

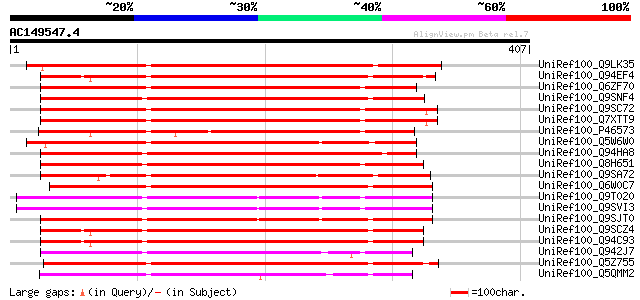
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149547.4 + phase: 0
(407 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LK35 Receptor-protein kinase-like protein [Arabidops... 248 2e-64
UniRef100_Q94EF4 Hypothetical protein P0665A11.8 [Oryza sativa] 238 3e-61
UniRef100_Q6ZF70 Putative PTH-2, resistance gene (PTO kinase) ho... 236 9e-61
UniRef100_Q9SNF4 Similar to putative Ser/Thr protein kinase [Ory... 236 1e-60
UniRef100_Q9SC72 L1332.5 protein [Oryza sativa] 236 1e-60
UniRef100_Q7XTT9 OSJNBa0058K23.13 protein [Oryza sativa] 236 1e-60
UniRef100_P46573 Protein kinase APK1B, chloroplast precursor [Ar... 235 1e-60
UniRef100_Q5W6W0 Hypothetical protein OSJNBb0059K16.2 [Oryza sat... 235 2e-60
UniRef100_Q94HA8 Hypothetical protein OSJNBb0048A17.15 [Oryza sa... 234 3e-60
UniRef100_Q8H651 Putative receptor-like protein kinase [Oryza sa... 234 3e-60
UniRef100_Q9SA72 T5I8.2 protein [Arabidopsis thaliana] 234 3e-60
UniRef100_Q6W0C7 Pto-like serine/threonine kinase [Capsicum chin... 234 4e-60
UniRef100_Q9T020 Putative receptor-like protein kinase [Arabidop... 233 6e-60
UniRef100_Q9SVI3 Putative receptor-like protein kinase [Arabidop... 233 6e-60
UniRef100_Q9SJT0 Hypothetical protein At2g21480 [Arabidopsis tha... 233 6e-60
UniRef100_Q9SCZ4 Receptor-protein kinase-like protein [Arabidops... 233 6e-60
UniRef100_Q94C93 Putative receptor-protein kinase [Arabidopsis t... 233 6e-60
UniRef100_Q942J7 Putative receptor protein kinase [Oryza sativa] 233 6e-60
UniRef100_Q5Z755 Putative receptor-like protein kinase [Oryza sa... 233 7e-60
UniRef100_Q5QMM2 Protein kinase-like [Oryza sativa] 233 1e-59
>UniRef100_Q9LK35 Receptor-protein kinase-like protein [Arabidopsis thaliana]
Length = 855
Score = 248 bits (634), Expect = 2e-64
Identities = 136/331 (41%), Positives = 202/331 (60%), Gaps = 12/331 (3%)
Query: 14 YKSSKYDKISL------RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIK 67
+KS+ ISL RCF +E+ AT F + LLG G F VYKG E +A+K
Sbjct: 479 HKSATASCISLASTHLGRCFMFQEIMDATNKFDESSLLGVGGFGRVYKGTLEDGTKVAVK 538
Query: 68 RPHSESFLSVEEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYM 127
R + S + EFR E+ +LS ++H++L+ L+GYC+E ILVYEY+ NG L ++
Sbjct: 539 RGNPRSEQGMAEFRTEIEMLSKLRHRHLVSLIGYCDERSE---MILVYEYMANGPLRSHL 595
Query: 128 MGNRRRSLTWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGL 187
G L+WKQR+ I IGAA+G+ YLH SIIHRD+K +NILL E+ AKV+DFGL
Sbjct: 596 YGADLPPLSWKQRLEICIGAARGLHYLHTGASQSIIHRDVKTTNILLDENLVAKVADFGL 655
Query: 188 VKSGPTGDQSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAE 247
K+GP+ DQ+HVS+ +KG+ GYLDP Y LT+ SDVYSFGV+L++++ RPA++
Sbjct: 656 SKTGPSLDQTHVSTAVKGSFGYLDPEYFRRQQLTEKSDVYSFGVVLMEVLCCRPALNPVL 715
Query: 248 NPSNQHIIDWARPSIEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPT 307
+I +WA +KG++ +IMD+NL + P +++ K G+ +C+A+ RP+
Sbjct: 716 PREQVNIAEWAMAWQKKGLLDQIMDSNLTGKVNPASLK---KFGETAEKCLAEYGVDRPS 772
Query: 308 MTQVCREIEHALYSDDSFTSKDSETLGSVQH 338
M V +E+AL +++ ++ S H
Sbjct: 773 MGDVLWNLEYALQLEETSSALMEPDDNSTNH 803
>UniRef100_Q94EF4 Hypothetical protein P0665A11.8 [Oryza sativa]
Length = 896
Score = 238 bits (606), Expect = 3e-61
Identities = 132/313 (42%), Positives = 197/313 (62%), Gaps = 13/313 (4%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEG---ILAIKRPHSESFLSVEEFR 81
R F+ E++ AT NF + LLG G F VY+G E++G +AIKR + S V EF+
Sbjct: 529 RHFSFAEIKAATNNFDESLLLGVGGFGKVYRG--EIDGGVTKVAIKRGNPLSEQGVHEFQ 586
Query: 82 NEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRI 141
E+ +LS ++H++L+ L+GYCEE ILVY+Y+ +G+L E++ + LTW+QR+
Sbjct: 587 TEIEMLSKLRHRHLVSLIGYCEEKNE---MILVYDYMAHGTLREHLYKTKNAPLTWRQRL 643
Query: 142 NIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSS 201
I IGAA+G+ YLH K +IIHRD+K +NILL E + AKVSDFGL K+GP+ D +HVS+
Sbjct: 644 EICIGAARGLHYLHTGAKHTIIHRDVKTTNILLDEKWVAKVSDFGLSKTGPSMDHTHVST 703
Query: 202 QIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPS 261
+KG+ GYLDP Y LT+ SDVYSFGV+L +++ ARPA++ + +WA
Sbjct: 704 VVKGSFGYLDPEYFRRQQLTEKSDVYSFGVVLFEVLCARPALNPTLAKEEVSLAEWALHC 763
Query: 262 IEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYS 321
+KGI+ +I+D +L + P + K + +CV+ E RP+M V +E AL
Sbjct: 764 QKKGILDQIVDPHLKGKIAP---QCFKKFAETAEKCVSDEGIDRPSMGDVLWNLEFALQM 820
Query: 322 DDSFTSKDSETLG 334
+S ++DS ++G
Sbjct: 821 QES--AEDSGSIG 831
>UniRef100_Q6ZF70 Putative PTH-2, resistance gene (PTO kinase) homologs [Oryza
sativa]
Length = 849
Score = 236 bits (602), Expect = 9e-61
Identities = 122/295 (41%), Positives = 185/295 (62%), Gaps = 6/295 (2%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEV 84
R FT+ E+ AT NF ++G G F VYKG E ++AIKR H ES V+EF E+
Sbjct: 503 RQFTVAEIREATMNFDDSLVIGVGGFGKVYKGEMEDGKLVAIKRGHPESQQGVKEFETEI 562
Query: 85 RLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIA 144
+LS ++H++L+ L+GYC+E ILVYE++ NG+L ++ G +LTWKQR+ I
Sbjct: 563 EILSRLRHRHLVSLIGYCDEQNE---MILVYEHMANGTLRSHLYGTDLPALTWKQRLEIC 619
Query: 145 IGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIK 204
IGAA+G+ YLH + IIHRD+K +NILL ++F AK++DFG+ K GP D +HVS+ +K
Sbjct: 620 IGAARGLHYLHTGLDRGIIHRDVKTTNILLDDNFVAKMADFGISKDGPPLDHTHVSTAVK 679
Query: 205 GTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEK 264
G+ GYLDP Y LT+ SDVYSFGV+L +++ ARP ++ A ++ +WA ++
Sbjct: 680 GSFGYLDPEYYRRQQLTQSSDVYSFGVVLFEVLCARPVINPALPRDQINLAEWALKWQKQ 739
Query: 265 GIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHAL 319
++ I+D L +E + K ++ +C+A E + RP++ +V +E AL
Sbjct: 740 KLLETIIDPRL---EGNYTLESIRKFSEIAEKCLADEGRSRPSIGEVLWHLESAL 791
>UniRef100_Q9SNF4 Similar to putative Ser/Thr protein kinase [Oryza sativa]
Length = 898
Score = 236 bits (601), Expect = 1e-60
Identities = 126/303 (41%), Positives = 194/303 (63%), Gaps = 8/303 (2%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEV 84
R F+ E++ ATKNFS D +G G F VY+G+ + + +A+KR + S + EF+ EV
Sbjct: 532 RHFSFAEIKAATKNFSNDLAIGVGGFGVVYRGVVDGDVKVAVKRSNPSSEQGITEFQTEV 591
Query: 85 RLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRR-SLTWKQRINI 143
+LS ++H++L+ L+G+CEE DG +LVY+Y+ +G+L E++ N + +L+W+ R++I
Sbjct: 592 EMLSKLRHRHLVSLIGFCEE---DGEMVLVYDYMEHGTLREHLYHNGGKPTLSWRHRLDI 648
Query: 144 AIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTG-DQSHVSSQ 202
IGAA+G+ YLH K +IIHRD+K +NIL+ +++ AKVSDFGL KSGPT +QSHVS+
Sbjct: 649 CIGAARGLHYLHTGAKYTIIHRDVKTTNILVDDNWVAKVSDFGLSKSGPTTLNQSHVSTV 708
Query: 203 IKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSI 262
+KG+ GYLDP Y LT SDVYSFGV+L +++ ARPA+D A + D+A
Sbjct: 709 VKGSFGYLDPEYYRRQQLTDKSDVYSFGVVLFEVLMARPALDPALPRDQVSLADYALACK 768
Query: 263 EKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYSD 322
G + +++D + Q P E + K +C+++ RPTM V +E A++
Sbjct: 769 RGGALPDVVDPAIRDQIAP---ECLAKFADTAEKCLSENGTERPTMGDVLWNLESAMHFQ 825
Query: 323 DSF 325
D+F
Sbjct: 826 DAF 828
>UniRef100_Q9SC72 L1332.5 protein [Oryza sativa]
Length = 844
Score = 236 bits (601), Expect = 1e-60
Identities = 125/313 (39%), Positives = 192/313 (60%), Gaps = 8/313 (2%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEV 84
R F+I E+ ATKNF + L+G+G F VYKG + +AIKR + ++EF E+
Sbjct: 503 RRFSISEIRAATKNFDEALLIGTGGFGKVYKGEVDEGTTVAIKRANPLCGQGLKEFETEI 562
Query: 85 RLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIA 144
+LS ++H++L+ ++GYCEE + ILVYEY+ G+L ++ G+ LTWKQR++
Sbjct: 563 EMLSKLRHRHLVAMIGYCEEQKE---MILVYEYMAKGTLRSHLYGSDLPPLTWKQRVDAC 619
Query: 145 IGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIK 204
IGAA+G+ YLH IIHRD+K +NILL E+F AK++DFGL K+GPT DQ+HVS+ +K
Sbjct: 620 IGAARGLHYLHTGADRGIIHRDVKTTNILLDENFVAKIADFGLSKTGPTLDQTHVSTAVK 679
Query: 205 GTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEK 264
G+ GYLDP Y LT+ SDVYSFGV+L ++ RP +D ++ +WA +
Sbjct: 680 GSFGYLDPEYFRRQQLTQKSDVYSFGVVLFEVACGRPVIDPTLPKDQINLAEWAMRWQRQ 739
Query: 265 GIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYSDDS 324
+ I+D L + E + K G++ +C+A + + RP+M +V +E+ L ++
Sbjct: 740 RSLDAIVDPRL---DGDFSSESLKKFGEIAEKCLADDGRSRPSMGEVLWHLEYVLQLHEA 796
Query: 325 F--TSKDSETLGS 335
+ + D E+ GS
Sbjct: 797 YKRNNVDCESFGS 809
>UniRef100_Q7XTT9 OSJNBa0058K23.13 protein [Oryza sativa]
Length = 844
Score = 236 bits (601), Expect = 1e-60
Identities = 125/313 (39%), Positives = 192/313 (60%), Gaps = 8/313 (2%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEV 84
R F+I E+ ATKNF + L+G+G F VYKG + +AIKR + ++EF E+
Sbjct: 503 RRFSISEIRAATKNFDEALLIGTGGFGKVYKGEVDEGTTVAIKRANPLCGQGLKEFETEI 562
Query: 85 RLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIA 144
+LS ++H++L+ ++GYCEE + ILVYEY+ G+L ++ G+ LTWKQR++
Sbjct: 563 EMLSKLRHRHLVAMIGYCEEQKE---MILVYEYMAKGTLRSHLYGSDLPPLTWKQRVDAC 619
Query: 145 IGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIK 204
IGAA+G+ YLH IIHRD+K +NILL E+F AK++DFGL K+GPT DQ+HVS+ +K
Sbjct: 620 IGAARGLHYLHTGADRGIIHRDVKTTNILLDENFVAKIADFGLSKTGPTLDQTHVSTAVK 679
Query: 205 GTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEK 264
G+ GYLDP Y LT+ SDVYSFGV+L ++ RP +D ++ +WA +
Sbjct: 680 GSFGYLDPEYFRRQQLTQKSDVYSFGVVLFEVACGRPVIDPTLPKDQINLAEWAMRWQRQ 739
Query: 265 GIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYSDDS 324
+ I+D L + E + K G++ +C+A + + RP+M +V +E+ L ++
Sbjct: 740 RSLDAIVDPRL---DGDFSSESLKKFGEIAEKCLADDGRSRPSMGEVLWHLEYVLQLHEA 796
Query: 325 F--TSKDSETLGS 335
+ + D E+ GS
Sbjct: 797 YKRNNVDCESFGS 809
>UniRef100_P46573 Protein kinase APK1B, chloroplast precursor [Arabidopsis thaliana]
Length = 412
Score = 235 bits (600), Expect = 1e-60
Identities = 128/308 (41%), Positives = 190/308 (61%), Gaps = 20/308 (6%)
Query: 23 SLRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEG----------ILAIKRPHSE 72
+L+ FT EL+ AT+NF D +LG G F +V+KG + + ++A+K+ + +
Sbjct: 53 NLKSFTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQD 112
Query: 73 SFLSVEEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMM--GN 130
+ +E+ EV L H NL+ L+GYC E E ++LVYE++P GSL ++ G+
Sbjct: 113 GWQGHQEWLAEVNYLGQFSHPNLVKLIGYCLEDEH---RLLVYEFMPRGSLENHLFRRGS 169
Query: 131 RRRSLTWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKS 190
+ L+W R+ +A+GAAKG+A+LH + S+I+RD K SNILL + AK+SDFGL K
Sbjct: 170 YFQPLSWTLRLKVALGAAKGLAFLHN-AETSVIYRDFKTSNILLDSEYNAKLSDFGLAKD 228
Query: 191 GPTGDQSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPS 250
GPTGD+SHVS++I GT GY P Y ++ HLT SDVYS+GV+LL+++S R AVD P
Sbjct: 229 GPTGDKSHVSTRIMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPG 288
Query: 251 NQHIIDWARPSI-EKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMT 309
Q +++WARP + K + ++D L + +ME KV L +RC+ E K RP M
Sbjct: 289 EQKLVEWARPLLANKRKLFRVIDNRL---QDQYSMEEACKVATLALRCLTFEIKLRPNMN 345
Query: 310 QVCREIEH 317
+V +EH
Sbjct: 346 EVVSHLEH 353
>UniRef100_Q5W6W0 Hypothetical protein OSJNBb0059K16.2 [Oryza sativa]
Length = 846
Score = 235 bits (599), Expect = 2e-60
Identities = 136/311 (43%), Positives = 192/311 (61%), Gaps = 13/311 (4%)
Query: 14 YKSSKYDKISLRC--FTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGI-LAIKRPH 70
Y++ Y S C FT +E++ AT+NF + LLG G F +VY+G + G +AIKR +
Sbjct: 486 YRTEFYHSPSNLCRNFTFDEIQVATRNFDESLLLGRGGFGDVYRGEIDNNGENVAIKRSN 545
Query: 71 SESFLSVEEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGN 130
S V EF+ E+ LLS +++ +L+ L+GYC+E ILVYEY+ G+L E++ +
Sbjct: 546 PLSVQGVHEFQTEIELLSKLRYCHLVSLIGYCKEKNE---MILVYEYMAQGTLREHLYNS 602
Query: 131 RRRSLTWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKS 190
+ SL WKQR+ I IGAA+G+ YLH +IIHRD+K +NILL + + AKVSDFGL K+
Sbjct: 603 NKPSLPWKQRLKICIGAARGLHYLHMGANQTIIHRDVKTANILLDDKWVAKVSDFGLSKA 662
Query: 191 GPTGDQSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPS 250
P D +HVS+ +KGT GYLDP Y LT+ SDVYSFGV+L +++ ARPAV N E P
Sbjct: 663 NPDIDSTHVSTVVKGTFGYLDPEYYRRKQLTQKSDVYSFGVVLFEILCARPAV-NIELPE 721
Query: 251 NQ-HIIDWARPSIEKGIIAEIMDANLFCQ-SEPCNMEVMLKVGQLGIRCVAQEPKHRPTM 308
Q + DWA +KG++ +I+D +L + S PC + +CVA RP M
Sbjct: 722 EQASLRDWALSCQKKGMLGKIIDPHLHGEISPPC----LRMFADCAKQCVADRSIDRPLM 777
Query: 309 TQVCREIEHAL 319
+ V +E AL
Sbjct: 778 SDVLWSLEAAL 788
>UniRef100_Q94HA8 Hypothetical protein OSJNBb0048A17.15 [Oryza sativa]
Length = 843
Score = 234 bits (598), Expect = 3e-60
Identities = 124/296 (41%), Positives = 190/296 (63%), Gaps = 7/296 (2%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEV 84
R FT E+++ATKNF + ++G G F VY G+ E LAIKR + S + EF E+
Sbjct: 511 RYFTFVEIQKATKNFEEKAVIGVGGFGKVYLGVLEDGTKLAIKRGNPSSDQGMNEFLTEI 570
Query: 85 RLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRR-RSLTWKQRINI 143
++LS ++H++L+ L+G C+E + ILVYE++ NG L +++ G + L+WKQR+ I
Sbjct: 571 QMLSKLRHRHLVSLIGCCDE---NNEMILVYEFMSNGPLRDHLYGGTDIKPLSWKQRLEI 627
Query: 144 AIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQI 203
+IGAAKG+ YLH IIHRD+K +NILL E+F AKV+DFGL K+ P+ +Q+HVS+ +
Sbjct: 628 SIGAAKGLHYLHTGAAQGIIHRDVKTTNILLDENFVAKVADFGLSKAAPSLEQTHVSTAV 687
Query: 204 KGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIE 263
KG+ GYLDP Y LT+ SDVYSFGV+L +++ ARPA++ ++ +WAR
Sbjct: 688 KGSFGYLDPEYFRRQQLTEKSDVYSFGVVLFEVLCARPAINPTLPRDQVNLAEWARTWHR 747
Query: 264 KGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHAL 319
KG + +I+D ++ Q P ++E+ + + +C+A RP+M V ++E AL
Sbjct: 748 KGELNKIIDPHISGQIRPDSLEIFAEAAE---KCLADYGVDRPSMGDVLWKLEFAL 800
>UniRef100_Q8H651 Putative receptor-like protein kinase [Oryza sativa]
Length = 845
Score = 234 bits (598), Expect = 3e-60
Identities = 125/300 (41%), Positives = 182/300 (60%), Gaps = 6/300 (2%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEV 84
R F+ E++ ATKNF + ++G G F NVY G + +A+KR + +S + EF E+
Sbjct: 501 RFFSFAEIQAATKNFEESAIIGVGGFGNVYIGEIDDGTKVAVKRGNPQSEQGINEFNTEI 560
Query: 85 RLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIA 144
++LS ++H++L+ L+GYC+E + ILVYEY+ NG +++ G +LTWKQR+ I
Sbjct: 561 QMLSKLRHRHLVSLIGYCDE---NAEMILVYEYMHNGPFRDHIYGKDLPALTWKQRLEIC 617
Query: 145 IGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIK 204
IGAA+G+ YLH IIHRD+K +NILL ++F AKVSDFGL K GP +Q HVS+ +K
Sbjct: 618 IGAARGLHYLHTGTAQGIIHRDVKTTNILLDDNFVAKVSDFGLSKDGPGMNQLHVSTAVK 677
Query: 205 GTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEK 264
G+ GYLDP Y LT SDVYSFGV+LL+ + ARP +D + +W K
Sbjct: 678 GSFGYLDPEYFRCQQLTDKSDVYSFGVVLLETLCARPPIDPQLPREQVSLAEWGMQWKRK 737
Query: 265 GIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYSDDS 324
G+I +IMD L + N E + K + +C+A+ R +M V +E+AL D+
Sbjct: 738 GLIEKIMDPKL---AGTVNQESLNKFAEAAEKCLAEFGSDRISMGDVLWNLEYALQLQDA 794
>UniRef100_Q9SA72 T5I8.2 protein [Arabidopsis thaliana]
Length = 849
Score = 234 bits (597), Expect = 3e-60
Identities = 130/309 (42%), Positives = 196/309 (63%), Gaps = 12/309 (3%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKR--PHSESFLSVEEFRN 82
R FT+ E+ ATKNF +G G F VY+G E ++AIKR PHS+ L+ EF
Sbjct: 506 RKFTLAEIRAATKNFDDGLAIGVGGFGKVYRGELEDGTLIAIKRATPHSQQGLA--EFET 563
Query: 83 EVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRIN 142
E+ +LS ++H++L+ L+G+C+E ILVYEY+ NG+L ++ G+ L+WKQR+
Sbjct: 564 EIVMLSRLRHRHLVSLIGFCDEHNE---MILVYEYMANGTLRSHLFGSNLPPLSWKQRLE 620
Query: 143 IAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQ 202
IG+A+G+ YLH + IIHRD+K +NILL E+F AK+SDFGL K+GP+ D +HVS+
Sbjct: 621 ACIGSARGLHYLHTGSERGIIHRDVKTTNILLDENFVAKMSDFGLSKAGPSMDHTHVSTA 680
Query: 203 IKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQ-HIIDWARPS 261
+KG+ GYLDP Y LT+ SDVYSFGV+L + + AR AV N P +Q ++ +WA
Sbjct: 681 VKGSFGYLDPEYFRRQQLTEKSDVYSFGVVLFEAVCAR-AVINPTLPKDQINLAEWALSW 739
Query: 262 IEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYS 321
++ + I+D+NL P ++E K G++ +C+A E K+RP M +V +E+ L
Sbjct: 740 QKQRNLESIIDSNLRGNYSPESLE---KYGEIAEKCLADEGKNRPMMGEVLWSLEYVLQI 796
Query: 322 DDSFTSKDS 330
+++ K +
Sbjct: 797 HEAWLRKQN 805
>UniRef100_Q6W0C7 Pto-like serine/threonine kinase [Capsicum chinense]
Length = 359
Score = 234 bits (596), Expect = 4e-60
Identities = 124/300 (41%), Positives = 185/300 (61%), Gaps = 6/300 (2%)
Query: 32 LERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEVRLLSAVK 91
L AT NF + ++G G F VYKG+ LA+KR + +S + EFR E+ +LS +
Sbjct: 11 LLEATSNFDESLVIGIGGFGKVYKGVLYDGTKLAVKRGNPKSQQGLAEFRTEIEMLSQFR 70
Query: 92 HKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIAIGAAKGI 151
H++L+ L+GYC+E ILVYEY+ NG+L ++ G+ S++WKQR+ I IG+A+G+
Sbjct: 71 HRHLVSLMGYCDEKNE---MILVYEYMENGTLKSHLYGSDLPSMSWKQRLEICIGSARGL 127
Query: 152 AYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIKGTPGYLD 211
YLH ++IHRD+K +NILL ESF AKV+DFGL K+GP DQ+HVS+ +KG+ GYLD
Sbjct: 128 HYLHTGYAKAVIHRDVKSANILLDESFMAKVADFGLSKTGPELDQTHVSTAVKGSFGYLD 187
Query: 212 PAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEKGIIAEIM 271
P Y LT+ SDVYSFGV+L +++ ARP +D + ++ +WA +KG + +I+
Sbjct: 188 PEYFRRQQLTEKSDVYSFGVVLFEVLCARPVIDPSLPREMVNLAEWAMKWQKKGQLEQII 247
Query: 272 DANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYSDDSFTSKDSE 331
D L + P + + K G+ +C+A RP+M V E+AL ++ D E
Sbjct: 248 DPTLVGKIRP---DSLRKFGETAEKCLADFGVDRPSMGDVLWNXEYALQLQEAVIQDDPE 304
>UniRef100_Q9T020 Putative receptor-like protein kinase [Arabidopsis thaliana]
Length = 878
Score = 233 bits (595), Expect = 6e-60
Identities = 134/329 (40%), Positives = 199/329 (59%), Gaps = 11/329 (3%)
Query: 6 FSFDAGKEYKSSKYDK-ISL-RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGI 63
F G KS+ Y+ + L R F++ EL+ ATKNF ++G G F NVY G +
Sbjct: 491 FMTSKGGSQKSNFYNSTLGLGRYFSLSELQEATKNFEASQIIGVGGFGNVYIGTLDDGTK 550
Query: 64 LAIKRPHSESFLSVEEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSL 123
+A+KR + +S + EF+ E+++LS ++H++L+ L+GYC+E + ILVYE++ NG
Sbjct: 551 VAVKRGNPQSEQGITEFQTEIQMLSKLRHRHLVSLIGYCDE---NSEMILVYEFMSNGPF 607
Query: 124 LEYMMGNRRRSLTWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVS 183
+++ G LTWKQR+ I IG+A+G+ YLH IIHRD+K +NILL E+ AKV+
Sbjct: 608 RDHLYGKNLAPLTWKQRLEICIGSARGLHYLHTGTAQGIIHRDVKSTNILLDEALVAKVA 667
Query: 184 DFGLVKSGPTGDQSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAV 243
DFGL K G Q+HVS+ +KG+ GYLDP Y LT SDVYSFGV+LL+ + ARPA+
Sbjct: 668 DFGLSKDVAFG-QNHVSTAVKGSFGYLDPEYFRRQQLTDKSDVYSFGVVLLEALCARPAI 726
Query: 244 DNAENPSNQ-HIIDWARPSIEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEP 302
N + P Q ++ +WA KG++ +I+D +L + N E M K + +C+
Sbjct: 727 -NPQLPREQVNLAEWAMQWKRKGLLEKIIDPHL---AGTINPESMKKFAEAAEKCLEDYG 782
Query: 303 KHRPTMTQVCREIEHALYSDDSFTSKDSE 331
RPTM V +E+AL ++FT +E
Sbjct: 783 VDRPTMGDVLWNLEYALQLQEAFTQGKAE 811
>UniRef100_Q9SVI3 Putative receptor-like protein kinase [Arabidopsis thaliana]
Length = 573
Score = 233 bits (595), Expect = 6e-60
Identities = 134/329 (40%), Positives = 199/329 (59%), Gaps = 11/329 (3%)
Query: 6 FSFDAGKEYKSSKYDK-ISL-RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGI 63
F G KS+ Y+ + L R F++ EL+ ATKNF ++G G F NVY G +
Sbjct: 186 FMTSKGGSQKSNFYNSTLGLGRYFSLSELQEATKNFEASQIIGVGGFGNVYIGTLDDGTK 245
Query: 64 LAIKRPHSESFLSVEEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSL 123
+A+KR + +S + EF+ E+++LS ++H++L+ L+GYC+E + ILVYE++ NG
Sbjct: 246 VAVKRGNPQSEQGITEFQTEIQMLSKLRHRHLVSLIGYCDE---NSEMILVYEFMSNGPF 302
Query: 124 LEYMMGNRRRSLTWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVS 183
+++ G LTWKQR+ I IG+A+G+ YLH IIHRD+K +NILL E+ AKV+
Sbjct: 303 RDHLYGKNLAPLTWKQRLEICIGSARGLHYLHTGTAQGIIHRDVKSTNILLDEALVAKVA 362
Query: 184 DFGLVKSGPTGDQSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAV 243
DFGL K G Q+HVS+ +KG+ GYLDP Y LT SDVYSFGV+LL+ + ARPA+
Sbjct: 363 DFGLSKDVAFG-QNHVSTAVKGSFGYLDPEYFRRQQLTDKSDVYSFGVVLLEALCARPAI 421
Query: 244 DNAENPSNQ-HIIDWARPSIEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEP 302
N + P Q ++ +WA KG++ +I+D +L + N E M K + +C+
Sbjct: 422 -NPQLPREQVNLAEWAMQWKRKGLLEKIIDPHL---AGTINPESMKKFAEAAEKCLEDYG 477
Query: 303 KHRPTMTQVCREIEHALYSDDSFTSKDSE 331
RPTM V +E+AL ++FT +E
Sbjct: 478 VDRPTMGDVLWNLEYALQLQEAFTQGKAE 506
>UniRef100_Q9SJT0 Hypothetical protein At2g21480 [Arabidopsis thaliana]
Length = 871
Score = 233 bits (595), Expect = 6e-60
Identities = 130/308 (42%), Positives = 189/308 (61%), Gaps = 9/308 (2%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEV 84
R F++ EL+ TKNF ++G G F NVY G + +AIKR + +S + EF E+
Sbjct: 511 RYFSLSELQEVTKNFDASEIIGVGGFGNVYIGTIDDGTQVAIKRGNPQSEQGITEFHTEI 570
Query: 85 RLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIA 144
++LS ++H++L+ L+GYC+E + ILVYEY+ NG +++ G LTWKQR+ I
Sbjct: 571 QMLSKLRHRHLVSLIGYCDE---NAEMILVYEYMSNGPFRDHLYGKNLSPLTWKQRLEIC 627
Query: 145 IGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIK 204
IGAA+G+ YLH IIHRD+K +NILL E+ AKV+DFGL K G Q+HVS+ +K
Sbjct: 628 IGAARGLHYLHTGTAQGIIHRDVKSTNILLDEALVAKVADFGLSKDVAFG-QNHVSTAVK 686
Query: 205 GTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQ-HIIDWARPSIE 263
G+ GYLDP Y LT SDVYSFGV+LL+ + ARPA+ N + P Q ++ +WA +
Sbjct: 687 GSFGYLDPEYFRRQQLTDKSDVYSFGVVLLEALCARPAI-NPQLPREQVNLAEWAMLWKQ 745
Query: 264 KGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYSDD 323
KG++ +I+D +L P E M K + +C+A RPTM V +E+AL +
Sbjct: 746 KGLLEKIIDPHLVGAVNP---ESMKKFAEAAEKCLADYGVDRPTMGDVLWNLEYALQLQE 802
Query: 324 SFTSKDSE 331
+F+ +E
Sbjct: 803 AFSQGKAE 810
>UniRef100_Q9SCZ4 Receptor-protein kinase-like protein [Arabidopsis thaliana]
Length = 895
Score = 233 bits (595), Expect = 6e-60
Identities = 129/303 (42%), Positives = 190/303 (62%), Gaps = 11/303 (3%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEG---ILAIKRPHSESFLSVEEFR 81
R F+ E++ ATKNF + +LG G F VY+G E++G +AIKR + S V EF+
Sbjct: 522 RHFSFAEIKAATKNFDESRVLGVGGFGKVYRG--EIDGGTTKVAIKRGNPMSEQGVHEFQ 579
Query: 82 NEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRI 141
E+ +LS ++H++L+ L+GYCEE + ILVY+Y+ +G++ E++ + SL WKQR+
Sbjct: 580 TEIEMLSKLRHRHLVSLIGYCEE---NCEMILVYDYMAHGTMREHLYKTQNPSLPWKQRL 636
Query: 142 NIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSS 201
I IGAA+G+ YLH K +IIHRD+K +NILL E + AKVSDFGL K+GPT D +HVS+
Sbjct: 637 EICIGAARGLHYLHTGAKHTIIHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDHTHVST 696
Query: 202 QIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPS 261
+KG+ GYLDP Y LT+ SDVYSFGV+L + + ARPA++ + +WA
Sbjct: 697 VVKGSFGYLDPEYFRRQQLTEKSDVYSFGVVLFEALCARPALNPTLAKEQVSLAEWAPYC 756
Query: 262 IEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYS 321
+KG++ +I+D L + P E K + ++CV + RP+M V +E AL
Sbjct: 757 YKKGMLDQIVDPYLKGKITP---ECFKKFAETAMKCVLDQGIERPSMGDVLWNLEFALQL 813
Query: 322 DDS 324
+S
Sbjct: 814 QES 816
>UniRef100_Q94C93 Putative receptor-protein kinase [Arabidopsis thaliana]
Length = 895
Score = 233 bits (595), Expect = 6e-60
Identities = 129/303 (42%), Positives = 190/303 (62%), Gaps = 11/303 (3%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEG---ILAIKRPHSESFLSVEEFR 81
R F+ E++ ATKNF + +LG G F VY+G E++G +AIKR + S V EF+
Sbjct: 522 RHFSFAEIKAATKNFDESRVLGVGVFGKVYRG--EIDGGTTKVAIKRGNPMSEQGVHEFQ 579
Query: 82 NEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRI 141
E+ +LS ++H++L+ L+GYCEE + ILVY+Y+ +G++ E++ + SL WKQR+
Sbjct: 580 TEIEMLSKLRHRHLVSLIGYCEE---NCEMILVYDYMAHGTMREHLYKTQNPSLPWKQRL 636
Query: 142 NIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSS 201
I IGAA+G+ YLH K +IIHRD+K +NILL E + AKVSDFGL K+GPT D +HVS+
Sbjct: 637 EICIGAARGLHYLHTGAKHTIIHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDHTHVST 696
Query: 202 QIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPS 261
+KG+ GYLDP Y LT+ SDVYSFGV+L + + ARPA++ + +WA
Sbjct: 697 VVKGSFGYLDPEYFRRQQLTEKSDVYSFGVVLFEALCARPALNPTLAKEQVSLAEWAPYC 756
Query: 262 IEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYS 321
+KG++ +I+D L + P E K + ++CV + RP+M V +E AL
Sbjct: 757 YKKGMLDQIVDPYLKGKITP---ECFKKFAETAMKCVLDQGIERPSMGDVLWNLEFALQL 813
Query: 322 DDS 324
+S
Sbjct: 814 QES 816
>UniRef100_Q942J7 Putative receptor protein kinase [Oryza sativa]
Length = 856
Score = 233 bits (595), Expect = 6e-60
Identities = 130/297 (43%), Positives = 180/297 (59%), Gaps = 16/297 (5%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEV 84
R F EEL+R T NFS+ +GSG + VYKG+ + AIKR S EF+NE+
Sbjct: 510 RYFAFEELKRCTNNFSETQEIGSGGYGKVYKGMLANGQMAAIKRAQQGSMQGAAEFKNEI 569
Query: 85 RLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIA 144
LLS V HKNL+ LVG+C E G ++LVYEY+PNG+L E + G L WK+R+ IA
Sbjct: 570 ELLSRVHHKNLVSLVGFCYE---QGEQMLVYEYIPNGTLRENLKGKGGMHLDWKKRLQIA 626
Query: 145 IGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIK 204
+G+AKG+AYLHE P IIHRDIK +NILL ES AKV+DFGL K + HVS+Q+K
Sbjct: 627 VGSAKGLAYLHELADPPIIHRDIKSTNILLDESLNAKVADFGLSKLVSDTKKGHVSTQVK 686
Query: 205 GTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEK 264
GT GYLDP Y + L++ SDVYSFGV++L+LI++R ++ +I+ R +I++
Sbjct: 687 GTLGYLDPEYYMTQQLSEKSDVYSFGVVMLELITSRQPIE-----KGTYIVREIRTAIDQ 741
Query: 265 GI-----IAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIE 316
+ ++D + + M + QL + CV + RPTM V +E+E
Sbjct: 742 YDQEYYGLKSLIDPTI---RDSAKMVGFRRFVQLAMECVEESAADRPTMNDVVKELE 795
>UniRef100_Q5Z755 Putative receptor-like protein kinase [Oryza sativa]
Length = 859
Score = 233 bits (594), Expect = 7e-60
Identities = 125/310 (40%), Positives = 188/310 (60%), Gaps = 10/310 (3%)
Query: 27 FTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEVRL 86
F L+ AT NF ++ ++G G F VYKG+ + +A+KR + +S + EFR E+ L
Sbjct: 503 FAFSVLQEATNNFDENWVIGVGGFGKVYKGVLRDDTKVAVKRGNPKSQQGLNEFRTEIEL 562
Query: 87 LSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIAIG 146
LS ++H++L+ L+GYC+E ILVYEY+ G+L ++ G+ SL WKQR+ I IG
Sbjct: 563 LSRLRHRHLVSLIGYCDERNE---MILVYEYMEKGTLKSHLYGSDNPSLNWKQRLEICIG 619
Query: 147 AAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIKGT 206
AA+G+ YLH +IIHRD+K +NILL E+ AKV+DFGL K+GP DQ+HVS+ +KG+
Sbjct: 620 AARGLHYLHTGSAKAIIHRDVKSANILLDENLLAKVADFGLSKTGPELDQTHVSTAVKGS 679
Query: 207 PGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEKGI 266
GYLDP Y LT+ SDVYSFGV+LL+++ ARP +D ++ +W ++G
Sbjct: 680 FGYLDPEYFRRQQLTEKSDVYSFGVVLLEVLCARPVIDPTLPREMVNLAEWGMKWQKRGE 739
Query: 267 IAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYSDDSFT 326
+ +I+D + P + + K G+ +C+A RP+M V +E+ L D+
Sbjct: 740 LHQIVDQRVSGSIRP---DSLRKFGETVEKCLADYGVERPSMGDVLWNLEYVLQLQDA-- 794
Query: 327 SKDSETLGSV 336
DS T+ V
Sbjct: 795 --DSSTVSDV 802
>UniRef100_Q5QMM2 Protein kinase-like [Oryza sativa]
Length = 478
Score = 233 bits (593), Expect = 1e-59
Identities = 123/298 (41%), Positives = 179/298 (59%), Gaps = 16/298 (5%)
Query: 24 LRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNE 83
+RCFT +E+ AT +F+ +G G + VYKG +AIKR H S +EF E
Sbjct: 127 VRCFTFDEMAAATNDFTDSAQVGQGGYGKVYKGNLTDGTAVAIKRAHEGSLQGSKEFCTE 186
Query: 84 VRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINI 143
+ LLS + H+NL+ LVGYC+E + ++LVYE++PNG+L +++ RR L + QRI+I
Sbjct: 187 IELLSRLHHRNLVSLVGYCDEEDE---QMLVYEFMPNGTLRDHLSAKSRRPLNFSQRIHI 243
Query: 144 AIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGD-----QSH 198
A+GAAKGI YLH + P I HRD+K SNILL F AKV+DFGL + P D +H
Sbjct: 244 ALGAAKGILYLHTEADPPIFHRDVKASNILLDSKFVAKVADFGLSRLAPVPDVDGTMPAH 303
Query: 199 VSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWA 258
+S+ +KGTPGYLDP Y + LT SDVYS GV+LL+L++ + + +N I+
Sbjct: 304 ISTVVKGTPGYLDPEYFLTHKLTDKSDVYSLGVVLLELLTGMKPIQHGKN-----IVREV 358
Query: 259 RPSIEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIE 316
+ + G IA ++D + S P E + ++ L ++C E RP+M V RE++
Sbjct: 359 NTAYQSGEIAGVIDERISSSSSP---ECVARLASLAVKCCKDETDARPSMADVVRELD 413
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 626,884,867
Number of Sequences: 2790947
Number of extensions: 25463886
Number of successful extensions: 99818
Number of sequences better than 10.0: 17885
Number of HSP's better than 10.0 without gapping: 7622
Number of HSP's successfully gapped in prelim test: 10264
Number of HSP's that attempted gapping in prelim test: 67407
Number of HSP's gapped (non-prelim): 19788
length of query: 407
length of database: 848,049,833
effective HSP length: 130
effective length of query: 277
effective length of database: 485,226,723
effective search space: 134407802271
effective search space used: 134407802271
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC149547.4


