

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149576.7 + phase: 1 /pseudo
(576 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
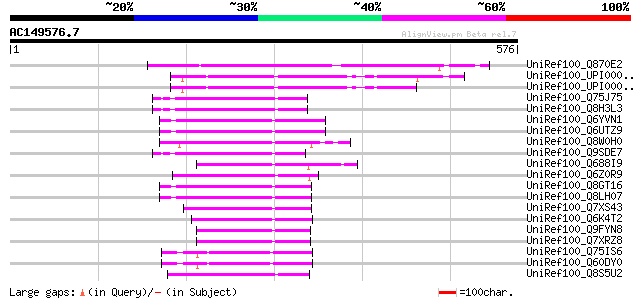
UniRef100_Q870E2 Transposase [Fusarium oxysporum f. sp. melonis] 162 2e-38
UniRef100_UPI000042CB10 UPI000042CB10 UniRef100 entry 129 2e-28
UniRef100_UPI000042F827 UPI000042F827 UniRef100 entry 126 2e-27
UniRef100_Q75J75 Putative far-red impaired response protein [Ory... 88 6e-16
UniRef100_Q8H3L3 Putative far-red impaired response protein [Ory... 86 2e-15
UniRef100_Q6YVN1 Putative far-red impaired response protein [Ory... 86 4e-15
UniRef100_Q6UTZ9 Putative transposase [Oryza sativa] 86 4e-15
UniRef100_Q8W0H0 P0529E05.9 protein [Oryza sativa] 85 7e-15
UniRef100_Q9SDE7 Similar to Arabidopsis thaliana DNA chromosome ... 82 3e-14
UniRef100_Q688I9 Hypothetical protein OSJNBb0012G21.8 [Oryza sat... 82 4e-14
UniRef100_Q6Z0R9 Putative far-red impaired response protein [Ory... 82 6e-14
UniRef100_Q8GT16 Putative far-red impaired response protein [Ory... 81 7e-14
UniRef100_Q8LH07 Far-red impaired response-like protein [Oryza s... 81 1e-13
UniRef100_Q7XS43 OSJNBa0074L08.5 protein [Oryza sativa] 81 1e-13
UniRef100_Q6K4T2 Putative far-red impaired response protein [Ory... 80 2e-13
UniRef100_Q9FYN8 P0702D12.15 protein [Oryza sativa] 79 3e-13
UniRef100_Q7XRZ8 OSJNBb0002N06.8 protein [Oryza sativa] 79 3e-13
UniRef100_Q75IS6 Putative Mutator-like transposase [Oryza sativa] 79 4e-13
UniRef100_Q60DY0 Hypothetical protein OSJNBa0090H02.8 [Oryza sat... 79 4e-13
UniRef100_Q8S5U2 Putative far-red impaired response protein [Ory... 79 5e-13
>UniRef100_Q870E2 Transposase [Fusarium oxysporum f. sp. melonis]
Length = 836
Score = 162 bits (411), Expect = 2e-38
Identities = 116/395 (29%), Positives = 200/395 (50%), Gaps = 24/395 (6%)
Query: 157 GETKKYLTNLKKKKKESITNIKRVYNECHKFKKAKRGDLTEMQYLISKLEENEYVNYVRE 216
G K + + + ++ + +YN K ++ + + L +L E + N +
Sbjct: 144 GIAPKEIGSFLRITSNTLATQQDIYNCIAKGRRDLSKGQSNIHALADQLNEEGFWNRIC- 202
Query: 217 KPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGF 276
ES V + + HP S++ T+P VLI+DSTYKTN ++MPL +IVGV + T+ + F
Sbjct: 203 LDESSRVTAVLFAHPKSLEYLKTYPEVLILDSTYKTNRFKMPLLDIVGVDACQRTFCIAF 262
Query: 277 AFMMSKKEDNFTWALQMLLKLLKP-NSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFH 335
AF+ ++E +FTWALQ L + + N +P V++TDR + MNAV++ P S+ LC +H
Sbjct: 263 AFLSGEEEGDFTWALQALRSVYEDHNIGLPSVILTDRCLACMNAVSSCFPGSALFLCLWH 322
Query: 336 VGKNIRSRI-ITDCKVKQNVVVVDGQKKIVDEESHSKLVDTIFDASEKLVESHTQELYAG 394
+ K ++S + K N + G+ + E F+ ++V S T+++Y
Sbjct: 323 INKAVQSYCRPAFTRGKDNPQGLGGESEEWKE---------FFNFWHEIVASTTEDIYNE 373
Query: 395 NLVEFQDA-CKDHPKFLEYVETTTLKPFKDKLVRAWMELVLHLGCRTTNRVEGAHGVVKE 453
L +F+ D+ + Y+ T L +K V+AW+ LH T+RVEG H ++K
Sbjct: 374 RLEKFKKRYIPDYINEVGYILETWLDLYKKSFVKAWVNTHLHFEQYATSRVEGIHSLIKL 433
Query: 454 YLSTSKGDLGTCSQKIDEMLANQFGEIQSSFGR---SVTVLEHRYKDVTLYSGLGGHMSR 510
+L+ S+ DL + I +L NQ +++++ R S + E R LYS + G +S
Sbjct: 434 HLNHSQVDLFEAWRVIKLVLMNQLSQLEANQARQHISNPIRESR----VLYSNIRGWISH 489
Query: 511 QAMNFIFVEEARSRKTLCIKKKTCGCVQRMSYGLP 545
+A+ + + R K + + C V + GLP
Sbjct: 490 EALRKVETQRERLLKEVPV----CTGVFTRTLGLP 520
>UniRef100_UPI000042CB10 UPI000042CB10 UniRef100 entry
Length = 932
Score = 129 bits (324), Expect = 2e-28
Identities = 99/345 (28%), Positives = 169/345 (48%), Gaps = 37/345 (10%)
Query: 183 ECHKFKKAKRGDL----TEMQYLISKLEENEYVNYVREKPESQIVQDIFWTHPTSVKLFN 238
+ H+ +R DL + I +L N+ ++ +++ + +T T+ L +
Sbjct: 260 DLHRLAFRRRADLRAGSSASAACIRQLVSNDELHRPFVNTRDELI-GLVYTKRTARALIH 318
Query: 239 TFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLL 298
FP VL D TYKTNLYRMP+ IVG TST +TY+ G M+ + + +T AL +L+
Sbjct: 319 RFPEVLFFDCTYKTNLYRMPMLHIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELV 378
Query: 299 -KPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGKNIRSRIITDCKVKQNVVVV 357
KP+ KVV+TDRDP+++NA+ +VLP + C++H+ +N++S I ++N+V+
Sbjct: 379 GKPD---VKVVITDRDPALINALMSVLPKAYRFSCFWHLQENVKSNIRPCFVDEENIVMA 435
Query: 358 DGQ--KKIVDEESHSKLVDTIFDASEKLVESHTQELYAGNLVEFQDACKDHPKFLEYVET 415
K V H+K + + + K+ +ELY G + + + + Y+
Sbjct: 436 VSNFCKAWVRYCVHAKSDEQLEEGYRKM-----EELYPG---------QKYARAISYIR- 480
Query: 416 TTLKPFKDKLVRAWMELVLHLGCRTTNRVEGAHGVVKEYLSTSKGDL----GTCSQKIDE 471
L K++ V A++ LH G +R+EG H +K+ + T GDL G+ S D+
Sbjct: 481 -GLDEIKERFVHAYINKQLHFGQTGNSRLEGQHATLKKSIDTKYGDLLLVIGSLSTYFDQ 539
Query: 472 MLANQFGEIQSSFGRSVTVLEHRYKDVTLYSGLGGHMSRQAMNFI 516
I+ R T + + + L G +SR AM +
Sbjct: 540 QWLKILKRIELERTRCATHIP------STFVRLKGSISRAAMKLL 578
>UniRef100_UPI000042F827 UPI000042F827 UniRef100 entry
Length = 580
Score = 126 bits (317), Expect = 2e-27
Identities = 87/287 (30%), Positives = 149/287 (51%), Gaps = 27/287 (9%)
Query: 183 ECHKFKKAKRGDL----TEMQYLISKLEENEYVNYVREKPESQIVQDIFWTHPTSVKLFN 238
+ H+ +R DL + I +L N+ ++ +++ + +T T+ L +
Sbjct: 260 DLHRLAFRRRADLRAGSSASAACIRQLVSNDELHRPFVNTRDELI-GLVYTKRTARALIH 318
Query: 239 TFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLL 298
FP VL D TYKTNLYRMP+ IVG TST +TY+ G M+ + + +T AL +L+
Sbjct: 319 RFPEVLFFDCTYKTNLYRMPMLHIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELV 378
Query: 299 -KPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGKNIRSRIITDCKVKQNVVVV 357
KP+ KVV+TDRDP+++NA+ +VLP + C++H+ +N++S I ++N+V+
Sbjct: 379 GKPD---VKVVITDRDPALINALMSVLPKAYRFSCFWHLQENVKSNIRPCFVDEENIVMA 435
Query: 358 DGQ--KKIVDEESHSKLVDTIFDASEKLVESHTQELYAGNLVEFQDACKDHPKFLEYVET 415
K V H+K + + + K+ +ELY G + + + + Y+
Sbjct: 436 VSNFCKAWVRYCVHAKSDEQLEEGYRKM-----EELYPG---------QKYARAISYIR- 480
Query: 416 TTLKPFKDKLVRAWMELVLHLGCRTTNRVEGAHGVVKEYLSTSKGDL 462
L K++ V A++ LH G +R+EG H +K+ + T GDL
Sbjct: 481 -GLDEIKERFVHAYINKQLHFGQTGNSRLEGQHATLKKSIDTKYGDL 526
>UniRef100_Q75J75 Putative far-red impaired response protein [Oryza sativa]
Length = 888
Score = 88.2 bits (217), Expect = 6e-16
Identities = 51/177 (28%), Positives = 94/177 (52%), Gaps = 9/177 (5%)
Query: 163 LTNLKKKKKESITNIKRVYNECHKFKKAKRGDLTE-MQYLISKLEENEYVNYVREKPESQ 221
LT+L KK+ I+N++ N K+ D+ + +++ K E++ + Y + ES+
Sbjct: 460 LTSLPYTKKD-ISNVRTTIN-----KETSSNDMMKTLEFFRRKKEKDPHFFYEFDLDESK 513
Query: 222 IVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMS 281
V+++FWT S + + + V+ D+TY TN Y +P VG++ T G AF+
Sbjct: 514 KVKNLFWTDGRSREWYEKYGDVVSFDTTYFTNRYNLPFAPFVGISGHGNTIVFGCAFLHD 573
Query: 282 KKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGK 338
+ + F W + LK + PK+++TD+D +M +A+A V ++ C+FH+ K
Sbjct: 574 ETSETFKWMFRTFLKAMSQKE--PKIIITDQDGAMRSAIAQVFQNAKHGNCFFHIVK 628
>UniRef100_Q8H3L3 Putative far-red impaired response protein [Oryza sativa]
Length = 1148
Score = 86.3 bits (212), Expect = 2e-15
Identities = 51/177 (28%), Positives = 92/177 (51%), Gaps = 9/177 (5%)
Query: 163 LTNLKKKKKESITNIKRVYNECHKFKKAKRGDLTE-MQYLISKLEENEYVNYVREKPESQ 221
LT+L KK+ I+N++ N K+ D+ + +++ K E++ + Y + ES+
Sbjct: 519 LTSLPYTKKD-ISNVRTTIN-----KETSSNDIMKTLEFFRRKKEKDPHFFYEFDLDESK 572
Query: 222 IVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMS 281
V+++FWT S + + + V+ D+TY TN Y +P VG+ T G AF+
Sbjct: 573 KVKNLFWTDGRSREWYEKYGDVVSFDTTYFTNKYNLPFAPFVGIYGHGNTIVFGCAFLHD 632
Query: 282 KKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGK 338
+ + F W + LK + PK ++TD+D +M +A+A V ++ C+FH+ K
Sbjct: 633 ETSETFKWLFRTFLKAMSQKE--PKTIITDQDGAMRSAIAQVFQNAKHRNCFFHIVK 687
>UniRef100_Q6YVN1 Putative far-red impaired response protein [Oryza sativa]
Length = 1132
Score = 85.5 bits (210), Expect = 4e-15
Identities = 51/189 (26%), Positives = 89/189 (46%), Gaps = 8/189 (4%)
Query: 171 KESITNIKRVYNECHKFKKAKRGDLTE-MQYLISKLEENEYVNYVREKPESQIVQDIFWT 229
K+ ++N+ N + + D+ + + YL K E+ ++Y + E+ V +FWT
Sbjct: 562 KKDVSNVGTAINS-----ETRNNDMKQVLAYLRKKEIEDPGISYKFKLDENNKVTSMFWT 616
Query: 230 HPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTW 289
S +L+ + + D+TY+TN Y MP VGV T G AF+ + + F W
Sbjct: 617 DGRSTQLYEEYGDCISFDTTYRTNRYNMPFAPFVGVIGHGTTCLFGCAFLGDETAETFKW 676
Query: 290 ALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGKNIRSRIITDCK 349
+ + PK ++TD+D +M +A+A V ++ C FH+ KN R + +
Sbjct: 677 VFETFATAM--GGKHPKTIITDQDNAMRSAIAQVFQNTKHRNCLFHIKKNCREKTGSMFS 734
Query: 350 VKQNVVVVD 358
K N + D
Sbjct: 735 QKSNKNLYD 743
>UniRef100_Q6UTZ9 Putative transposase [Oryza sativa]
Length = 1037
Score = 85.5 bits (210), Expect = 4e-15
Identities = 51/189 (26%), Positives = 89/189 (46%), Gaps = 8/189 (4%)
Query: 171 KESITNIKRVYNECHKFKKAKRGDLTE-MQYLISKLEENEYVNYVREKPESQIVQDIFWT 229
K+ ++N+ N + + D+ + + YL K E+ ++Y + E+ V +FWT
Sbjct: 467 KKDVSNVGTAINS-----ETRNNDMKQVLAYLRKKEIEDPGISYKFKLDENNKVTSMFWT 521
Query: 230 HPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTW 289
S +L+ + + D+TY+TN Y MP VGV T G AF+ + + F W
Sbjct: 522 DGRSTQLYEEYGDCISFDTTYRTNRYNMPFAPFVGVIGHGTTCLFGCAFLGDETAETFKW 581
Query: 290 ALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGKNIRSRIITDCK 349
+ + PK ++TD+D +M +A+A V ++ C FH+ KN R + +
Sbjct: 582 VFETFATAM--GGKHPKTIITDQDNAMRSAIAQVFQNTKHRNCLFHIKKNCREKTGSMFS 639
Query: 350 VKQNVVVVD 358
K N + D
Sbjct: 640 QKSNKNLYD 648
>UniRef100_Q8W0H0 P0529E05.9 protein [Oryza sativa]
Length = 773
Score = 84.7 bits (208), Expect = 7e-15
Identities = 63/224 (28%), Positives = 104/224 (46%), Gaps = 17/224 (7%)
Query: 171 KESITNIKRVYNECHKFKKAK---RGDLTE-MQYLISKLEENEYVNYVREKPESQIVQDI 226
++S+T ++ Y + K+ K GD +QYL K EN Y + E +++ +I
Sbjct: 297 RQSLTFTRKDYKNYLRSKRMKSIQEGDTGAILQYLQDKQMENPSFFYAIQVDEDEMMTNI 356
Query: 227 FWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDN 286
FW SV F+ F V+ D+TY+TN Y P VGV T G A + +
Sbjct: 357 FWADARSVLDFDYFGDVICFDTTYRTNNYGRPFALFVGVNHHKQTVVFGAALLYDETTST 416
Query: 287 FTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGKNIR---SR 343
F W + + + + P+ ++TD+ +++NA+ V P+S+ LC +H+ +N S
Sbjct: 417 FEWLFETFKRAM--SGKEPRTILTDQCAAIINAIGTVFPNSTHRLCVWHMYQNAAVHLSH 474
Query: 344 IITDCKVKQNVVVVDGQKKIVDEESHSKLVDTIFDASEKLVESH 387
I K +N D K + D E VD A K++E +
Sbjct: 475 IFQGSKTFKN----DFGKCVFDFEE----VDEFISAWNKMIEEY 510
>UniRef100_Q9SDE7 Similar to Arabidopsis thaliana DNA chromosome 4 [Oryza sativa]
Length = 861
Score = 82.4 bits (202), Expect = 3e-14
Identities = 51/175 (29%), Positives = 87/175 (49%), Gaps = 9/175 (5%)
Query: 163 LTNLKKKKKESITNIKRVYNECHKFKKAKRGDLTE-MQYLISKLEENEYVNYVREKPESQ 221
LT+L KK+ I+N++ N ++ D+ + + YL K EEN Y + E++
Sbjct: 226 LTSLPYTKKD-ISNVRTEIN-----RETSSNDMMQCLTYLKKKQEENPNFFYEFDLDENR 279
Query: 222 IVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMS 281
++ FWT S ++ F + D+T+ TN Y +P VGV+ T G AF+
Sbjct: 280 KGRNFFWTDGRSRDWYHKFGDCISFDTTFLTNRYNLPFAPFVGVSGHGNTILFGCAFLHD 339
Query: 282 KKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHV 336
+ + F W LK + N PK ++TD+D +M A+ N+ ++ C+FH+
Sbjct: 340 ETTETFEWLFITFLKCM--NGKQPKTIITDQDGAMRAAIKNIFREAKHRNCFFHI 392
>UniRef100_Q688I9 Hypothetical protein OSJNBb0012G21.8 [Oryza sativa]
Length = 822
Score = 82.0 bits (201), Expect = 4e-14
Identities = 58/190 (30%), Positives = 88/190 (45%), Gaps = 11/190 (5%)
Query: 213 YVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTY 272
Y + E+ V++IFW+H S F F V+ D+TYKTN Y MP VGV + + +
Sbjct: 310 YSIQVDEACRVKNIFWSHAVSRLNFEHFGDVITFDTTYKTNKYNMPFAPFVGVNNHFQST 369
Query: 273 SVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILC 332
G A + + E++FTW + + N +P ++TD PSM A+ V P++ +C
Sbjct: 370 FFGCALLREETEESFTWLFNTFKECM--NGKVPIGILTDNCPSMAAAIRTVFPNTIHRVC 427
Query: 333 YFHVGK-------NIRSRIITDCKVKQNVVVVDGQKKIVDEESHSKLVDTIFDASEKLVE 385
+HV K NI S+ T K V+ D K D F +EKL +
Sbjct: 428 KWHVLKKAKEFMGNIYSKRHTFKKAFHKVLCWDDYYTENSMNGFVKRYDRFF--NEKLQK 485
Query: 386 SHTQELYAGN 395
++E N
Sbjct: 486 EDSEEFQTSN 495
>UniRef100_Q6Z0R9 Putative far-red impaired response protein [Oryza sativa]
Length = 690
Score = 81.6 bits (200), Expect = 6e-14
Identities = 50/170 (29%), Positives = 88/170 (51%), Gaps = 7/170 (4%)
Query: 186 KFKKAKRGDLTEM-QYLISKLEENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVL 244
K K+ GD+ + +Y+ ++ +++ Y E E+ ++ +FW P S ++ F V+
Sbjct: 160 KKKQIVGGDVDRVIKYMQARQKDDMEFFYEYETDEAGRLKRLFWADPQSRIDYDAFGDVV 219
Query: 245 IMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDM 304
+ DSTY+ N Y +P VGV T A + +K + W L+ L +
Sbjct: 220 VFDSTYRVNKYNLPFIPFVGVNHHGSTIIFACAIVADEKIATYEWVLKRFLDCMCQKH-- 277
Query: 305 PKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGKN----IRSRIITDCKV 350
PK ++TD D SM A+A V+P+S LC +H+ +N +R ++I+D +V
Sbjct: 278 PKGLITDSDNSMRRAIATVMPNSEHRLCTWHIEQNMARHLRPKMISDFRV 327
>UniRef100_Q8GT16 Putative far-red impaired response protein [Oryza sativa]
Length = 1114
Score = 81.3 bits (199), Expect = 7e-14
Identities = 49/174 (28%), Positives = 82/174 (46%), Gaps = 8/174 (4%)
Query: 171 KESITNIKRVYNECHKFKKAKRGDLTE-MQYLISKLEENEYVNYVREKPESQIVQDIFWT 229
++ ++N+ N + + D+ + M+YL K ++ Y E+ V+ +FWT
Sbjct: 512 RKDVSNVGTAINS-----ETRNTDMNQVMEYLRQKEAKDPGYYYKLTLDENNKVKSMFWT 566
Query: 230 HPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTW 289
S +L+ + + D+TYKTN Y MP IVGVT AF+ + + F W
Sbjct: 567 DGRSTQLYEQYGEFVSFDTTYKTNRYNMPFAPIVGVTGHGNICIFACAFLGDETTETFKW 626
Query: 290 ALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGKNIRSR 343
+ L + P+ ++TD+D +M A+ V P+S C FH+ K R R
Sbjct: 627 VFETFLTAM--GGKHPETIITDQDLAMRAAIRQVFPNSKHRNCLFHILKKCRER 678
>UniRef100_Q8LH07 Far-red impaired response-like protein [Oryza sativa]
Length = 772
Score = 80.9 bits (198), Expect = 1e-13
Identities = 49/174 (28%), Positives = 82/174 (46%), Gaps = 8/174 (4%)
Query: 171 KESITNIKRVYNECHKFKKAKRGDLTE-MQYLISKLEENEYVNYVREKPESQIVQDIFWT 229
++ ++N+ N + + D+ + M+YL K ++ Y E+ V+ +FWT
Sbjct: 197 RKDVSNVGTTINS-----ETRNTDMNQVMEYLRQKEAKDPGYYYKLTLDENNKVKSMFWT 251
Query: 230 HPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTW 289
S +L+ + + D+TYKTN Y MP IVGVT AF+ + + F W
Sbjct: 252 DGRSTQLYEQYGEFVSFDTTYKTNRYNMPFAPIVGVTGHGNICIFACAFLGDETTETFKW 311
Query: 290 ALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGKNIRSR 343
+ L + P+ ++TD+D +M A+ V P+S C FH+ K R R
Sbjct: 312 VFETFLTAM--GRKHPETIITDQDLAMRAAIRQVFPNSKHRNCLFHILKKYRER 363
>UniRef100_Q7XS43 OSJNBa0074L08.5 protein [Oryza sativa]
Length = 1061
Score = 80.9 bits (198), Expect = 1e-13
Identities = 46/146 (31%), Positives = 70/146 (47%), Gaps = 2/146 (1%)
Query: 198 MQYLISKLEENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRM 257
M+YL K ++ Y E+ V+ +FWT S +L+ + + D+TYKTN Y M
Sbjct: 482 MEYLRQKEAKDPGYYYKLTLDENNKVKSMFWTDGRSTQLYEQYGEFVSFDTTYKTNRYNM 541
Query: 258 PLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMM 317
P IVGVT AF+ + + F W + L + P+ ++TD+D +M
Sbjct: 542 PFAPIVGVTGHGNICIFACAFLGDETTETFKWVFETFLTAM--GGKHPETIITDQDLAMR 599
Query: 318 NAVANVLPDSSAILCYFHVGKNIRSR 343
A+ V P+S C FH+ K R R
Sbjct: 600 AAIRQVFPNSKHRNCLFHILKKCRER 625
>UniRef100_Q6K4T2 Putative far-red impaired response protein [Oryza sativa]
Length = 934
Score = 79.7 bits (195), Expect = 2e-13
Identities = 40/138 (28%), Positives = 70/138 (49%), Gaps = 2/138 (1%)
Query: 207 ENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVT 266
+N YV +K E V IFWT ++ F +++ D+ Y T++Y MP I+G+
Sbjct: 459 QNPEFLYVMQKDEDNTVTSIFWTDARLRIEYDIFGDLIMFDAAYSTDMYNMPFVPIIGIN 518
Query: 267 STYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPD 326
S + +G A + +K + F W L+ L+++ MP+ V+T++D SM A A ++P
Sbjct: 519 SHATPFLLGCALLKDEKVETFEWMLRTFLQVM--GGKMPRAVITNQDTSMEKAFAELMPH 576
Query: 327 SSAILCYFHVGKNIRSRI 344
C HV + ++
Sbjct: 577 VRLRFCKRHVMSKAQEKL 594
>UniRef100_Q9FYN8 P0702D12.15 protein [Oryza sativa]
Length = 832
Score = 79.3 bits (194), Expect = 3e-13
Identities = 43/129 (33%), Positives = 68/129 (52%), Gaps = 2/129 (1%)
Query: 213 YVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTY 272
Y + E+ V++IFW+H S F F V+ D+TYKTN Y MP VGV + + +
Sbjct: 257 YSIQVDEACRVKNIFWSHAVSRLNFEHFGDVITFDTTYKTNKYNMPFAPFVGVNNHFQST 316
Query: 273 SVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILC 332
G A + + E++FTW + + N +P ++TD PSM A+ V P++ +C
Sbjct: 317 FFGCALLREETEESFTWLFNTFKECM--NGKVPIGILTDNCPSMAAAIRTVFPNTIHRVC 374
Query: 333 YFHVGKNIR 341
+HV K +
Sbjct: 375 KWHVLKKAK 383
>UniRef100_Q7XRZ8 OSJNBb0002N06.8 protein [Oryza sativa]
Length = 885
Score = 79.3 bits (194), Expect = 3e-13
Identities = 43/129 (33%), Positives = 68/129 (52%), Gaps = 2/129 (1%)
Query: 213 YVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTY 272
Y + E+ V++IFW+H S F F V+ D+TYKTN Y MP VGV + + +
Sbjct: 310 YSIQVDEACRVKNIFWSHAVSRLNFEHFGDVITFDTTYKTNKYNMPFAPFVGVNNHFQST 369
Query: 273 SVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILC 332
G A + + E++FTW + + N +P ++TD PSM A+ V P++ +C
Sbjct: 370 FFGCALLREETEESFTWLFNTFKECM--NGKVPIGILTDNCPSMAAAIRTVFPNTIHRVC 427
Query: 333 YFHVGKNIR 341
+HV K +
Sbjct: 428 KWHVLKKAK 436
>UniRef100_Q75IS6 Putative Mutator-like transposase [Oryza sativa]
Length = 1510
Score = 79.0 bits (193), Expect = 4e-13
Identities = 49/179 (27%), Positives = 91/179 (50%), Gaps = 17/179 (9%)
Query: 173 SITNIKRVYNECH-KFKKAKRGDLTEMQYLISKLEENEYVN------YVREKPESQIVQD 225
++ N++ Y++ K K A + Q + +LE + VN ++ +K + ++V+
Sbjct: 867 TLRNLQNYYHDLRCKIKDA------DAQMFVGQLERKKEVNPAFFYEFMVDK-QGRLVR- 918
Query: 226 IFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKED 285
+FW K ++ F VL +DSTY TN Y M GV + +G +F+ +K +
Sbjct: 919 VFWADAICRKNYSVFGDVLSVDSTYSTNQYNMKFVPFTGVNHHLQSVFLGASFLADEKIE 978
Query: 286 NFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGKNIRSRI 344
+F W Q LK P++++TD D SM A+A +LP++ LC +H+ + + ++
Sbjct: 979 SFVWLFQTFLK--ATGGVAPRLIITDEDASMKAAIAQILPNTVHRLCMWHIMEKVPEKV 1035
>UniRef100_Q60DY0 Hypothetical protein OSJNBa0090H02.8 [Oryza sativa]
Length = 896
Score = 79.0 bits (193), Expect = 4e-13
Identities = 49/179 (27%), Positives = 91/179 (50%), Gaps = 17/179 (9%)
Query: 173 SITNIKRVYNECH-KFKKAKRGDLTEMQYLISKLEENEYVN------YVREKPESQIVQD 225
++ N++ Y++ K K A + Q + +LE + VN ++ +K + ++V+
Sbjct: 258 TLRNLQNYYHDLRCKIKDA------DAQMFVGQLERKKEVNPAFFYEFMVDK-QGRLVR- 309
Query: 226 IFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKED 285
+FW K ++ F VL +DSTY TN Y M GV + +G +F+ +K +
Sbjct: 310 VFWADAICRKNYSVFGDVLSVDSTYSTNQYNMKFVPFTGVNHHLQSVFLGASFLADEKIE 369
Query: 286 NFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGKNIRSRI 344
+F W Q LK P++++TD D SM A+A +LP++ LC +H+ + + ++
Sbjct: 370 SFVWLFQTFLK--ATGGVAPRLIITDEDASMKAAIAQILPNTVHRLCMWHIMEKVPEKV 426
>UniRef100_Q8S5U2 Putative far-red impaired response protein [Oryza sativa]
Length = 611
Score = 78.6 bits (192), Expect = 5e-13
Identities = 48/164 (29%), Positives = 82/164 (49%), Gaps = 5/164 (3%)
Query: 180 VYNECHKFKKAK--RGDLTEM-QYLISKLEENEYVNYVREKPESQIVQDIFWTHPTSVKL 236
+YN K KK + GD + +Y+ ++ +++ Y E E+ ++ +FW P S
Sbjct: 177 LYNFFVKMKKKRIDGGDADRVIKYMQARQKDDMDFYYEYETDEAGCLKRLFWADPQSRID 236
Query: 237 FNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLK 296
++ F V++ DSTY+ N Y +P VGV T G A + ++ + W L+ L
Sbjct: 237 YDAFGDVVVFDSTYRVNKYNLPFIPFVGVNHHGSTVIFGCAVVSDERVGTYEWVLKQFLS 296
Query: 297 LLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGKNI 340
+ PK V+TD D +M A+ V P+S LC +H+ +N+
Sbjct: 297 CMCQKH--PKSVITDGDNAMRRAILLVFPNSDHRLCTWHIEQNM 338
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.334 0.144 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 881,740,557
Number of Sequences: 2790947
Number of extensions: 35046090
Number of successful extensions: 150713
Number of sequences better than 10.0: 305
Number of HSP's better than 10.0 without gapping: 140
Number of HSP's successfully gapped in prelim test: 165
Number of HSP's that attempted gapping in prelim test: 150374
Number of HSP's gapped (non-prelim): 386
length of query: 576
length of database: 848,049,833
effective HSP length: 133
effective length of query: 443
effective length of database: 476,853,882
effective search space: 211246269726
effective search space used: 211246269726
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 78 (34.7 bits)
Medicago: description of AC149576.7


