Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001043A_C01 KMC001043A_c01
(773 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
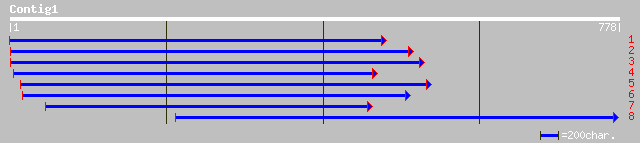
ref|NP_201065.1| unknown protein; protein id: At5g62590.1 [Arabi... 167 2e-40
gb|AAO00768.1| unknown protein [Arabidopsis thaliana] 167 2e-40
ref|NP_010708.1| cell surface protein that may regulate cell wal... 35 0.92
sp|P41809|HKR1_YEAST HANSENULA MRAKII KILLER TOXIN-RESISTANT PRO... 35 0.92
ref|NP_717268.1| conserved hypothetical protein [Shewanella onei... 35 0.92
>ref|NP_201065.1| unknown protein; protein id: At5g62590.1 [Arabidopsis thaliana]
gi|10178090|dbj|BAB11509.1| gene_id:K19B1.20~unknown
protein [Arabidopsis thaliana]
Length = 406
Score = 167 bits (423), Expect = 2e-40
Identities = 84/135 (62%), Positives = 106/135 (78%)
Frame = -1
Query: 773 DLGNSTFGERFIPYRDSVIIPRGPSITRILIASLTGALPKSRVDVVAYTLLSLTRSYGMQ 594
DL S E+F+ RD++IIPRG +ITRILIASL GALP SR+D V Y+LL+LTR+Y +Q
Sbjct: 268 DLEKSVNEEQFVRIRDNIIIPRGATITRILIASLAGALPSSRLDTVTYSLLALTRTYRLQ 327
Query: 593 ALEWAKKCVVLIPSTAATDVERSRFLKALSDVASGGDTNGLSVPIEELADVCRRNRAVQE 414
A+ WAK+ V LIP TA T+ E ++FL+ALSD+A G D N L +EEL+DVCRRNR VQE
Sbjct: 328 AVSWAKESVSLIPRTALTETESTKFLQALSDIAYGADVNSLIGQVEELSDVCRRNRTVQE 387
Query: 413 IVQEALRPLELNLVS 369
+VQ AL+PLELNLV+
Sbjct: 388 LVQAALKPLELNLVT 402
>gb|AAO00768.1| unknown protein [Arabidopsis thaliana]
Length = 958
Score = 167 bits (423), Expect = 2e-40
Identities = 84/135 (62%), Positives = 106/135 (78%)
Frame = -1
Query: 773 DLGNSTFGERFIPYRDSVIIPRGPSITRILIASLTGALPKSRVDVVAYTLLSLTRSYGMQ 594
DL S E+F+ RD++IIPRG +ITRILIASL GALP SR+D V Y+LL+LTR+Y +Q
Sbjct: 820 DLEKSVNEEQFVRIRDNIIIPRGATITRILIASLAGALPSSRLDTVTYSLLALTRTYRLQ 879
Query: 593 ALEWAKKCVVLIPSTAATDVERSRFLKALSDVASGGDTNGLSVPIEELADVCRRNRAVQE 414
A+ WAK+ V LIP TA T+ E ++FL+ALSD+A G D N L +EEL+DVCRRNR VQE
Sbjct: 880 AVSWAKESVSLIPRTALTETESTKFLQALSDIAYGADVNSLIGQVEELSDVCRRNRTVQE 939
Query: 413 IVQEALRPLELNLVS 369
+VQ AL+PLELNLV+
Sbjct: 940 LVQAALKPLELNLVT 954
>ref|NP_010708.1| cell surface protein that may regulate cell wall beta-glucan
synthesis and bud site selection; Hanenula mrakii killer
toxin-resistance protein; Hkr1p [Saccharomyces
cerevisiae] gi|2131284|pir||S69703 HKR1 protein
precursor - yeast (Saccharomyces cerevisiae)
gi|927691|gb|AAB64857.1| Hkr1p; YDR420W; CAI: 0.10
[Saccharomyces cerevisiae]
Length = 1802
Score = 35.4 bits (80), Expect = 0.92
Identities = 22/76 (28%), Positives = 35/76 (45%), Gaps = 10/76 (13%)
Frame = +1
Query: 469 DSPLVSPPEATSESAFKNLDRSTSVAAVDGIKTTHFFAHSRA----------CIPYDRVN 618
D P S TS A + + R ++ AV I TT+F S + +PY V+
Sbjct: 349 DRPATSSTAETSSEASQGVSRESNTFAVSSISTTNFIVSSASDTVVSTSSTNTVPYSSVH 408
Query: 619 DSNVYATTSTRDFGSA 666
+ V+AT+S+ S+
Sbjct: 409 STFVHATSSSTYISSS 424
>sp|P41809|HKR1_YEAST HANSENULA MRAKII KILLER TOXIN-RESISTANT PROTEIN 1 PRECURSOR
gi|545660|gb|AAB30051.1| Hkr1p [Saccharomyces
cerevisiae]
Length = 1802
Score = 35.4 bits (80), Expect = 0.92
Identities = 22/76 (28%), Positives = 35/76 (45%), Gaps = 10/76 (13%)
Frame = +1
Query: 469 DSPLVSPPEATSESAFKNLDRSTSVAAVDGIKTTHFFAHSRA----------CIPYDRVN 618
D P S TS A + + R ++ AV I TT+F S + +PY V+
Sbjct: 349 DRPATSSTAETSSEASQGVSRESNTFAVSSISTTNFIVSSASDTVVSTSSTNTVPYSSVH 408
Query: 619 DSNVYATTSTRDFGSA 666
+ V+AT+S+ S+
Sbjct: 409 STFVHATSSSTYISSS 424
>ref|NP_717268.1| conserved hypothetical protein [Shewanella oneidensis MR-1]
gi|24347454|gb|AAN54712.1|AE015612_1 conserved
hypothetical protein [Shewanella oneidensis MR-1]
Length = 244
Score = 35.4 bits (80), Expect = 0.92
Identities = 28/86 (32%), Positives = 44/86 (50%), Gaps = 1/86 (1%)
Frame = -1
Query: 725 SVIIPRGPSITRILIA-SLTGALPKSRVDVVAYTLLSLTRSYGMQALEWAKKCVVLIPST 549
S+II R P + + IA + T L S V Y + L Y + A +CVV++ +
Sbjct: 25 SLIIWRRPRLAKWAIAGAFTLLLVLSSQFSVDYLVKPLETQYPINASSITGQCVVMVLGS 84
Query: 548 AATDVERSRFLKALSDVASGGDTNGL 471
A +D+E + +++LS VA T GL
Sbjct: 85 AHSDIEDATAVQSLSAVALARLTEGL 110
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 693,042,498
Number of Sequences: 1393205
Number of extensions: 15847919
Number of successful extensions: 40174
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 38179
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 40005
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 38095156112
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)