

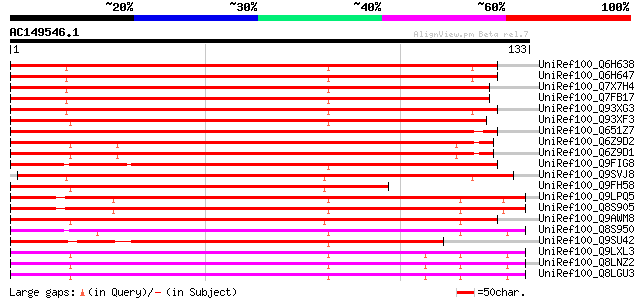
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149546.1 - phase: 0
(133 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6H638 Putative kinesin heavy chain [Oryza sativa] 140 8e-33
UniRef100_Q6H647 Putative kinesin heavy chain [Oryza sativa] 140 8e-33
UniRef100_Q7X7H4 OSJNBa0011L07.1 protein [Oryza sativa] 136 1e-31
UniRef100_Q7FB17 OSJNBa0091D06.23 protein [Oryza sativa] 136 1e-31
UniRef100_Q93XG3 Kinesin heavy chain [Zea mays] 135 2e-31
UniRef100_Q93XF3 Kinesin heavy chain [Zea mays] 133 1e-30
UniRef100_Q651Z7 Putative kinesin heavy chain [Oryza sativa] 128 2e-29
UniRef100_Q6Z9D2 Putative kinesin heavy chain [Oryza sativa] 120 7e-27
UniRef100_Q6Z9D1 Putative kinesin heavy chain [Oryza sativa] 120 7e-27
UniRef100_Q9FIG8 Kinesin heavy chain-like protein [Arabidopsis t... 117 7e-26
UniRef100_Q9SVJ8 Kinesin like protein [Arabidopsis thaliana] 112 1e-24
UniRef100_Q9FH58 Kinesin heavy chain DNA binding protein-like [A... 106 1e-22
UniRef100_Q9LPQ5 F15H18.12 [Arabidopsis thaliana] 101 4e-21
UniRef100_Q8S905 AtNACK1 kinesin-like protein [Arabidopsis thali... 101 4e-21
UniRef100_Q9AWM8 Putative KIF3 protein [Oryza sativa] 97 6e-20
UniRef100_Q8S950 Kinesin-like protein NACK1 [Nicotiana tabacum] 97 1e-19
UniRef100_Q9SU42 Hypothetical protein AT4g24170 [Arabidopsis tha... 92 3e-18
UniRef100_Q9LXL3 Kinesin-like protein [Arabidopsis thaliana] 91 4e-18
UniRef100_Q8LNZ2 Kinesin-like protein [Arabidopsis thaliana] 91 4e-18
UniRef100_Q8LGU3 Putative kinesin-like protein [Arabidopsis thal... 91 4e-18
>UniRef100_Q6H638 Putative kinesin heavy chain [Oryza sativa]
Length = 884
Score = 140 bits (352), Expect = 8e-33
Identities = 72/137 (52%), Positives = 100/137 (72%), Gaps = 12/137 (8%)
Query: 1 MEVELRRLSYLKD--------NQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSER 52
MEVELRRLS+L+D + + + +P +S + L+RER+ML+RQMQ++LS ER
Sbjct: 740 MEVELRRLSFLRDTYSRGSTPSNAIVGSLSTSPVASAKKLQREREMLARQMQKRLSTEER 799
Query: 53 DNMYLKWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAFKEMFGL 111
++ Y KWG+S+ SK R+LQ+A RLW+ET D+ HVRESA++VAKL+G EP Q KEMFGL
Sbjct: 800 EHTYTKWGVSLDSKRRKLQVARRLWTETKDLEHVRESASLVAKLIGLQEPGQVLKEMFGL 859
Query: 112 NFAPRR---RRKKSFGW 125
+FAP++ RR+ S GW
Sbjct: 860 SFAPQQQPTRRRSSNGW 876
>UniRef100_Q6H647 Putative kinesin heavy chain [Oryza sativa]
Length = 650
Score = 140 bits (352), Expect = 8e-33
Identities = 72/137 (52%), Positives = 100/137 (72%), Gaps = 12/137 (8%)
Query: 1 MEVELRRLSYLKD--------NQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSER 52
MEVELRRLS+L+D + + + +P +S + L+RER+ML+RQMQ++LS ER
Sbjct: 506 MEVELRRLSFLRDTYSRGSTPSNAIVGSLSTSPVASAKKLQREREMLARQMQKRLSTEER 565
Query: 53 DNMYLKWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAFKEMFGL 111
++ Y KWG+S+ SK R+LQ+A RLW+ET D+ HVRESA++VAKL+G EP Q KEMFGL
Sbjct: 566 EHTYTKWGVSLDSKRRKLQVARRLWTETKDLEHVRESASLVAKLIGLQEPGQVLKEMFGL 625
Query: 112 NFAPRR---RRKKSFGW 125
+FAP++ RR+ S GW
Sbjct: 626 SFAPQQQPTRRRSSNGW 642
>UniRef100_Q7X7H4 OSJNBa0011L07.1 protein [Oryza sativa]
Length = 945
Score = 136 bits (342), Expect = 1e-31
Identities = 70/127 (55%), Positives = 98/127 (77%), Gaps = 4/127 (3%)
Query: 1 MEVELRRLSYLKD---NQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYL 57
MEVELRRLS+LKD N + + SS + L+RER+ML RQMQR+LS ER++MY
Sbjct: 808 MEVELRRLSFLKDTYSNGAIASIPNTSLVSSAKKLQREREMLCRQMQRRLSIEERESMYT 867
Query: 58 KWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPR 116
KWG+S++SK RRLQ+A LW+ET D+ HVRESA++VA+L+G +EP +A +EMFGL+FAP+
Sbjct: 868 KWGVSLASKRRRLQVARCLWTETKDLEHVRESASLVARLIGLLEPGKALREMFGLSFAPQ 927
Query: 117 RRRKKSF 123
+ ++S+
Sbjct: 928 QFTRRSY 934
>UniRef100_Q7FB17 OSJNBa0091D06.23 protein [Oryza sativa]
Length = 915
Score = 136 bits (342), Expect = 1e-31
Identities = 70/127 (55%), Positives = 98/127 (77%), Gaps = 4/127 (3%)
Query: 1 MEVELRRLSYLKD---NQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYL 57
MEVELRRLS+LKD N + + SS + L+RER+ML RQMQR+LS ER++MY
Sbjct: 778 MEVELRRLSFLKDTYSNGAIASIPNTSLVSSAKKLQREREMLCRQMQRRLSIEERESMYT 837
Query: 58 KWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPR 116
KWG+S++SK RRLQ+A LW+ET D+ HVRESA++VA+L+G +EP +A +EMFGL+FAP+
Sbjct: 838 KWGVSLASKRRRLQVARCLWTETKDLEHVRESASLVARLIGLLEPGKALREMFGLSFAPQ 897
Query: 117 RRRKKSF 123
+ ++S+
Sbjct: 898 QFTRRSY 904
>UniRef100_Q93XG3 Kinesin heavy chain [Zea mays]
Length = 766
Score = 135 bits (340), Expect = 2e-31
Identities = 70/138 (50%), Positives = 98/138 (70%), Gaps = 13/138 (9%)
Query: 1 MEVELRRLSYLKD--------NQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSER 52
MEVELRRLS+L+D + + +P +S + L+RER+ML+RQMQ++L+ ER
Sbjct: 621 MEVELRRLSFLRDTYSRGNTPSNAVVGSLNSSPAASAKKLQREREMLARQMQKRLTAEER 680
Query: 53 DNMYLKWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAFKEMFGL 111
+ +Y KWGIS+ SK R+LQ+A RLW+E D+ HVRESA++VAKL+G EP Q +EMFGL
Sbjct: 681 ERLYTKWGISLDSKKRKLQVARRLWTEAEDLEHVRESASLVAKLIGLQEPGQVLREMFGL 740
Query: 112 NFAPRR----RRKKSFGW 125
+FAP++ RR+ S GW
Sbjct: 741 SFAPQQQPPPRRRSSNGW 758
>UniRef100_Q93XF3 Kinesin heavy chain [Zea mays]
Length = 897
Score = 133 bits (334), Expect = 1e-30
Identities = 67/131 (51%), Positives = 99/131 (75%), Gaps = 9/131 (6%)
Query: 1 MEVELRRLSYLKDN--------QILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSER 52
MEVELRRLS+LK+ ++ + + SS + L+RER+ML RQMQ++L+ ER
Sbjct: 755 MEVELRRLSFLKNTYSNGSMGYDMVAGSLSTSLVSSAKKLQREREMLCRQMQKRLTIQER 814
Query: 53 DNMYLKWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAFKEMFGL 111
+++Y KWG+S+SSK RRLQ+A RLW+ET D+ HVRESA++VA+L+G +EP +A +EMFGL
Sbjct: 815 ESLYTKWGVSLSSKRRRLQVARRLWTETRDLEHVRESASLVARLIGLLEPGKALREMFGL 874
Query: 112 NFAPRRRRKKS 122
+FAP++ ++S
Sbjct: 875 SFAPQQSTRRS 885
>UniRef100_Q651Z7 Putative kinesin heavy chain [Oryza sativa]
Length = 862
Score = 128 bits (322), Expect = 2e-29
Identities = 68/125 (54%), Positives = 90/125 (71%), Gaps = 2/125 (1%)
Query: 1 MEVELRRLSYLKDNQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWG 60
MEVE RRLS++K++ I + T SS R LR ER ML RQM RKL +E++ +Y KWG
Sbjct: 733 MEVEHRRLSFIKNSLIADGELHATTASSLRNLRHERDMLYRQMVRKLHLAEKERLYGKWG 792
Query: 61 ISMSSKHRRLQLAHRLWSETDINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPRRRRK 120
I MS+K RRLQL+ R+W++T ++HVRESA +VAKLV +E QA +EMFGL+F+ + RR
Sbjct: 793 IDMSTKQRRLQLSRRIWTQTGMDHVRESAALVAKLVEHLEKGQAIREMFGLSFSFKPRR- 851
Query: 121 KSFGW 125
SF W
Sbjct: 852 -SFSW 855
>UniRef100_Q6Z9D2 Putative kinesin heavy chain [Oryza sativa]
Length = 1003
Score = 120 bits (301), Expect = 7e-27
Identities = 67/129 (51%), Positives = 91/129 (69%), Gaps = 6/129 (4%)
Query: 1 MEVELRRLSYLKDN-QILEDGRTLTPE--SSKRYLRRERQMLSRQMQRKLSKSERDNMYL 57
MEVE RRLS+++ + G L SS + LRRER ML +QM +KL+ E++ +Y
Sbjct: 872 MEVEHRRLSFIRRSFSASPAGGELNSAVVSSLKNLRRERDMLYKQMLKKLTNGEKERVYA 931
Query: 58 KWGISMSSKHRRLQLAHRLWSETDINHVRESATIVAKLVGTVEPDQAFKEMFGLNF--AP 115
+WGI +SSK RRLQL+ +W++TD+ H+RESA++VAKL+ +EP QA KEMFGLNF AP
Sbjct: 932 RWGIDLSSKQRRLQLSRLVWTQTDMEHIRESASLVAKLIELLEPAQALKEMFGLNFTLAP 991
Query: 116 RRRRKKSFG 124
R R +SFG
Sbjct: 992 RSER-RSFG 999
>UniRef100_Q6Z9D1 Putative kinesin heavy chain [Oryza sativa]
Length = 986
Score = 120 bits (301), Expect = 7e-27
Identities = 67/129 (51%), Positives = 91/129 (69%), Gaps = 6/129 (4%)
Query: 1 MEVELRRLSYLKDN-QILEDGRTLTPE--SSKRYLRRERQMLSRQMQRKLSKSERDNMYL 57
MEVE RRLS+++ + G L SS + LRRER ML +QM +KL+ E++ +Y
Sbjct: 855 MEVEHRRLSFIRRSFSASPAGGELNSAVVSSLKNLRRERDMLYKQMLKKLTNGEKERVYA 914
Query: 58 KWGISMSSKHRRLQLAHRLWSETDINHVRESATIVAKLVGTVEPDQAFKEMFGLNF--AP 115
+WGI +SSK RRLQL+ +W++TD+ H+RESA++VAKL+ +EP QA KEMFGLNF AP
Sbjct: 915 RWGIDLSSKQRRLQLSRLVWTQTDMEHIRESASLVAKLIELLEPAQALKEMFGLNFTLAP 974
Query: 116 RRRRKKSFG 124
R R +SFG
Sbjct: 975 RSER-RSFG 982
>UniRef100_Q9FIG8 Kinesin heavy chain-like protein [Arabidopsis thaliana]
Length = 1087
Score = 117 bits (292), Expect = 7e-26
Identities = 61/126 (48%), Positives = 88/126 (69%), Gaps = 3/126 (2%)
Query: 1 MEVELRRLSYLKDNQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWG 60
MEVELRRLS+LK I D T ++ K L RE++ +S+Q+ +K ++R +Y KWG
Sbjct: 951 MEVELRRLSFLKQT-ISNDMETSRMQTVKA-LTREKEWISKQLPKKFPWNQRIGLYQKWG 1008
Query: 61 ISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPRRRR 119
+ ++SK R LQ+AH+LW+ T D++H++ESA++VAKL+G VEP + KEMFGL+ PR
Sbjct: 1009 VEVNSKQRSLQVAHKLWTNTQDMDHIKESASLVAKLLGFVEPSRMPKEMFGLSLLPRTEN 1068
Query: 120 KKSFGW 125
KS GW
Sbjct: 1069 VKSSGW 1074
>UniRef100_Q9SVJ8 Kinesin like protein [Arabidopsis thaliana]
Length = 834
Score = 112 bits (281), Expect = 1e-24
Identities = 66/135 (48%), Positives = 86/135 (62%), Gaps = 8/135 (5%)
Query: 3 VELRRLSYLKD-----NQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYL 57
VELRRL ++KD NQ LE G TLT SS++ L RER+MLS+ + ++ S ER +Y
Sbjct: 696 VELRRLLFMKDSFSQGNQALEGGETLTLASSRKELHRERKMLSKLVGKRFSGEERKRIYH 755
Query: 58 KWGISMSSKHRRLQLAHRLWSE-TDINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPR 116
K+GI+++SK RRLQL + LWS D+ V ESA +VAKLV E +A KEMFGL F P
Sbjct: 756 KFGIAINSKRRRLQLVNELWSNPKDMTQVMESADVVAKLVRFAEQGRAMKEMFGLTFTPP 815
Query: 117 R--RRKKSFGWTSSM 129
++S W SM
Sbjct: 816 SFLTTRRSHSWRKSM 830
>UniRef100_Q9FH58 Kinesin heavy chain DNA binding protein-like [Arabidopsis thaliana]
Length = 1037
Score = 106 bits (265), Expect = 1e-22
Identities = 55/100 (55%), Positives = 74/100 (74%), Gaps = 3/100 (3%)
Query: 1 MEVELRRLSYLKDN--QILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLK 58
+EVELRRL Y++++ Q DG +T S R L RER LS+ MQRKLSK ER+N++L+
Sbjct: 925 LEVELRRLKYIRESFAQNSNDGNNMTLISCTRALTRERYKLSKLMQRKLSKEERENLFLR 984
Query: 59 WGISMSSKHRRLQLAHRLWSE-TDINHVRESATIVAKLVG 97
WGI +++ HRR+QLA RLWS+ D+ HVRESA++V KL G
Sbjct: 985 WGIGLNTNHRRVQLARRLWSDYKDMGHVRESASLVGKLNG 1024
>UniRef100_Q9LPQ5 F15H18.12 [Arabidopsis thaliana]
Length = 1003
Score = 101 bits (251), Expect = 4e-21
Identities = 61/147 (41%), Positives = 90/147 (60%), Gaps = 17/147 (11%)
Query: 1 MEVELRRLSYLKDNQILEDGRTLTP-----------ESSKRYLRRERQMLSRQMQRKLSK 49
MEVELRRL++L+ Q L + TP SS + LRRER+ L++++ +L+
Sbjct: 859 MEVELRRLTWLE--QHLAEVGNATPARNCDESVVSLSSSIKALRREREFLAKRVNSRLTP 916
Query: 50 SERDNMYLKWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAFKEM 108
ER+ +Y+KW + + K R+LQ ++LW++ D HV+ESA IVAKLVG E KEM
Sbjct: 917 EEREELYMKWDVPLEGKQRKLQFVNKLWTDPYDSRHVQESAEIVAKLVGFCESGNISKEM 976
Query: 109 FGLNFA-PRRRRKKSFGW--TSSMKHI 132
F LNFA P +R+ + GW S++ H+
Sbjct: 977 FELNFAVPSDKRQWNIGWDNISNLLHL 1003
>UniRef100_Q8S905 AtNACK1 kinesin-like protein [Arabidopsis thaliana]
Length = 974
Score = 101 bits (251), Expect = 4e-21
Identities = 61/147 (41%), Positives = 90/147 (60%), Gaps = 17/147 (11%)
Query: 1 MEVELRRLSYLKDNQILEDGRTLTP-----------ESSKRYLRRERQMLSRQMQRKLSK 49
MEVELRRL++L+ Q L + TP SS + LRRER+ L++++ +L+
Sbjct: 830 MEVELRRLTWLE--QHLAEVGNATPARNCDESVVSLSSSIKALRREREFLAKRVNSRLTP 887
Query: 50 SERDNMYLKWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAFKEM 108
ER+ +Y+KW + + K R+LQ ++LW++ D HV+ESA IVAKLVG E KEM
Sbjct: 888 EEREELYMKWDVPLEGKQRKLQFVNKLWTDPYDSRHVQESAEIVAKLVGFCESGNISKEM 947
Query: 109 FGLNFA-PRRRRKKSFGW--TSSMKHI 132
F LNFA P +R+ + GW S++ H+
Sbjct: 948 FELNFAVPSDKRQWNIGWDNISNLLHL 974
>UniRef100_Q9AWM8 Putative KIF3 protein [Oryza sativa]
Length = 954
Score = 97.4 bits (241), Expect = 6e-20
Identities = 55/135 (40%), Positives = 84/135 (61%), Gaps = 10/135 (7%)
Query: 1 MEVELRRLSYLKDN--------QILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSER 52
+EVE+RRL++L+ + D T++ SS + LR ER+ L+R+M +L++ ER
Sbjct: 806 IEVEVRRLTWLQQHFAEVGDASPAAGDDSTISLASSIKALRNEREFLARRMGSRLTEEER 865
Query: 53 DNMYLKWGISMSSKHRRLQLAHRLWSE-TDINHVRESATIVAKLVGTVEPDQAFKEMFGL 111
+ +++KW + + +K R+LQL +RLW++ D H+ ESA IVA+LVG E KEMF L
Sbjct: 866 ERLFIKWQVPLEAKQRKLQLVNRLWTDPNDQAHIDESADIVARLVGFCEGGNISKEMFEL 925
Query: 112 NFA-PRRRRKKSFGW 125
NFA P R+ GW
Sbjct: 926 NFAVPASRKPWLMGW 940
>UniRef100_Q8S950 Kinesin-like protein NACK1 [Nicotiana tabacum]
Length = 959
Score = 96.7 bits (239), Expect = 1e-19
Identities = 60/146 (41%), Positives = 86/146 (58%), Gaps = 15/146 (10%)
Query: 1 MEVELRRLSYLKDNQILEDGR----------TLTPESSKRYLRRERQMLSRQMQRKLSKS 50
+EVELRRL++L+ + + E G T++ SS R L+RER+ L++++ +L+
Sbjct: 815 LEVELRRLTWLQQH-LAELGNATPARVGNEPTVSLSSSIRALKREREFLAKRLTTRLTAE 873
Query: 51 ERDNMYLKWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAFKEMF 109
ERD +Y+KW + + K RR+Q ++LW+ D HV ESA IVAKLVG E +EMF
Sbjct: 874 ERDYLYIKWEVPLEGKQRRMQFINKLWTNPHDAKHVHESAEIVAKLVGFCEGGNMSREMF 933
Query: 110 GLNFA-PRRRRKKSFGWT--SSMKHI 132
LNF P RR GW S + HI
Sbjct: 934 ELNFVLPSDRRPWFAGWNQISDLLHI 959
>UniRef100_Q9SU42 Hypothetical protein AT4g24170 [Arabidopsis thaliana]
Length = 1263
Score = 91.7 bits (226), Expect = 3e-18
Identities = 50/113 (44%), Positives = 73/113 (64%), Gaps = 8/113 (7%)
Query: 1 MEVELRRLSYLKDNQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWG 60
MEVELRRLS+LKD+ E R T ++ + RER+ L++Q+ K K E++ +Y KWG
Sbjct: 870 MEVELRRLSFLKDST--ETSRKQTAKA----VTREREWLAKQIPNKFGKKEKEEVYKKWG 923
Query: 61 ISMSSKHRRLQLAHRLWSET--DINHVRESATIVAKLVGTVEPDQAFKEMFGL 111
+ +SSK R LQ+ H+LW+ DI H +ESA+++A LVG V+ KE+ L
Sbjct: 924 VELSSKRRSLQVTHKLWNNNTKDIEHCKESASLIATLVGFVDSTLTPKEISDL 976
>UniRef100_Q9LXL3 Kinesin-like protein [Arabidopsis thaliana]
Length = 932
Score = 91.3 bits (225), Expect = 4e-18
Identities = 58/146 (39%), Positives = 86/146 (58%), Gaps = 14/146 (9%)
Query: 1 MEVELRRLSYLKDN---------QILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSE 51
MEVELRRL++L+ + +L D SS R L++ER+ L++++ KL E
Sbjct: 787 MEVELRRLTWLEQHLAELGNASPALLGDEPASYVASSIRALKQEREYLAKRVNTKLGAEE 846
Query: 52 RDNMYLKWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAF-KEMF 109
R+ +YLKW + K RR Q ++LW++ ++ HVRESA IVAKLVG + + KEMF
Sbjct: 847 REMLYLKWDVPPVGKQRRQQFINKLWTDPHNMQHVRESAEIVAKLVGFCDSGETIRKEMF 906
Query: 110 GLNFA-PRRRRKKSFGWT--SSMKHI 132
LNFA P ++ GW S++ H+
Sbjct: 907 ELNFASPSDKKTWMMGWNFISNLLHL 932
>UniRef100_Q8LNZ2 Kinesin-like protein [Arabidopsis thaliana]
Length = 938
Score = 91.3 bits (225), Expect = 4e-18
Identities = 58/146 (39%), Positives = 86/146 (58%), Gaps = 14/146 (9%)
Query: 1 MEVELRRLSYLKDN---------QILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSE 51
MEVELRRL++L+ + +L D SS R L++ER+ L++++ KL E
Sbjct: 793 MEVELRRLTWLEQHLAELGNASPALLGDEPASYVASSIRALKQEREYLAKRVNTKLGAEE 852
Query: 52 RDNMYLKWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAF-KEMF 109
R+ +YLKW + K RR Q ++LW++ ++ HVRESA IVAKLVG + + KEMF
Sbjct: 853 REMLYLKWDVPPVGKQRRQQFINKLWTDPHNMQHVRESAEIVAKLVGFCDSGETIRKEMF 912
Query: 110 GLNFA-PRRRRKKSFGWT--SSMKHI 132
LNFA P ++ GW S++ H+
Sbjct: 913 ELNFASPSDKKTWMMGWNFISNLLHL 938
>UniRef100_Q8LGU3 Putative kinesin-like protein [Arabidopsis thaliana]
Length = 937
Score = 91.3 bits (225), Expect = 4e-18
Identities = 58/146 (39%), Positives = 86/146 (58%), Gaps = 14/146 (9%)
Query: 1 MEVELRRLSYLKDN---------QILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSE 51
MEVELRRL++L+ + +L D SS R L++ER+ L++++ KL E
Sbjct: 792 MEVELRRLTWLEQHLAELGNASPALLGDEPASYVASSIRALKQEREYLAKRVNTKLGAEE 851
Query: 52 RDNMYLKWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAF-KEMF 109
R+ +YLKW + K RR Q ++LW++ ++ HVRESA IVAKLVG + + KEMF
Sbjct: 852 REMLYLKWDVPPVGKQRRQQFINKLWTDPHNMQHVRESAEIVAKLVGFCDSGETIRKEMF 911
Query: 110 GLNFA-PRRRRKKSFGWT--SSMKHI 132
LNFA P ++ GW S++ H+
Sbjct: 912 ELNFASPSDKKTWMMGWNFISNLLHL 937
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.131 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 198,161,225
Number of Sequences: 2790947
Number of extensions: 6377511
Number of successful extensions: 20422
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 35
Number of HSP's successfully gapped in prelim test: 33
Number of HSP's that attempted gapping in prelim test: 20243
Number of HSP's gapped (non-prelim): 159
length of query: 133
length of database: 848,049,833
effective HSP length: 109
effective length of query: 24
effective length of database: 543,836,610
effective search space: 13052078640
effective search space used: 13052078640
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC149546.1


