Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002318A_C01 KMC002318A_c01
(715 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
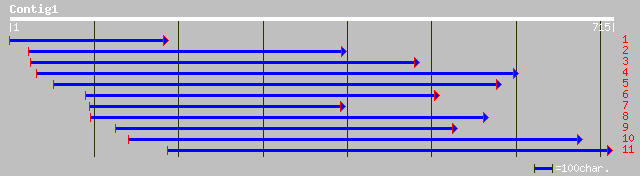
ref|NP_197190.1| TOM (target of myb1) -like protein; protein id:... 192 5e-48
gb|AAL58181.1|AC027037_3 hepatocyte growth factor-regulated tyro... 154 1e-36
ref|NP_563762.1| expressed protein; protein id: At1g06210.1, sup... 68 1e-10
gb|AAK27511.1|AF343969_1 ADP-ribosylation factor 60 [Coffea arab... 58 1e-07
gb|AAL65395.1| seed protein B32E [Oryza sativa] 53 5e-06
>ref|NP_197190.1| TOM (target of myb1) -like protein; protein id: At5g16880.1,
supported by cDNA: 1330., supported by cDNA:
gi_15983760, supported by cDNA: gi_17065127 [Arabidopsis
thaliana] gi|11358881|pir||T51543 TOM (target of
myb1)-like protein - Arabidopsis thaliana
gi|9755689|emb|CAC01701.1| TOM (target of myb1)-like
protein [Arabidopsis thaliana]
gi|15983761|gb|AAL10477.1| AT5g16880/F2K13_30
[Arabidopsis thaliana] gi|17065128|gb|AAL32718.1| TOM
(target of myb1)-like protein [Arabidopsis thaliana]
gi|21537352|gb|AAM61693.1| TOM (target of myb1)-like
protein [Arabidopsis thaliana]
gi|27311895|gb|AAO00913.1| TOM (target of myb1)-like
protein [Arabidopsis thaliana]
Length = 407
Score = 192 bits (487), Expect = 5e-48
Identities = 101/154 (65%), Positives = 120/154 (77%), Gaps = 6/154 (3%)
Frame = -2
Query: 714 SPQQDVLQDDLTTTLVQQCRRSQSTVQRIVETSGDNEALLFEALNVNDEIQKVLDKYEEV 535
SPQ D LQDDLTTTLVQQCR+SQ+TVQRI+ET+G+NEALLFEALNVNDE+ K L KYEE+
Sbjct: 254 SPQHDALQDDLTTTLVQQCRQSQTTVQRIIETAGENEALLFEALNVNDELVKTLSKYEEM 313
Query: 534 KKPTTAPLQPEPAMIPVAVEPDESPRHTKEDALIRKPAGSRTVVQ--GGSNDDMMDDLDE 361
KP+ EPAMIPVA EPD+SP H +E++L+RK +G R GGS DDMMDDLDE
Sbjct: 314 NKPSAPLTSHEPAMIPVAEEPDDSPIHGREESLVRKSSGVRGGFHGGGGSGDDMMDDLDE 373
Query: 360 MIFGKKSG--DASDAGHDTKKQQSS--KDDLISF 271
MIFGKK+G +++ HD KK+QSS DDLI F
Sbjct: 374 MIFGKKNGCDSSTNPDHDPKKEQSSSKNDDLIRF 407
>gb|AAL58181.1|AC027037_3 hepatocyte growth factor-regulated tyrosine kinase substrate-like
protein [Oryza sativa]
Length = 387
Score = 154 bits (389), Expect = 1e-36
Identities = 83/148 (56%), Positives = 108/148 (72%)
Frame = -2
Query: 714 SPQQDVLQDDLTTTLVQQCRRSQSTVQRIVETSGDNEALLFEALNVNDEIQKVLDKYEEV 535
SPQ++ L+DDLTTTLVQQC++ Q T+QRI+ET+GDNEA LFEAL+V+DE++KVL KY+E+
Sbjct: 254 SPQKEALKDDLTTTLVQQCQQCQRTIQRIIETAGDNEAQLFEALSVHDELEKVLSKYKEL 313
Query: 534 KKPTTAPLQPEPAMIPVAVEPDESPRHTKEDALIRKPAGSRTVVQGGSNDDMMDDLDEMI 355
K+P A + EPAMIPV VEP+ SPR +D + K AGS D+++ DLD+MI
Sbjct: 314 KEPVVAEPEAEPAMIPVTVEPENSPR--TKDGTVGKRAGS-------GADELLQDLDDMI 364
Query: 354 FGKKSGDASDAGHDTKKQQSSKDDLISF 271
FGKK G +S D K+Q KDD ISF
Sbjct: 365 FGKKGGTSSQ--QDRKEQ---KDDFISF 387
>ref|NP_563762.1| expressed protein; protein id: At1g06210.1, supported by cDNA:
gi_15450710 [Arabidopsis thaliana]
gi|25406904|pir||G86197 hypothetical protein [imported]
- Arabidopsis thaliana
gi|8844126|gb|AAF80218.1|AC025290_7 Contains similarity
to an ADP-ribosylation factor binding protein GGA1 from
Homo sapiens gb|AF190862 and contains a VHS PF|00790
domain. EST gb|BE037588 comes from this gene.
[Arabidopsis thaliana] gi|15450711|gb|AAK96627.1|
At1g06210/F9P14_4 [Arabidopsis thaliana]
gi|23308355|gb|AAN18147.1| At1g06210/F9P14_4
[Arabidopsis thaliana]
Length = 383
Score = 67.8 bits (164), Expect = 1e-10
Identities = 41/132 (31%), Positives = 72/132 (54%)
Frame = -2
Query: 693 QDDLTTTLVQQCRRSQSTVQRIVETSGDNEALLFEALNVNDEIQKVLDKYEEVKKPTTAP 514
+DDLT +L+++C++SQ +Q I+E++ D+E +LFEAL++NDE+Q+VL Y KKP
Sbjct: 256 EDDLTVSLMEKCKQSQPLIQMIIESTTDDEGVLFEALHLNDELQQVLSSY---KKPDETE 312
Query: 513 LQPEPAMIPVAVEPDESPRHTKEDALIRKPAGSRTVVQGGSNDDMMDDLDEMIFGKKSGD 334
+ + D P+ T+++ V + G++DD KK +
Sbjct: 313 KKASIVEQESSGSKDTGPKPTEQEE-------QEPVKKTGADDD-----------KKHSE 354
Query: 333 ASDAGHDTKKQQ 298
AS + + T K++
Sbjct: 355 ASGSSNKTVKEE 366
>gb|AAK27511.1|AF343969_1 ADP-ribosylation factor 60 [Coffea arabica]
Length = 135
Score = 58.2 bits (139), Expect = 1e-07
Identities = 22/60 (36%), Positives = 47/60 (77%)
Frame = -2
Query: 705 QDVLQDDLTTTLVQQCRRSQSTVQRIVETSGDNEALLFEALNVNDEIQKVLDKYEEVKKP 526
Q +++LT ++++ C++S +Q IVE++ D+E LLFEAL+++DE+Q+++ +Y++++ P
Sbjct: 70 QKPAKNELTDSMLENCKQSLPVIQGIVESTVDDEGLLFEALSIHDELQQIISRYQQMEDP 129
>gb|AAL65395.1| seed protein B32E [Oryza sativa]
Length = 287
Score = 52.8 bits (125), Expect = 5e-06
Identities = 26/65 (40%), Positives = 41/65 (63%)
Frame = -2
Query: 690 DDLTTTLVQQCRRSQSTVQRIVETSGDNEALLFEALNVNDEIQKVLDKYEEVKKPTTAPL 511
+++ LV QCR Q V +V +GD E+LLF+AL +NDE+Q+VL +++++ K
Sbjct: 1 EEVIVDLVGQCRSYQGRVMDLVSNTGD-ESLLFQALGLNDELQRVLQRHDDIAKGVPPGS 59
Query: 510 QPEPA 496
P PA
Sbjct: 60 GPAPA 64
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 585,137,569
Number of Sequences: 1393205
Number of extensions: 12313013
Number of successful extensions: 44147
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 41514
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44024
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 32936043699
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)