QS P Y P +QTGY+Q+ G
Sbjct: 468 YGQSHGPEYGHAAPYSQTGYQQTYG 492
>gb|AAN71223.1| GM32356p [Drosophila melanogaster]
Length = 391
Score = 63.2 bits (152), Expect = 2e-09
Identities = 46/115 (40%), Positives = 54/115 (46%), Gaps = 6/115 (5%)
Frame = -3
Query: 528 GQPTYSQPSAQPAASYAQGYGSYPSQ-QTYP--EQSAPNNAVYGYQAPQDPSYGSAA--A 364
GQ Y P+ Q + GYG P+ T P E + P + GY AP P+Y + A A
Sbjct: 143 GQNNYVAPAPQSPSYGGDGYGVLPNGGYTKPDTEYNGPGDGDAGYVAPAAPAYEAPAPPA 202
Query: 363 PAYSAAQSGQPGYAQPTPTQTGYEQSAGYAAVPAASAPAAAYPQYDT-AQVYGAP 202
PAY A P Y P P YE A A PA APA A P Y+T A Y AP
Sbjct: 203 PAYEAPAPPAPVYEAPAPAAPAYEAPA--PAAPAYEAPAPAAPAYETPATDYSAP 255
Score = 42.4 bits (98), Expect = 0.005
Identities = 44/141 (31%), Positives = 55/141 (38%), Gaps = 11/141 (7%)
Frame = -3
Query: 591 PAADGYNHP---------PAASGPVYAQPG-GQPTYSQPSAQPAASYAQGYGSYPSQQT- 445
PAA Y P PA PVY P P Y P A A +Y + P+ +T
Sbjct: 190 PAAPAYEAPAPPAPAYEAPAPPAPVYEAPAPAAPAYEAP-APAAPAYEAPAPAAPAYETP 248
Query: 444 YPEQSAPNNAVYGYQAPQDPSYGSAAAPAYSAAQSGQPGYAQPTPTQTGYEQSAGYAAVP 265
+ SAP Y+ P S+ YS Q QP Y P Q Y Q Y + P
Sbjct: 249 ATDYSAPAPPAPAYEPP-----ASSYTQGYS--QPAQPSYVGAPPAQIVY-QPIIYLSTP 300
Query: 264 AASAPAAAYPQYDTAQVYGAP 202
AS + + +YD Q Y P
Sbjct: 301 LASKSSTSQVEYDD-QKYVTP 320
Score = 34.3 bits (77), Expect = 1.2
Identities = 37/128 (28%), Positives = 44/128 (33%), Gaps = 35/128 (27%)
Frame = -3
Query: 591 PAADGYNHPPAASGPVYAQPGGQPTYSQPSAQ----PAASYAQGYGSYPSQQTY----PE 436
PAA Y PA + P Y P + P A PA+SY QGY S P+Q +Y P
Sbjct: 230 PAAPAYE-APAPAAPAYETPATDYSAPAPPAPAYEPPASSYTQGY-SQPAQPSYVGAPPA 287
Query: 435 QSAPNNAVY-------------------GYQAPQDPSYGSAAAPAYSA--------AQSG 337
Q +Y Y P P AP Y A A
Sbjct: 288 QIVYQPIIYLSTPLASKSSTSQVEYDDQKYVTPTAPPSPPPPAPVYEAPSQSCYQPAAPP 347
Query: 336 QPGYAQPT 313
P YA P+
Sbjct: 348 APNYATPS 355
>ref|NP_695271.1| narrowly conserved hypothetical protein [Bifidobacterium longum
NCC2705] gi|23325229|gb|AAN23907.1|AE014618_4 narrowly
conserved hypothetical protein [Bifidobacterium longum
NCC2705]
Length = 216
Score = 60.5 bits (145), Expect = 2e-08
Identities = 50/121 (41%), Positives = 57/121 (46%), Gaps = 3/121 (2%)
Frame = -3
Query: 555 SGPVYAQPG--GQPTYSQPSAQPAASYAQGYGSYPSQQTYPEQSAPNNAVYGYQAPQDPS 382
S P AQP QP+Y Q +AQ QGYG+ P Y + +A + YG Q PQ
Sbjct: 24 SVPQPAQPAQPAQPSYGQDAAQQP----QGYGA-PQPAAYAQPAAGS---YGQQQPQ--- 72
Query: 381 YGSAAAPAYSAAQSGQPGYAQPTPTQTGYEQSAGYAAVPAASAP-AAAYPQYDTAQVYGA 205
YG A AY+ Q GQP Y QP GY Q GY A P A A PQY A A
Sbjct: 73 YGQPAG-AYAQPQYGQPQYGQPAQPAAGYGQQPGYG--QAYQQPNAYAQPQYTAAPYAAA 129
Query: 204 P 202
P
Sbjct: 130 P 130
>gb|AAC47010.1| fibroin-3
Length = 636
Score = 60.5 bits (145), Expect = 2e-08
Identities = 49/120 (40%), Positives = 54/120 (44%)
Frame = -3
Query: 588 AADGYNHPPAASGPVYAQPGGQPTYSQPSAQPAASYAQGYGSYPSQQTYPEQSAPNNAVY 409
AA GY P + GP PGGQ Y P A AA+ A GYG QQ P Q P
Sbjct: 169 AAGGYG-PGSGQGPGQQGPGGQGPYG-PGASAAAAAAGGYGPGSGQQG-PGQQGPGQQGP 225
Query: 408 GYQAPQDPSYGSAAAPAYSAAQSGQPGYAQPTPTQTGYEQSAGYAAVPAASAPAAAYPQY 229
G Q P YG A+ A +AA PGY Q P Q G Y P ASA +AA Y
Sbjct: 226 GGQGP----YGPGASAAAAAAGGYGPGYGQQGPGQQGPGGQGPYG--PGASAASAASGGY 279
Score = 54.3 bits (129), Expect = 1e-06
Identities = 48/121 (39%), Positives = 50/121 (40%), Gaps = 1/121 (0%)
Frame = -3
Query: 588 AADGYNHPPAASGPVYAQPGGQPTYSQPSAQPAASYAQGYGSYPSQQTYPEQSAPNNAVY 409
AA GY GP PGGQ Y P A AA+ A GYG P P Q P
Sbjct: 135 AAGGYGPGSGQQGPGQQGPGGQGPYG-PGASAAAAAAGGYG--PGSGQGPGQQGP----- 186
Query: 408 GYQAPQDPSYGSAAAPAYS-AAQSGQPGYAQPTPTQTGYEQSAGYAAVPAASAPAAAYPQ 232
G Q P P +AAA A SGQ G Q P Q G Y P ASA AAA
Sbjct: 187 GGQGPYGPGASAAAAAAGGYGPGSGQQGPGQQGPGQQGPGGQGPYG--PGASAAAAAAGG 244
Query: 231 Y 229
Y
Sbjct: 245 Y 245
Score = 51.6 bits (122), Expect = 7e-06
Identities = 43/123 (34%), Positives = 46/123 (36%), Gaps = 5/123 (4%)
Frame = -3
Query: 588 AADGYNHPPAASGPVYAQPGGQPTYSQPSAQPAASYAQGYGSYPSQQ----TYPEQSAPN 421
A+ GY GP PGGQ Y P A AA+ A GYG QQ P Q P
Sbjct: 275 ASGGYGPGSGQQGPGQQGPGGQGPYG-PGASAAAAAAGGYGPGSGQQGPGQQGPGQQGPG 333
Query: 420 NAVYGYQAPQDPSYGSAAAPAYS-AAQSGQPGYAQPTPTQTGYEQSAGYAAVPAASAPAA 244
G Q P P +AAA A SGQ G Q P Q G Q P P
Sbjct: 334 QQGPGGQGPYGPGASAAAAAAGGYGPGSGQQGPGQQGPGQQGPGQQGPGQQGPGQQGPGQ 393
Query: 243 AYP 235
P
Sbjct: 394 QGP 396
Score = 47.0 bits (110), Expect = 2e-04
Identities = 47/126 (37%), Positives = 51/126 (40%), Gaps = 6/126 (4%)
Frame = -3
Query: 588 AADGYN-----HPPAASGPVYAQPGGQPTYSQPSAQPAASYAQGYGSYPSQQTYPEQSAP 424
AA GY P+ GP PGGQ Y P A AA+ A GYG QQ P
Sbjct: 36 AAGGYGPGSGQQGPSQQGPGQQGPGGQGPYG-PGASAAAAAAGGYGPGSGQQ------GP 88
Query: 423 NNAVYGYQAPQDPSYGSAAAPAY-SAAQSGQPGYAQPTPTQTGYEQSAGYAAVPAASAPA 247
G Q P P +AAA A + SGQ G Q P Q G P ASA A
Sbjct: 89 -----GGQGPYGPGSSAAAAAAGGNGPGSGQQGAGQQGPGQQG----------PGASAAA 133
Query: 246 AAYPQY 229
AA Y
Sbjct: 134 AAAGGY 139
Score = 46.2 bits (108), Expect = 3e-04
Identities = 40/116 (34%), Positives = 43/116 (36%)
Frame = -3
Query: 564 PAASGPVYAQPGGQPTYSQPSAQPAASYAQGYGSYPSQQTYPEQSAPNNAVYGYQAPQDP 385
P GP PG Q Y P A AA+ A GYG QQ P Q P G Q P
Sbjct: 10 PGQQGPGQQGPGQQGPYG-PGASAAAAAAGGYGPGSGQQG-PSQQGPGQQGPGGQGP--- 64
Query: 384 SYGSAAAPAYSAAQSGQPGYAQPTPTQTGYEQSAGYAAVPAASAPAAAYPQYDTAQ 217
YG A+ A +AA PG Q P G AA AA Q Q
Sbjct: 65 -YGPGASAAAAAAGGYGPGSGQQGPGGQGPYGPGSSAAAAAAGGNGPGSGQQGAGQ 119
Score = 45.4 bits (106), Expect = 5e-04
Identities = 49/154 (31%), Positives = 52/154 (32%), Gaps = 22/154 (14%)
Frame = -3
Query: 564 PAASGPVYAQPGGQPTYSQPSAQPAASYAQGYGSYPSQQ-------------------TY 442
P GP PGGQ Y P A AA A GYG QQ
Sbjct: 396 PGQQGPGQQGPGGQGAYG-PGASAAAGAAGGYGPGSGQQGPGQQGPGQQGPGQQGPGQQG 454
Query: 441 PEQSAPNNAVYGYQAPQDPSYGSAAAPAYS-AAQSGQPGYAQPTPTQTGYEQSAGY--AA 271
P Q P G Q P P +AAA A SGQ G Q P Q G Y A
Sbjct: 455 PGQQGPGQQGPGQQGPYGPGASAAAAAAGGYGPGSGQQGPGQQGPGQQGPGGQGPYGPGA 514
Query: 270 VPAASAPAAAYPQYDTAQVYGAPR*SL*EPNHSS 169
AA + PQ + V A L P SS
Sbjct: 515 ASAAVSVGGYGPQSSSVPVASAVASRLSSPAASS 548
Score = 41.2 bits (95), Expect = 0.010
Identities = 44/141 (31%), Positives = 47/141 (33%), Gaps = 31/141 (21%)
Frame = -3
Query: 564 PAASGPVYAQPGGQPTYSQPSAQPAASYAQGYGSYPSQQTYPEQSAPNNAVYGYQAP--- 394
P GP PGGQ Y P A AA+ A GYG QQ P Q P G Q P
Sbjct: 327 PGQQGPGQQGPGGQGPYG-PGASAAAAAAGGYGPGSGQQG-PGQQGPGQQGPGQQGPGQQ 384
Query: 393 -------------------QDP----SYGSAAAPAYSAAQ-----SGQPGYAQPTPTQTG 298
Q P +YG A+ A AA SGQ G Q P Q G
Sbjct: 385 GPGQQGPGQQGPGQQGPGQQGPGGQGAYGPGASAAAGAAGGYGPGSGQQGPGQQGPGQQG 444
Query: 297 YEQSAGYAAVPAASAPAAAYP 235
Q P P P
Sbjct: 445 PGQQGPGQQGPGQQGPGQQGP 465
Score = 36.2 bits (82), Expect = 0.32
Identities = 30/77 (38%), Positives = 32/77 (40%), Gaps = 1/77 (1%)
Frame = -3
Query: 456 SQQTYPEQSAPNNAVYGYQAPQDPSYGSAAAPAYS-AAQSGQPGYAQPTPTQTGYEQSAG 280
S Q P Q P G Q P P +AAA A SGQ G +Q P Q G
Sbjct: 5 SGQQGPGQQGPGQQGPGQQGPYGPGASAAAAAAGGYGPGSGQQGPSQQGPGQQGPGGQGP 64
Query: 279 YAAVPAASAPAAAYPQY 229
Y P ASA AAA Y
Sbjct: 65 YG--PGASAAAAAAGGY 79
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 535,327,507
Number of Sequences: 1393205
Number of extensions: 11905392
Number of successful extensions: 57557
Number of sequences better than 10.0: 1906
Number of HSP's better than 10.0 without gapping: 43890
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 52573
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22854740960
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)