Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000031A_C01 KMC000031A_c01
(527 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
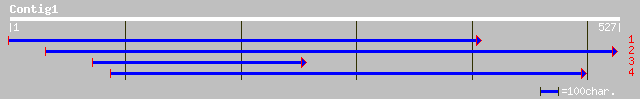
ref|NP_189060.2| unknown protein; protein id: At3g24180.1, suppo... 81 9e-15
dbj|BAB01359.1| emb|CAB39630.1~gene_id:MUJ8.8~strong similarity ... 81 9e-15
gb|AAG16864.1|AC069145_13 unknown protein [Oryza sativa] 72 5e-12
gb|AAL31035.1|AC078948_19 unknown protein [Oryza sativa] 72 5e-12
ref|NP_174631.1| hypothetical protein; protein id: At1g33700.1 [... 33 2.1
>ref|NP_189060.2| unknown protein; protein id: At3g24180.1, supported by cDNA:
gi_17529231 [Arabidopsis thaliana]
gi|17529232|gb|AAL38843.1| unknown protein [Arabidopsis
thaliana]
Length = 950
Score = 80.9 bits (198), Expect = 9e-15
Identities = 37/69 (53%), Positives = 53/69 (76%)
Frame = -1
Query: 527 LIYMRPLXIWGMPYALTLPKAILEAPKMNIMDRIPISPLNGGFSHH*TGVRKIATKAKCF 348
LIYMRPL IWGM +AL+LPKAIL+AP++N+MDR+ +SP + FS++ + + KAKCF
Sbjct: 885 LIYMRPLAIWGMQWALSLPKAILDAPQINMMDRVHLSPRSRRFSNN---FKVVKHKAKCF 941
Query: 347 SHSVFNCAC 321
+S +C+C
Sbjct: 942 GNSALSCSC 950
>dbj|BAB01359.1| emb|CAB39630.1~gene_id:MUJ8.8~strong similarity to unknown protein
[Arabidopsis thaliana]
Length = 937
Score = 80.9 bits (198), Expect = 9e-15
Identities = 37/69 (53%), Positives = 53/69 (76%)
Frame = -1
Query: 527 LIYMRPLXIWGMPYALTLPKAILEAPKMNIMDRIPISPLNGGFSHH*TGVRKIATKAKCF 348
LIYMRPL IWGM +AL+LPKAIL+AP++N+MDR+ +SP + FS++ + + KAKCF
Sbjct: 872 LIYMRPLAIWGMQWALSLPKAILDAPQINMMDRVHLSPRSRRFSNN---FKVVKHKAKCF 928
Query: 347 SHSVFNCAC 321
+S +C+C
Sbjct: 929 GNSALSCSC 937
>gb|AAG16864.1|AC069145_13 unknown protein [Oryza sativa]
Length = 444
Score = 71.6 bits (174), Expect = 5e-12
Identities = 37/69 (53%), Positives = 45/69 (64%)
Frame = -1
Query: 527 LIYMRPLXIWGMPYALTLPKAILEAPKMNIMDRIPISPLNGGFSHH*TGVRKIATKAKCF 348
LIYMRPL IW M +A + PKAIL+APK+N+MDRI +SP + VRKIA +CF
Sbjct: 377 LIYMRPLAIWAMQWARSPPKAILDAPKVNLMDRIHLSPQMIRAMNE-INVRKIAPDNRCF 435
Query: 347 SHSVFNCAC 321
S F C C
Sbjct: 436 PSSAFRCEC 444
>gb|AAL31035.1|AC078948_19 unknown protein [Oryza sativa]
Length = 967
Score = 71.6 bits (174), Expect = 5e-12
Identities = 37/69 (53%), Positives = 45/69 (64%)
Frame = -1
Query: 527 LIYMRPLXIWGMPYALTLPKAILEAPKMNIMDRIPISPLNGGFSHH*TGVRKIATKAKCF 348
LIYMRPL IW M +A + PKAIL+APK+N+MDRI +SP + VRKIA +CF
Sbjct: 900 LIYMRPLAIWAMQWARSPPKAILDAPKVNLMDRIHLSPQMIRAMNE-INVRKIAPDNRCF 958
Query: 347 SHSVFNCAC 321
S F C C
Sbjct: 959 PSSAFRCEC 967
>ref|NP_174631.1| hypothetical protein; protein id: At1g33700.1 [Arabidopsis
thaliana]
Length = 905
Score = 33.1 bits (74), Expect = 2.1
Identities = 13/20 (65%), Positives = 16/20 (80%)
Frame = -1
Query: 527 LIYMRPLXIWGMPYALTLPK 468
L YMRPL IWG+ +A T+PK
Sbjct: 827 LCYMRPLAIWGIQWAHTMPK 846
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 440,513,319
Number of Sequences: 1393205
Number of extensions: 9298795
Number of successful extensions: 17186
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 16866
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 17175
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17308240320
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)