Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000011A_C01 KMC000011A_c01
(670 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
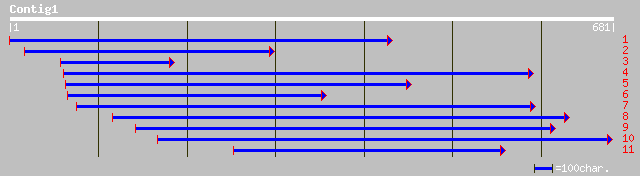
gb|AAL66981.1| unknown protein [Arabidopsis thaliana] gi|2505501... 189 4e-47
ref|NP_176226.2| RRM-containing protein; protein id: At1g60200.1... 189 4e-47
gb|AAM22757.1| unknown [Deschampsia antarctica] 183 2e-45
pir||T02272 hypothetical protein T13D8.9 - Arabidopsis thaliana ... 175 6e-43
dbj|BAC40049.1| unnamed protein product [Mus musculus] 112 6e-24
>gb|AAL66981.1| unknown protein [Arabidopsis thaliana] gi|25055015|gb|AAN71971.1|
unknown protein [Arabidopsis thaliana]
Length = 899
Score = 189 bits (479), Expect = 4e-47
Identities = 93/122 (76%), Positives = 108/122 (88%)
Frame = -2
Query: 666 KSKTHDNRRLLDAKQLIDMIPKTKEELFSYEIDWTVYDKHQLHERMRPWISKKIQEFLGE 487
K+K D + LDAKQLID IPKTKE+LFSYEI+W +YDKHQ+HERMRPWISKKI EFLGE
Sbjct: 779 KAKIIDTK-FLDAKQLIDTIPKTKEDLFSYEINWAMYDKHQVHERMRPWISKKIMEFLGE 837
Query: 486 EENTLIDYIVSSTQEHVKASPMLERLQIILDEEAEMFVLKMWRMLIFEIKRVETGLALRS 307
EE TL+D+IVS+TQ+HV+AS MLE LQ ILDEEAEMFVLKMWRMLIFEIKRVE G+ ++S
Sbjct: 838 EEATLVDFIVSNTQQHVQASQMLELLQSILDEEAEMFVLKMWRMLIFEIKRVEAGVPVKS 897
Query: 306 KS 301
K+
Sbjct: 898 KA 899
>ref|NP_176226.2| RRM-containing protein; protein id: At1g60200.1 [Arabidopsis
thaliana]
Length = 750
Score = 189 bits (479), Expect = 4e-47
Identities = 93/122 (76%), Positives = 108/122 (88%)
Frame = -2
Query: 666 KSKTHDNRRLLDAKQLIDMIPKTKEELFSYEIDWTVYDKHQLHERMRPWISKKIQEFLGE 487
K+K D + LDAKQLID IPKTKE+LFSYEI+W +YDKHQ+HERMRPWISKKI EFLGE
Sbjct: 630 KAKIIDTK-FLDAKQLIDTIPKTKEDLFSYEINWAMYDKHQVHERMRPWISKKIMEFLGE 688
Query: 486 EENTLIDYIVSSTQEHVKASPMLERLQIILDEEAEMFVLKMWRMLIFEIKRVETGLALRS 307
EE TL+D+IVS+TQ+HV+AS MLE LQ ILDEEAEMFVLKMWRMLIFEIKRVE G+ ++S
Sbjct: 689 EEATLVDFIVSNTQQHVQASQMLELLQSILDEEAEMFVLKMWRMLIFEIKRVEAGVPVKS 748
Query: 306 KS 301
K+
Sbjct: 749 KA 750
>gb|AAM22757.1| unknown [Deschampsia antarctica]
Length = 192
Score = 183 bits (464), Expect = 2e-45
Identities = 86/117 (73%), Positives = 105/117 (89%)
Frame = -2
Query: 651 DNRRLLDAKQLIDMIPKTKEELFSYEIDWTVYDKHQLHERMRPWISKKIQEFLGEEENTL 472
+N+++ DAKQLIDMIP+TKE+LFSY+I+W +Y+KH+LHERMRPWISKKI EFLGEEE+TL
Sbjct: 76 ENKKIADAKQLIDMIPRTKEDLFSYDINWAIYEKHELHERMRPWISKKIIEFLGEEESTL 135
Query: 471 IDYIVSSTQEHVKASPMLERLQIILDEEAEMFVLKMWRMLIFEIKRVETGLALRSKS 301
++YIVS T++HV AS MLE LQ ILD EAEMFVLKMWRMLIFEIK+VE GL+ R K+
Sbjct: 136 VEYIVSCTKDHVHASKMLELLQSILDVEAEMFVLKMWRMLIFEIKKVEAGLSGRGKA 192
>pir||T02272 hypothetical protein T13D8.9 - Arabidopsis thaliana
gi|3249070|gb|AAC24054.1| Contains similarity to siah
binding protein 1 (SiahBP1) gb|U51586 from Homo sapiens.
ESTs gb|T43314, gb|T43315 and gb|R90521, gb|T75905
[Arabidopsis thaliana]
Length = 781
Score = 175 bits (443), Expect = 6e-43
Identities = 93/147 (63%), Positives = 108/147 (73%), Gaps = 25/147 (17%)
Frame = -2
Query: 666 KSKTHDNRRLLDAKQLIDMIPKTKEELFSYEIDWTVYDK--------------------- 550
K+K D + LDAKQLID IPKTKE+LFSYEI+W +YDK
Sbjct: 636 KAKIIDTK-FLDAKQLIDTIPKTKEDLFSYEINWAMYDKINVFFYLDCEFVLVLTCFTFF 694
Query: 549 ----HQLHERMRPWISKKIQEFLGEEENTLIDYIVSSTQEHVKASPMLERLQIILDEEAE 382
HQ+HERMRPWISKKI EFLGEEE TL+D+IVS+TQ+HV+AS MLE LQ ILDEEAE
Sbjct: 695 NIEKHQVHERMRPWISKKIMEFLGEEEATLVDFIVSNTQQHVQASQMLELLQSILDEEAE 754
Query: 381 MFVLKMWRMLIFEIKRVETGLALRSKS 301
MFVLKMWRMLIFEIKRVE G+ ++SK+
Sbjct: 755 MFVLKMWRMLIFEIKRVEAGVPVKSKA 781
>dbj|BAC40049.1| unnamed protein product [Mus musculus]
Length = 315
Score = 112 bits (279), Expect = 6e-24
Identities = 48/103 (46%), Positives = 73/103 (70%)
Frame = -2
Query: 627 KQLIDMIPKTKEELFSYEIDWTVYDKHQLHERMRPWISKKIQEFLGEEENTLIDYIVSST 448
K LI+ IP K ELF+Y +DW++ D + R+RPWI+KKI E++GEEE TL+D++ S
Sbjct: 211 KSLIEKIPTAKPELFAYPLDWSIVDSILMERRIRPWINKKIIEYIGEEEATLVDFVCSKV 270
Query: 447 QEHVKASPMLERLQIILDEEAEMFVLKMWRMLIFEIKRVETGL 319
H +L+ + ++LDEEAE+F++KMWR+LI+E + + GL
Sbjct: 271 MAHSSPQSILDDVAMVLDEEAEVFIVKMWRLLIYETEAKKIGL 313
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 587,042,719
Number of Sequences: 1393205
Number of extensions: 12559250
Number of successful extensions: 25832
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 24964
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25820
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29138478756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)