

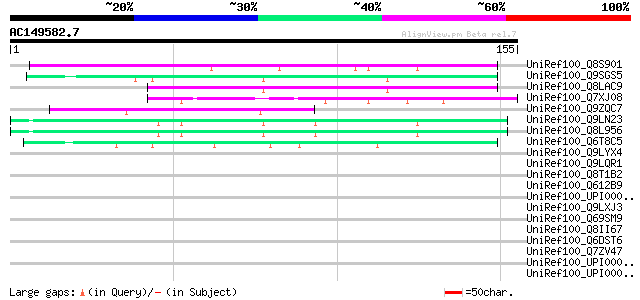
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149582.7 - phase: 0
(155 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8S901 Syringolide-induced protein 14-1-1 [Glycine max] 73 2e-12
UniRef100_Q9SGS5 T23E18.1 [Arabidopsis thaliana] 63 2e-09
UniRef100_Q8LAC9 Hypothetical protein [Arabidopsis thaliana] 56 3e-07
UniRef100_Q7XJ08 Hypothetical protein P0478E02.19 [Oryza sativa] 53 3e-06
UniRef100_Q9ZQC7 Hypothetical protein At2g37100 [Arabidopsis tha... 50 2e-05
UniRef100_Q9LN23 F14O10.9 protein [Arabidopsis thaliana] 49 3e-05
UniRef100_Q8L956 Hypothetical protein [Arabidopsis thaliana] 49 3e-05
UniRef100_Q6T8C5 Putative syringolide-induced protein 14-1-1 [He... 45 4e-04
UniRef100_Q9LYX4 Hypothetical protein F15A17_140 [Arabidopsis th... 42 0.005
UniRef100_Q9LQR1 T4O12.28 [Arabidopsis thaliana] 42 0.006
UniRef100_Q8T1B2 Hypothetical protein [Dictyostelium discoideum] 41 0.011
UniRef100_Q612B9 Hypothetical protein CBG16810 [Caenorhabditis b... 40 0.014
UniRef100_UPI000046D28E UPI000046D28E UniRef100 entry 38 0.068
UniRef100_Q9LXJ3 Hypothetical protein F3C22_110 [Arabidopsis tha... 38 0.068
UniRef100_Q69SM9 Hypothetical protein OSJNBa0022O02.23 [Oryza sa... 38 0.068
UniRef100_Q8II67 Hypothetical protein [Plasmodium falciparum] 38 0.068
UniRef100_Q6DST6 Hypothetical protein [Arabidopsis thaliana] 37 0.12
UniRef100_Q7ZV47 Similar to PRP4 pre-mRNA processing factor 4 ho... 37 0.15
UniRef100_UPI00001CF128 UPI00001CF128 UniRef100 entry 37 0.20
UniRef100_UPI0000432B0F UPI0000432B0F UniRef100 entry 37 0.20
>UniRef100_Q8S901 Syringolide-induced protein 14-1-1 [Glycine max]
Length = 257
Score = 73.2 bits (178), Expect = 2e-12
Identities = 57/178 (32%), Positives = 86/178 (48%), Gaps = 35/178 (19%)
Query: 7 MKNKFFKFLPKRPVASVSSYQNPTLSPNASVTTARKVSIIPKEARRKHRSISFS------ 60
MK+K K LPK A ++QN SP + VS+IP EARRK +
Sbjct: 1 MKSKILKALPKAVAAVTVTFQNHPFSPGRDHKSKPIVSMIPHEARRKTHDRGNNGIDEIY 60
Query: 61 AREPSSPKVSCMGQVKSKKKK-RKAKRIHQSSTKNNDSVTSHENK--------------- 104
++EP+SPK+SCMGQ+K KKK+ +K+K +S TK +V S +
Sbjct: 61 SQEPTSPKISCMGQIKHKKKQIKKSKSKSKSMTKEARNVASKKTSSASRDIEVKKHVSTF 120
Query: 105 -KILL-----------QSEKALVVEENQEFA-PSMLIVPSLGTMKKFESGRGSLSDFD 149
K+LL +S + + N++ A P + M++F SGR +L++FD
Sbjct: 121 HKMLLFHGAKPKSEGRKSNASAPGDINKDLALADAARAPRVSQMRRFSSGRDALAEFD 178
>UniRef100_Q9SGS5 T23E18.1 [Arabidopsis thaliana]
Length = 272
Score = 63.2 bits (152), Expect = 2e-09
Identities = 56/187 (29%), Positives = 75/187 (39%), Gaps = 46/187 (24%)
Query: 6 KMKNKFFKFLPKRPVASVSSYQNPTLSPNASV--------TTARK-------VSIIPKEA 50
K KNK K LPK A ++ P SP + T A K V ++P A
Sbjct: 13 KNKNKLLKMLPK---AMSFGHRVPPFSPGRDLHHNNHHNYTAANKMFFSGPMVPLVPNAA 69
Query: 51 RRKHRSISFSAREPSSPKVSCMGQVK--------SKKKKRKAKRIHQSSTKNNDSVTSHE 102
R + EP+SPKVSC+GQ+K KK K + I + S + S+T +
Sbjct: 70 RVRRNKSDAVWDEPTSPKVSCIGQIKLGKSKCPTGKKNKAPSSLIPKISKTSTSSLTKED 129
Query: 103 NKKILLQSEKAL--------------------VVEENQEFAPSMLIVPSLGTMKKFESGR 142
K L + + +E+ S VPSLG MKKF S R
Sbjct: 130 EKGRLSKIKSIFSFSPASGRNTSRKSHPTAVSAADEHPVTVVSTAAVPSLGQMKKFASSR 189
Query: 143 GSLSDFD 149
+L DFD
Sbjct: 190 DALGDFD 196
>UniRef100_Q8LAC9 Hypothetical protein [Arabidopsis thaliana]
Length = 252
Score = 55.8 bits (133), Expect = 3e-07
Identities = 41/135 (30%), Positives = 57/135 (41%), Gaps = 28/135 (20%)
Query: 43 VSIIPKEARRKHRSISFSAREPSSPKVSCMGQVK--------SKKKKRKAKRIHQSSTKN 94
V ++P AR + EP+SPKVSC+GQ+K KK K + I + S +
Sbjct: 42 VPLVPNAARVRRNKSDAVWDEPTSPKVSCIGQIKLGKSKCPTGKKNKAPSSLIPKISKTS 101
Query: 95 NDSVTSHENKKILLQSEKAL--------------------VVEENQEFAPSMLIVPSLGT 134
S+T + K L + + +E+ S VPSLG
Sbjct: 102 TSSLTKEDEKGRLSKIKSIFSFSPASGRNTSRKSHPTAVSAADEHPVTVVSTAAVPSLGQ 161
Query: 135 MKKFESGRGSLSDFD 149
MKKF S R +L DFD
Sbjct: 162 MKKFASSRDALGDFD 176
>UniRef100_Q7XJ08 Hypothetical protein P0478E02.19 [Oryza sativa]
Length = 254
Score = 52.8 bits (125), Expect = 3e-06
Identities = 50/151 (33%), Positives = 68/151 (44%), Gaps = 44/151 (29%)
Query: 43 VSIIPKEAR-------RKHRSISFSAREPSSPKVSCMGQVKSKKKKRKAKRIHQSSTKNN 95
VSI+P EAR R+HRS + EPSSPKVSC+GQ+ KK AK++ ++S KN
Sbjct: 61 VSIVPPEARGGGGGSRREHRS-GYRTPEPSSPKVSCIGQI----KKANAKKV-KASCKNG 114
Query: 96 ----------DSVTSHENKKILL--------QSEKALVVEENQ----------EFAPSML 127
D+ + K L+ +S KA ++
Sbjct: 115 ACPLPPRPPADAAAARRQKSSLVRRMLFRRSRSRKASSSSSRDGGFFKGRTAGRAGAAVA 174
Query: 128 IVPS---LGTMKKFESGRGSLSDFDATLAER 155
P+ LG MK+F SGR + DFD AER
Sbjct: 175 AAPAPAGLGQMKRFTSGRAAFEDFDWREAER 205
>UniRef100_Q9ZQC7 Hypothetical protein At2g37100 [Arabidopsis thaliana]
Length = 297
Score = 49.7 bits (117), Expect = 2e-05
Identities = 33/88 (37%), Positives = 48/88 (54%), Gaps = 7/88 (7%)
Query: 13 KFLPKRPVASVSSYQNPTLSPN----ASVTTARKVSIIPKEARRKHRSISFSAREPSSPK 68
+F RPV+S +NP L S + +R S P RRK+ S + +EP+SPK
Sbjct: 2 RFSSSRPVSSPGRTENPPLLMRFLRTKSRSRSRSRSRRPIFFRRKNASAAAETQEPTSPK 61
Query: 69 VSCMGQV---KSKKKKRKAKRIHQSSTK 93
V+CMGQV +SKK K + R+ +T+
Sbjct: 62 VTCMGQVRINRSKKPKPETARVSGGATE 89
>UniRef100_Q9LN23 F14O10.9 protein [Arabidopsis thaliana]
Length = 313
Score = 49.3 bits (116), Expect = 3e-05
Identities = 53/188 (28%), Positives = 74/188 (39%), Gaps = 37/188 (19%)
Query: 1 MEKLSKMKNKFFKFLPKRPVASVSSYQNPTLSPNASVTTARKVS-----------IIPKE 49
MEK S NK KF A + SP R S ++P
Sbjct: 1 MEKASN-SNKSTKFSKMLQRAMSIGHSAAPFSPRRDFHQHRTTSTANRGIFFSSPLVPTA 59
Query: 50 AR-RKHRSISFSAREPSSPKVSCMGQVK------SKKKKRKAKRIHQSST------KNND 96
AR R++ EP+SPKVSC+GQVK +KK + K + +S+ K D
Sbjct: 60 ARVRRNTKYEAVFAEPTSPKVSCIGQVKLARPKCPEKKNKAPKNLKTASSLSSCVIKEED 119
Query: 97 SVTSHENKKILLQSEKALVVEENQEFA------------PSMLIVPSLGTMKKFESGRGS 144
+ + + K+I + FA ++ PSLG MKKF S R +
Sbjct: 120 NGSFSKLKRIFSMRSYPSRKSNSTAFAAAAAREHPIAEVDAVTAAPSLGAMKKFASSREA 179
Query: 145 LSDFDATL 152
L FD T+
Sbjct: 180 LGGFDWTV 187
>UniRef100_Q8L956 Hypothetical protein [Arabidopsis thaliana]
Length = 240
Score = 49.3 bits (116), Expect = 3e-05
Identities = 53/188 (28%), Positives = 74/188 (39%), Gaps = 37/188 (19%)
Query: 1 MEKLSKMKNKFFKFLPKRPVASVSSYQNPTLSPNASVTTARKVS-----------IIPKE 49
MEK S NK KF A + SP R S ++P
Sbjct: 1 MEKASN-SNKSTKFSKMLQRAMSIGHSAAPFSPRRDFHQHRTTSTANRGIFFSSPLVPTA 59
Query: 50 AR-RKHRSISFSAREPSSPKVSCMGQVK------SKKKKRKAKRIHQSST------KNND 96
AR R++ EP+SPKVSC+GQVK +KK + K + +S+ K D
Sbjct: 60 ARVRRNTKYEAVFAEPTSPKVSCIGQVKLARPKCPEKKNKAPKNLKTASSLSSCVIKEED 119
Query: 97 SVTSHENKKILLQSEKALVVEENQEFA------------PSMLIVPSLGTMKKFESGRGS 144
+ + + K+I + FA ++ PSLG MKKF S R +
Sbjct: 120 NGSFSKLKRIFSMRSYPSRKSNSTAFAAAAAREHPIAEVDAVTAAPSLGAMKKFASSREA 179
Query: 145 LSDFDATL 152
L FD T+
Sbjct: 180 LGGFDWTV 187
>UniRef100_Q6T8C5 Putative syringolide-induced protein 14-1-1 [Helianthus annuus]
Length = 289
Score = 45.4 bits (106), Expect = 4e-04
Identities = 52/188 (27%), Positives = 72/188 (37%), Gaps = 45/188 (23%)
Query: 5 SKMKNKFFKFLPKRPVASVSSYQNPTL-SPNASVTTARK--------------VSIIPKE 49
+K K K+LP+ S S+QNP L SP + K +S +P +
Sbjct: 9 TKSMTKILKYLPR--ATSSLSFQNPPLYSPVKDKKPSEKPHKSNLGIGFSGPILSTLPSD 66
Query: 50 ARRKHRSISFSA--REPSSPKVSCMGQVKSK-------KKKRKAKRI------------- 87
RK + S EP+SP+VSC+G VK K K K R
Sbjct: 67 THRKINNDSKLTLLHEPTSPRVSCVGTVKCKHYRKLTNKPTNKFSRTTSYTPVRTYKTEE 126
Query: 88 HQSSTKNNDSVTSHENKKILLQSE------KALVVEENQEFAPSMLIVPSLGTMKKFESG 141
Q D V KK ++S A+ +Q + + P L TMK+F SG
Sbjct: 127 DQEKVMKVDPVVVKSKKKCXIRSLFSGGRISAMQSMRSQRWRLTGQKGPCLSTMKRFSSG 186
Query: 142 RGSLSDFD 149
R + FD
Sbjct: 187 RDAFMSFD 194
>UniRef100_Q9LYX4 Hypothetical protein F15A17_140 [Arabidopsis thaliana]
Length = 283
Score = 42.0 bits (97), Expect = 0.005
Identities = 30/84 (35%), Positives = 46/84 (54%), Gaps = 13/84 (15%)
Query: 15 LPKRPVAS---VSSYQNPTL----SPNASVTTARKVSIIPKEAR------RKHRSISFSA 61
+ +RPV+S V Y P + S + +T+R S +R R+++S + A
Sbjct: 3 ISRRPVSSPGRVEKYPPPFMGFLRSKSNGGSTSRSRSRSRGRSRASPLFVRRNKSAAAVA 62
Query: 62 REPSSPKVSCMGQVKSKKKKRKAK 85
+EPSSPKV+CMGQV+ + K K K
Sbjct: 63 QEPSSPKVTCMGQVRVNRSKPKIK 86
>UniRef100_Q9LQR1 T4O12.28 [Arabidopsis thaliana]
Length = 96
Score = 41.6 bits (96), Expect = 0.006
Identities = 30/86 (34%), Positives = 39/86 (44%), Gaps = 18/86 (20%)
Query: 6 KMKNKFFKFLPKRPVASVSSYQNPTLSPNASV--------TTARK-------VSIIPKEA 50
K KNK K LPK A ++ P SP + T A K V ++P A
Sbjct: 13 KNKNKLLKMLPK---AMSFGHRVPPFSPGRDLHHNNHHNYTAANKMFFSGPMVPLVPNAA 69
Query: 51 RRKHRSISFSAREPSSPKVSCMGQVK 76
R + EP+SPKVSC+GQ+K
Sbjct: 70 RVRRNKSDAVWDEPTSPKVSCIGQIK 95
>UniRef100_Q8T1B2 Hypothetical protein [Dictyostelium discoideum]
Length = 199
Score = 40.8 bits (94), Expect = 0.011
Identities = 29/97 (29%), Positives = 46/97 (46%), Gaps = 5/97 (5%)
Query: 5 SKMKNKFFKFLPKRPVASVSSYQNPTLSPNASVTTARKVSIIPKEARRKHRSISFSAREP 64
S++KN L ++ SS+ +P SP + + IPKE +R SISF ++
Sbjct: 76 SELKNSGNNILNLSTGSNGSSHLSPPHSPQSVHNSINSSGEIPKEKKR--ASISFGSKSK 133
Query: 65 SSPKVSCMGQVKSKKKKRKAKRIHQSSTKNNDSVTSH 101
K + + +K+K K K + KNN+S SH
Sbjct: 134 DKEKEK---EKEKEKEKEKEKEKEKEKEKNNNSAASH 167
>UniRef100_Q612B9 Hypothetical protein CBG16810 [Caenorhabditis briggsae]
Length = 823
Score = 40.4 bits (93), Expect = 0.014
Identities = 29/103 (28%), Positives = 53/103 (51%), Gaps = 7/103 (6%)
Query: 9 NKFFKFLPKRPVASVSSYQNPTLSPNASVTTARKVSIIPKEARRKHRSISFSAREPSSPK 68
NK PKR + + ++ + P S+TT+R+VS +PK+ +S S+ + +S
Sbjct: 6 NKNGSGAPKRAQKAADAPKSTSSRPRRSITTSRRVSYVPKDHLAGIEEMSSSSSDEASDS 65
Query: 69 VSCMG------QVKSKKKKRKAKRIHQSSTKNNDSVTSHENKK 105
M ++K KK+ KA +I +S ++DS S E+++
Sbjct: 66 DDVMDSEEPKKELKLPKKRSKAFKI-ESEESSSDSYESGESEE 107
>UniRef100_UPI000046D28E UPI000046D28E UniRef100 entry
Length = 4810
Score = 38.1 bits (87), Expect = 0.068
Identities = 32/139 (23%), Positives = 67/139 (48%), Gaps = 19/139 (13%)
Query: 2 EKLSKMKNKF-----FKFLP---KRPVASVSSYQNPTLSPNASVTTARKVSIIPKEARRK 53
+K+ K+K F F LP K + +S++ + + N +V S++ K +
Sbjct: 766 KKIKKLKYSFKNIFQFNSLPVHLKNIINDLSTHNSKQIKKNETVDNIYIQSLLKKRKKET 825
Query: 54 H-RSISFSAREPSS-------PKV---SCMGQVKSKKKKRKAKRIHQSSTKNNDSVTSHE 102
+++S + +S KV S MG +K KKK K+ ++ S+ N +++ +
Sbjct: 826 DIKNVSSDCNDGNSNNSKKGVSKVNENSYMGNIKKKKKNFKSGMMNDSNKTNENNLKYYL 885
Query: 103 NKKILLQSEKALVVEENQE 121
N K +++E L++E+ ++
Sbjct: 886 NNKSEIENELQLIIEKRKD 904
>UniRef100_Q9LXJ3 Hypothetical protein F3C22_110 [Arabidopsis thaliana]
Length = 289
Score = 38.1 bits (87), Expect = 0.068
Identities = 33/116 (28%), Positives = 54/116 (46%), Gaps = 29/116 (25%)
Query: 9 NKFFKFLPKRPVASVSSYQNPTLSPNAS---VTTARKVSII--PK-------EARRKH-- 54
N FF+ + S + NP +PN+S ++ K SII PK EA+++
Sbjct: 64 NAFFERKESQKGNSSAPISNPNTNPNSSSHRISLKSKASIIRLPKPQKTCFNEAKKRRNC 123
Query: 55 ---RSISFSAR------------EPSSPKVSCMGQVKSKKKKRKAKRIHQSSTKNN 95
R++ R EP SPKVSC+G+V+SK+ + + + +S N+
Sbjct: 124 RIARTLMIPKRIGSRLKSDPTLSEPCSPKVSCIGRVRSKRDRSRRMQRQKSGRTNS 179
>UniRef100_Q69SM9 Hypothetical protein OSJNBa0022O02.23 [Oryza sativa]
Length = 845
Score = 38.1 bits (87), Expect = 0.068
Identities = 17/36 (47%), Positives = 24/36 (66%)
Query: 59 FSAREPSSPKVSCMGQVKSKKKKRKAKRIHQSSTKN 94
F EPSSPKV+C+GQV+ K KRK K ++ ++
Sbjct: 109 FEPAEPSSPKVTCIGQVRVKGGKRKPKHASAAALRS 144
>UniRef100_Q8II67 Hypothetical protein [Plasmodium falciparum]
Length = 1338
Score = 38.1 bits (87), Expect = 0.068
Identities = 32/104 (30%), Positives = 53/104 (50%), Gaps = 7/104 (6%)
Query: 21 ASVSSYQNPTLSPNASVTTARKVSI-IPKEARRKHRSISFSAREPSSPKVSCMGQVKSKK 79
+++SS + +S N S + +S I RKH S SFS+ SS K S +K KK
Sbjct: 849 SNISSNISSNISNNISSNISNNISSNISSNISRKHNSYSFSSLSSSSTK-SLYNTMKKKK 907
Query: 80 KKRK--AKRIHQSSTKNNDSVTSHENKKILLQSEKALVVEENQE 121
KK+K +RI+ S+ + + S K ++ Q++ + N+E
Sbjct: 908 KKKKYINERIYDSAHEIQKDLKS---KGLIPQNDNNKYIYHNKE 948
>UniRef100_Q6DST6 Hypothetical protein [Arabidopsis thaliana]
Length = 182
Score = 37.4 bits (85), Expect = 0.12
Identities = 19/55 (34%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query: 26 YQNPTLSPNASVTTARKVSIIPKEARRKHRSISFSAREPSSPKVSCMGQVKSKKK 80
+Q PT P + K +P ++ H+ S+ +P SPK+SC+GQVK K
Sbjct: 11 FQRPTFLPMLCSRPSIKNVTLPAKS---HQDDSYQQADPLSPKISCIGQVKRSNK 62
>UniRef100_Q7ZV47 Similar to PRP4 pre-mRNA processing factor 4 homolog B [Brachydanio
rerio]
Length = 490
Score = 37.0 bits (84), Expect = 0.15
Identities = 23/65 (35%), Positives = 34/65 (51%)
Query: 33 PNASVTTARKVSIIPKEARRKHRSISFSAREPSSPKVSCMGQVKSKKKKRKAKRIHQSST 92
P++S T+ RK + P+E HRS S S++ S + + + +V KK KA SS
Sbjct: 229 PSSSATSNRKSNTQPEEQAVDHRSSSRSSKSGSKERSTSLPEVDRDKKPAKAPLKEVSSG 288
Query: 93 KNNDS 97
K N S
Sbjct: 289 KENRS 293
>UniRef100_UPI00001CF128 UPI00001CF128 UniRef100 entry
Length = 997
Score = 36.6 bits (83), Expect = 0.20
Identities = 24/63 (38%), Positives = 36/63 (57%), Gaps = 1/63 (1%)
Query: 51 RRKHRSISFSAREPSSPKVSCMGQVKSKKKKRKA-KRIHQSSTKNNDSVTSHENKKILLQ 109
+++ RS S SA E SS S KSKK K+K+ KR H+S + +D+ + K+ +
Sbjct: 883 KKRQRSESHSASEHSSSAESERSYKKSKKHKKKSKKRRHKSDSPESDTEREKDKKEKDRE 942
Query: 110 SEK 112
SEK
Sbjct: 943 SEK 945
>UniRef100_UPI0000432B0F UPI0000432B0F UniRef100 entry
Length = 971
Score = 36.6 bits (83), Expect = 0.20
Identities = 36/127 (28%), Positives = 56/127 (43%), Gaps = 18/127 (14%)
Query: 30 TLSPNASVTTA-------RKVSIIPKEARRKHRSISFSAR-------EPSSPKVSCMGQV 75
T SP T+A R S I K+ +KH+ S + R SPK S +
Sbjct: 170 TPSPAPDTTSAPEKKKRKRSASAIKKKKGKKHKKESETFRITMNEFPRSESPKASKKKEK 229
Query: 76 KSKKKKRKAKRIHQSSTKNNDSVTSHENKKILLQSEKALVVEENQEFAPSMLIVPSLGTM 135
KKKK K + ++S+ ++DS + K ++ +KA+ +N E + S +
Sbjct: 230 SKKKKKEKKHKKTEASSSDSDS----KRPKKSMKHDKAVEKNKNDEGENTTKESRSNRSP 285
Query: 136 KKFESGR 142
KK E GR
Sbjct: 286 KKQEKGR 292
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.310 0.123 0.323
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 235,437,665
Number of Sequences: 2790947
Number of extensions: 8558298
Number of successful extensions: 38789
Number of sequences better than 10.0: 278
Number of HSP's better than 10.0 without gapping: 78
Number of HSP's successfully gapped in prelim test: 205
Number of HSP's that attempted gapping in prelim test: 38344
Number of HSP's gapped (non-prelim): 512
length of query: 155
length of database: 848,049,833
effective HSP length: 116
effective length of query: 39
effective length of database: 524,299,981
effective search space: 20447699259
effective search space used: 20447699259
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC149582.7


