

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149496.4 - phase: 0
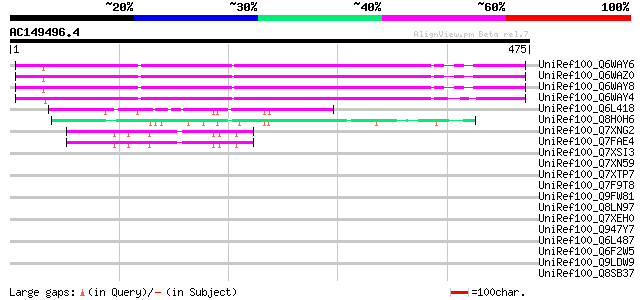
(475 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6WAY6 Hypothetical protein [Pisum sativum] 281 4e-74
UniRef100_Q6WAZ0 Hypothetical protein [Pisum sativum] 279 1e-73
UniRef100_Q6WAY8 Hypothetical protein [Pisum sativum] 276 1e-72
UniRef100_Q6WAY4 Hypothetical protein [Pisum sativum] 258 3e-67
UniRef100_Q6L418 Hypothetical protein PGEC989P08.9 [Solanum demi... 65 3e-09
UniRef100_Q8H0H6 Hypothetical protein B27 [Nicotiana tabacum] 56 2e-06
UniRef100_Q7XNG2 OSJNBa0096F01.11 protein [Oryza sativa] 48 5e-04
UniRef100_Q7FAE4 OSJNBa0052P16.6 protein [Oryza sativa] 48 5e-04
UniRef100_Q7XSI3 OSJNBa0073L13.4 protein [Oryza sativa] 46 0.002
UniRef100_Q7XN59 OSJNBa0089N06.23 protein [Oryza sativa] 45 0.003
UniRef100_Q7XTP7 OSJNBa0041A02.27 protein [Oryza sativa] 45 0.006
UniRef100_Q7F9T8 OSJNBb0015N08.1 protein [Oryza sativa] 45 0.006
UniRef100_Q9FW81 Mutator-like transposase [Oryza sativa] 44 0.013
UniRef100_Q8LN97 Putative Mu transposable element [Oryza sativa] 43 0.017
UniRef100_Q7XEH0 Putative mutator-like transposase [Oryza sativa] 43 0.017
UniRef100_Q947Y7 Putative mutator-like transposase [Oryza sativa] 43 0.023
UniRef100_Q6L487 Putative mutator transposase [Oryza sativa] 43 0.023
UniRef100_Q6F2W5 Putative MuDR family transposase [Oryza sativa] 43 0.023
UniRef100_Q9LDW9 EST C28952(C62945) corresponds to a region of t... 42 0.029
UniRef100_Q8SB37 Putative transposon protein [Oryza sativa] 42 0.029
>UniRef100_Q6WAY6 Hypothetical protein [Pisum sativum]
Length = 562
Score = 281 bits (718), Expect = 4e-74
Identities = 174/470 (37%), Positives = 247/470 (52%), Gaps = 22/470 (4%)
Query: 6 RSIKKYTFKKPDLTRLRELASHVT--NPRDFRERHGRLLDLLKVKVEDGILETLVQFYDP 63
R Y+F + LT L EL++ +T N + F ++HG +L LLK+ V+ L+TL+QFYDP
Sbjct: 25 RKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQHGDILTLLKMVVDPVPLQTLLQFYDP 84
Query: 64 ICHCFTFPDYQLVPTLEEYSFWVGLPVSEGEPFNGLEPSPKNATIAEALHLKPSDLVHPH 123
HCFTF DYQL PTLEEYS + +PV PF + +A ALHL + V
Sbjct: 85 ELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFGVVARALHLGIKE-VSDS 143
Query: 124 FTIKNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLIYGLFLFPNMDNFVDLNAIKIF 183
+ ++ GL KFL + A + + AF + ++IYG+ LFP+M NFVD A+ IF
Sbjct: 144 WKYSGDVVGLPLKFLLRVAREEAEKGSWEAFRAQLAVMIYGIVLFPSMPNFVDYAAVSIF 203
Query: 184 LAKNPVPTLLANTYHSIHHRNIREGGLIVCCAPLLYRWYASHLTKSVFSKENSGKASWSE 243
+ NPVPTLLA+TY++IH R+ +GG I CC PLL RW+ S L S + W++
Sbjct: 204 IGGNPVPTLLADTYYAIHSRH-GKGGAIRCCLPLLLRWFLSLLPVSGPFVDAQSTHKWTQ 262
Query: 244 RIMPLTPADIVWVHAGTNTAGIIGSCGEFENVPLIGTCGGITYNPTLAMRQYGYPMKGRP 303
R+M LT DI W + +I SCG F NVPL+GT G I YNP L++RQ G+ MKGRP
Sbjct: 263 RVMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINYNPVLSLRQLGFVMKGRP 322
Query: 304 DSLSLSNEFYLNKNDHTNLRMRFVQAWHSIRKFDGIQLGRKQSFAHESYTQWVIDRATAF 363
++ Y K + +AW SI DG LG+K A YT WV +R
Sbjct: 323 LEAEVAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFVVAMPDYTDWVKERVETL 382
Query: 364 GMPYTLPRFLSSTVPEIPLPLLPQTKEEYQERLAEAEREIHMWKRECQKKDKDYETVMGL 423
+PY L P I +P E Y++ L E R + K++D
Sbjct: 383 LLPYDRMEPLQEQPPSILAESVP--AEHYKQALME--------NRRLRGKEQD------- 425
Query: 424 LEQEAYDSRQKDVIIAKLNERIKEKDAALDRIPGRKKKRMD-LFDGPHSD 472
+ E Y ++ + +A ++E+DA+ R R + M+ + D H +
Sbjct: 426 TQLELYKAKADKLNLAHQLRGVREEDASRLRSKKRSYEEMESMLDAEHRE 475
>UniRef100_Q6WAZ0 Hypothetical protein [Pisum sativum]
Length = 562
Score = 279 bits (713), Expect = 1e-73
Identities = 173/470 (36%), Positives = 249/470 (52%), Gaps = 22/470 (4%)
Query: 6 RSIKKYTFKKPDLTRLRELASHVT--NPRDFRERHGRLLDLLKVKVEDGILETLVQFYDP 63
R Y+F + LT L EL++ +T N + F +++G +L LLK+ V+ L+TL+QFYDP
Sbjct: 25 RKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQYGDILTLLKMVVDPVPLQTLLQFYDP 84
Query: 64 ICHCFTFPDYQLVPTLEEYSFWVGLPVSEGEPFNGLEPSPKNATIAEALHLKPSDLVHPH 123
HCFTF DYQL PTLEEYS + +PV PF + +A ALHL + V +
Sbjct: 85 ELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFRVVARALHLGIKE-VSDN 143
Query: 124 FTIKNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLIYGLFLFPNMDNFVDLNAIKIF 183
+ ++ GL KFL + A + + AF + ++IYG+ LFP+M NFVD A+ IF
Sbjct: 144 WKSSGDVVGLPLKFLLRVAREEAEKGSWEAFHAQLAVMIYGIVLFPSMPNFVDYAAVSIF 203
Query: 184 LAKNPVPTLLANTYHSIHHRNIREGGLIVCCAPLLYRWYASHLTKSVFSKENSGKASWSE 243
+ NPVPTLLA+TY++IH R+ +GG I CC PLL RW+ S L S + W++
Sbjct: 204 IGGNPVPTLLADTYYAIHSRH-GKGGAIRCCLPLLLRWFLSLLPVSGPFVDAQSTHKWTQ 262
Query: 244 RIMPLTPADIVWVHAGTNTAGIIGSCGEFENVPLIGTCGGITYNPTLAMRQYGYPMKGRP 303
R+M LT DI W + +I SCG F NVPL+GT G I YNP L++RQ G+ MKGRP
Sbjct: 263 RVMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINYNPVLSLRQLGFVMKGRP 322
Query: 304 DSLSLSNEFYLNKNDHTNLRMRFVQAWHSIRKFDGIQLGRKQSFAHESYTQWVIDRATAF 363
++ Y K + +AW SI DG LG+K A YT WV +R
Sbjct: 323 LEAEIAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFVVAMPDYTDWVKERVETL 382
Query: 364 GMPYTLPRFLSSTVPEIPLPLLPQTKEEYQERLAEAEREIHMWKRECQKKDKDYETVMGL 423
+PY L P I +P E Y++ L E R ++K++D
Sbjct: 383 LLPYDRMEPLQEQPPLILAESVP--AEHYKQALME--------NRRLREKEQD------- 425
Query: 424 LEQEAYDSRQKDVIIAKLNERIKEKDAALDRIPGRKKKRMD-LFDGPHSD 472
+ E Y ++ + +A ++E+DA+ R R + M+ + D H +
Sbjct: 426 TQMELYKAKAGRLNLAHQLRGVREEDASRLRSKKRSYEEMESMLDAEHRE 475
>UniRef100_Q6WAY8 Hypothetical protein [Pisum sativum]
Length = 562
Score = 276 bits (705), Expect = 1e-72
Identities = 172/470 (36%), Positives = 248/470 (52%), Gaps = 22/470 (4%)
Query: 6 RSIKKYTFKKPDLTRLRELASHVT--NPRDFRERHGRLLDLLKVKVEDGILETLVQFYDP 63
R Y+F + LT L EL++ +T N + F +++G +L LLK+ V+ L+TL+QFYDP
Sbjct: 25 RKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQYGDILTLLKMVVDPVPLQTLLQFYDP 84
Query: 64 ICHCFTFPDYQLVPTLEEYSFWVGLPVSEGEPFNGLEPSPKNATIAEALHLKPSDLVHPH 123
HCFTF DYQL PTLEEYS + +PV PF + +A ALHL + V
Sbjct: 85 ELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFRVVARALHLGIKE-VSDS 143
Query: 124 FTIKNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLIYGLFLFPNMDNFVDLNAIKIF 183
+ ++ GL KFL + A + + AF + ++IYG+ LFP+M NFVD A+ IF
Sbjct: 144 WKSSGDVVGLPLKFLLRVAREEAEKGSWEAFHAQLAVMIYGIVLFPSMPNFVDYAAVSIF 203
Query: 184 LAKNPVPTLLANTYHSIHHRNIREGGLIVCCAPLLYRWYASHLTKSVFSKENSGKASWSE 243
+ NPVPTLLA+TY++IH R+ +GG I CC PLL RW+ S L S + W++
Sbjct: 204 IGGNPVPTLLADTYYAIHSRH-GKGGAIRCCLPLLLRWFLSLLPVSGPFVDAQSTHKWTQ 262
Query: 244 RIMPLTPADIVWVHAGTNTAGIIGSCGEFENVPLIGTCGGITYNPTLAMRQYGYPMKGRP 303
R+M LT DI W + +I SCG F NVPLIGT G I YNP L++RQ G+ MKGRP
Sbjct: 263 RVMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLIGTRGCINYNPVLSLRQLGFVMKGRP 322
Query: 304 DSLSLSNEFYLNKNDHTNLRMRFVQAWHSIRKFDGIQLGRKQSFAHESYTQWVIDRATAF 363
++ K + +AW SI DG LG+K + A YT WV +R
Sbjct: 323 LEAEVAESVCFEKRSDPARLEQIGRAWKSIGMKDGSVLGKKFAVAMPDYTDWVKERVETL 382
Query: 364 GMPYTLPRFLSSTVPEIPLPLLPQTKEEYQERLAEAEREIHMWKRECQKKDKDYETVMGL 423
+PY L P I +P E Y++ L E R ++K++D
Sbjct: 383 LLPYDRMEPLQEQPPLILAESVP--AEHYKQALME--------NRRLREKEQD------- 425
Query: 424 LEQEAYDSRQKDVIIAKLNERIKEKDAALDRIPGRKKKRMD-LFDGPHSD 472
+ E Y ++ + +A ++E+DA+ R R + ++ + D H +
Sbjct: 426 TQMELYKAKADSLNLAHQLRGVREEDASRLRSKKRSYEEVESMLDAEHRE 475
>UniRef100_Q6WAY4 Hypothetical protein [Pisum sativum]
Length = 546
Score = 258 bits (659), Expect = 3e-67
Identities = 166/470 (35%), Positives = 236/470 (49%), Gaps = 22/470 (4%)
Query: 6 RSIKKYTFKKPDLTRLRELASHVTNP--RDFRERHGRLLDLLKVKVEDGILETLVQFYDP 63
R Y+F + LT L EL S + + + F + G +L LLK V+ L+TL+QFYDP
Sbjct: 9 RKTCSYSFYREPLTSLIELGSLMPSDQLKGFVGQFGDILTLLKTVVDPVPLQTLLQFYDP 68
Query: 64 ICHCFTFPDYQLVPTLEEYSFWVGLPVSEGEPFNGLEPSPKNATIAEALHLKPSDLVHPH 123
CFTF DYQL PTLEEYS + +P+ PF + IA AL + + V +
Sbjct: 69 ELRCFTFQDYQLAPTLEEYSILMSIPIQHQVPFLDVPKEVDFRVIARALRMSVKE-VCDN 127
Query: 124 FTIKNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLIYGLFLFPNMDNFVDLNAIKIF 183
+ + G+ KFL + A + + F + +IYG+ LFP+M NF+D AI IF
Sbjct: 128 WKPSGEVVGMPLKFLLRVAREEAEKGNWEVFHAQLAAMIYGIVLFPSMPNFIDHAAISIF 187
Query: 184 LAKNPVPTLLANTYHSIHHRNIREGGLIVCCAPLLYRWYASHLTKSVFSKENSGKASWSE 243
+ NPVPTLLA+TY++IH R+ +GG I CC PLL RW+ S L S + W++
Sbjct: 188 IGGNPVPTLLADTYYAIHSRH-GKGGAIRCCLPLLLRWFMSLLPASGPFMDTQSTLKWTQ 246
Query: 244 RIMPLTPADIVWVHAGTNTAGIIGSCGEFENVPLIGTCGGITYNPTLAMRQYGYPMKGRP 303
R+M LT DI W + ++ SCGEF NVPL+GT G I YNP L++RQ G+ M RP
Sbjct: 247 RVMSLTSYDIRWQSYRMDVKDVVMSCGEFRNVPLVGTKGCINYNPVLSLRQLGFIMSRRP 306
Query: 304 DSLSLSNEFYLNKNDHTNLRMRFVQAWHSIRKFDGIQLGRKQSFAHESYTQWVIDRATAF 363
+ Y K + +AW SI DG LG+K + A YT WV R
Sbjct: 307 LEAEIVESVYFEKRTDPVRLEQIGRAWKSIGVKDGSVLGKKFAIAMPDYTDWVKKRVETL 366
Query: 364 GMPYTLPRFLSSTVPEIPLPLLPQTKEEYQERLAEAEREIHMWKRECQKKDKDYETVMGL 423
+PY L P I +P E Y++ L E R ++K+ +T M
Sbjct: 367 LLPYDRMEPLQEQPPLILADSVP--AEHYKQALMENRR----------LREKEQDTRM-- 412
Query: 424 LEQEAYDSRQKDVIIAKLNERIKEKDAALDRIPGRKKKRMD-LFDGPHSD 472
E Y ++ + +A ++ +DA+ R R K M+ + D H +
Sbjct: 413 ---ELYKAKADKLNLAHQLREMQGEDASRARSKKRSYKEMESMLDTEHRE 459
>UniRef100_Q6L418 Hypothetical protein PGEC989P08.9 [Solanum demissum]
Length = 607
Score = 65.5 bits (158), Expect = 3e-09
Identities = 76/304 (25%), Positives = 126/304 (41%), Gaps = 51/304 (16%)
Query: 36 ERHGRLLDLLKVKVEDGILETLVQFYDPICHCFTFPDYQLVPTLEEYSFWV--GLPVSEG 93
+R G L LL V+ ++E LV F+DPI + F F D++L PTLEE ++ G + EG
Sbjct: 46 KRLGFLRSLLYVEPRRDLIEALVHFWDPIKNVFHFSDFELTPTLEEIGAFIGRGKNLHEG 105
Query: 94 EPFNGLEPSPKNATIAEALHLK-------------PSDLVHPHFTIKNNLQGLTAKFLYQ 140
EP + E LH+ P + ++ + K+ + L K L+
Sbjct: 106 EPM--IPKHINGRRFLELLHINENEIGGCLDNGWVPLEFLYKRYGRKDGFE-LYGKKLHN 162
Query: 141 KASDFVKAKKTNAFESIFTLLIYGLFLFPNMDNFVDLNAIKIFLA--KNP----VPTLLA 194
+T+++++I T+ G+ +F +++N + A K P VP +LA
Sbjct: 163 NGCRL--TWETHSYDAI-TVAFLGIMVFLKRGGKININLAAVITAFEKKPNITLVPMILA 219
Query: 195 NTYHSIHHRNIREGG-LIVCCAPLLYRWYASHLTKSVF------------------SKEN 235
+ Y ++ + GG C LL W H+ + K++
Sbjct: 220 DIYRAL--TICKNGGDYFEGCNMLLQLWMIEHIRHHPYVVDFKVECNDYIGGHEERIKDH 277
Query: 236 S---GKASWSERIMPLTPADIVWVHAGTNTAGIIGSCGEFENVPLIGTCGGITYNPTLAM 292
S G +W + + LT IVW + +A +I + L+G G Y P M
Sbjct: 278 SFPKGIEAWKKYLNNLTADKIVWNYHWFPSAEVIYMSTFRSFIVLMGLRGVQPYMPLRVM 337
Query: 293 RQYG 296
RQ G
Sbjct: 338 RQLG 341
>UniRef100_Q8H0H6 Hypothetical protein B27 [Nicotiana tabacum]
Length = 464
Score = 56.2 bits (134), Expect = 2e-06
Identities = 99/446 (22%), Positives = 167/446 (37%), Gaps = 100/446 (22%)
Query: 39 GRLLDLLKVKVEDGILETLVQFYDPICHCFTFPDYQLVPTLEEYSFWVGLPVSEGEPFNG 98
G L D++K+K D ++E LV F+DP+ + F F D++L PTLEE + G G
Sbjct: 39 GALTDIMKIKPRDDLIEALVTFWDPVHNVFCFSDFELTPTLEEIDGYSGF---------G 89
Query: 99 LEPSPKNATIAEALHL-KPSDLVHPHFTIK--NNLQG------LTAKF-------LYQKA 142
+ + AL + + DL++ I+ N ++G L ++F +++K
Sbjct: 90 RDLRNQELIFPRALSVHRFFDLLNISKQIRKTNVVKGCCSFYFLYSRFGQPNGFEMHEKG 149
Query: 143 SDFVKAKKTNAFESIFTLLI--YGLFLFPNMDNFVD---LNAIKIFLAKNP---VPTLLA 194
+ + K T F ++ G+ +FPN + +D +++ K P +L+
Sbjct: 150 LNNKQNKDTWHIHRCFAFIMAFLGIMVFPNRERTIDTRIARVVQVLTTKEHHTLAPIILS 209
Query: 195 NTYHSIHHRNIREGG--LIVCCAPLLYRWYASHLTKSVF------SKEN----------- 235
+ Y ++ + + G C LL W HL SK+N
Sbjct: 210 DIYRAL---TLCKSGAKFFEGCNILLQMWLIEHLRHHPKFMSYGPSKDNFIDSYEERVKE 266
Query: 236 ----SGKASWSERIMPLTPADIVWVHAGTNTAGIIGSCGEFENVPLIGTCGGITYNPTLA 291
G +W + L I W +I ++ L+G Y P
Sbjct: 267 YNSPEGVETWISHLRSLNACQIEWTLGWLPLREVIHMSALKSHLLLLGLRSVQPYMPHRV 326
Query: 292 MRQYG-YPMKGRPDSLSLSNEFYLNKNDHTNLRMRFV-QAWHSIR--KFDGIQLGRKQSF 347
+RQ G Y + + + LS+ + + L + Q W+ R K D +
Sbjct: 327 LRQLGRYQVVPKDEDLSVQ---VIELHPKAPLPEALIQQIWNGCRYLKGDTQVPDPARGE 383
Query: 348 AHESYTQWVIDRATAFGMPYTLPRFLSSTVPEIPLPLLPQTK-------EEYQERLAEAE 400
Y W FG S V ++P P P + ++ QERLA E
Sbjct: 384 VDPGYAIW-------FGK--------RSRVDDVPEPKRPTKRPHVQAFDDKIQERLAWGE 428
Query: 401 REIHMWKRECQKKDKDYETVMGLLEQ 426
RE K Y+T + LE+
Sbjct: 429 RE------------KGYKTTIHALEE 442
>UniRef100_Q7XNG2 OSJNBa0096F01.11 protein [Oryza sativa]
Length = 1365
Score = 48.1 bits (113), Expect = 5e-04
Identities = 49/195 (25%), Positives = 86/195 (43%), Gaps = 28/195 (14%)
Query: 53 ILETLVQFYDPICHCFTFPDYQLVPTLEEYSFWVGLPVSEGE--PFNGLEPSPKNAT--- 107
+L LV + P H F P ++ PTL++ S+ +GLP++ P +G+ ++ T
Sbjct: 808 LLAALVDHWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGVFGWKEDITARF 867
Query: 108 --IAEALHLKPSDLVHPHFTI---KNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLI 162
+ HL P++ + P+ T+ K L TA L+ A D++ + A + L +
Sbjct: 868 EQVMRLPHLGPANTLPPYSTVGPSKAWLLQFTADLLHPDADDYLVRRSLEA----YLLWL 923
Query: 163 YGLFLFPNM-DNFVDLNAIKIFLA-----KNPVP------TLLANTYHSIHHRNIR--EG 208
+G +F + + VD + + VP +LA TYH++ + G
Sbjct: 924 FGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYHALCEACTKTDAG 983
Query: 209 GLIVCCAPLLYRWYA 223
+I C LL W A
Sbjct: 984 AIIAGCPMLLQLWAA 998
>UniRef100_Q7FAE4 OSJNBa0052P16.6 protein [Oryza sativa]
Length = 1489
Score = 48.1 bits (113), Expect = 5e-04
Identities = 49/195 (25%), Positives = 86/195 (43%), Gaps = 28/195 (14%)
Query: 53 ILETLVQFYDPICHCFTFPDYQLVPTLEEYSFWVGLPVSEGE--PFNGLEPSPKNAT--- 107
+L LV + P H F P ++ PTL++ S+ +GLP++ P +G+ ++ T
Sbjct: 808 LLAALVDHWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGVFGWKEDITARF 867
Query: 108 --IAEALHLKPSDLVHPHFTI---KNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLI 162
+ HL P++ + P+ T+ K L TA L+ A D++ + A + L +
Sbjct: 868 EQVMRLPHLGPANTLPPYSTVGPSKAWLLQFTADLLHPDADDYLVRRSLEA----YLLWL 923
Query: 163 YGLFLFPNM-DNFVDLNAIKIFLA-----KNPVP------TLLANTYHSIHHRNIR--EG 208
+G +F + + VD + + VP +LA TYH++ + G
Sbjct: 924 FGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYHALCEACTKTDAG 983
Query: 209 GLIVCCAPLLYRWYA 223
+I C LL W A
Sbjct: 984 AIIAGCPMLLQLWAA 998
>UniRef100_Q7XSI3 OSJNBa0073L13.4 protein [Oryza sativa]
Length = 1342
Score = 46.2 bits (108), Expect = 0.002
Identities = 53/220 (24%), Positives = 93/220 (42%), Gaps = 31/220 (14%)
Query: 53 ILETLVQFYDPICHCFTFPDYQLVPTLEEYSFWVGLPVSEGE--PFNGLEPSPKNAT--- 107
+L LV + P H F P ++ PTL++ S+ +GLP++ P +G+ ++ T
Sbjct: 830 LLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGVFGWKEDITARF 889
Query: 108 --IAEALHLKPSDLVHPHFTI---KNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLI 162
+ HL P++ + P+ T+ K L TA L+ A D+ + A + L +
Sbjct: 890 EQVMRLPHLGPANTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEA----YLLWL 945
Query: 163 YGLFLFPNM-DNFVDLNAIKIFLA-----KNPVP------TLLANTYHSIHHRNIR--EG 208
+G +F + + VD + + VP +LA TY ++ + G
Sbjct: 946 FGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKTDAG 1005
Query: 209 GLIVCCAPLLYRWYASH---LTKSVFSKENSGKASWSERI 245
+I C LL W A LT + + A W+ R+
Sbjct: 1006 AIIAGCPMLLQLWAAESVVTLTSRLTRRGQLAGALWAPRV 1045
>UniRef100_Q7XN59 OSJNBa0089N06.23 protein [Oryza sativa]
Length = 1597
Score = 45.4 bits (106), Expect = 0.003
Identities = 50/196 (25%), Positives = 86/196 (43%), Gaps = 30/196 (15%)
Query: 53 ILETLVQFYDPICHCFTFPDYQLVPTLEEYSFWVGLPVSEGEPFNGL--------EPSPK 104
+L LV + P H F P ++ PTL++ S+ +GLP++ G P + + + +
Sbjct: 779 LLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLA-GAPVGPVDCVFGWKEDITAR 837
Query: 105 NATIAEALHLKPSDLVHPHFTI---KNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLL 161
+ HL P++ + P+ T+ K L TA L+ A D+ + A + LL
Sbjct: 838 FEQVMRLPHLGPANTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEA----YLLL 893
Query: 162 IYGLFLFPNM-DNFVDLNAIKI--FLA---KNPVP------TLLANTYHSIHHRNIR--E 207
++G +F + + VD + F+A VP +LA TY ++ +
Sbjct: 894 LFGWVMFTSTHGHAVDFRLVHYARFIADAQPQDVPQWSWGSAVLAATYRALCEACTKTDA 953
Query: 208 GGLIVCCAPLLYRWYA 223
G +I C LL W A
Sbjct: 954 GAIIAGCPMLLQLWAA 969
>UniRef100_Q7XTP7 OSJNBa0041A02.27 protein [Oryza sativa]
Length = 1473
Score = 44.7 bits (104), Expect = 0.006
Identities = 51/195 (26%), Positives = 87/195 (44%), Gaps = 28/195 (14%)
Query: 53 ILETLVQFYDPICHCFTFPDYQLVPTLEEYSFWVGLPVSEGE--PFNGLEPSPKNAT--- 107
+L LV + P H F P ++ PTL++ S+ +GLP++ P +G+ ++ T
Sbjct: 827 LLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGVFGWKEDITARF 886
Query: 108 --IAEALHLKPSDLVHPHFTI---KNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLI 162
+ HL P++ + P+ T+ K L TA L+ A D+ + A + L +
Sbjct: 887 EQVMRLPHLGPANTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEA----YLLWL 942
Query: 163 YGLFLFPN-----MDNFVDLNAIKIFLAK-NPVP------TLLANTYHSIHH--RNIREG 208
+G +F + +D ++ A I A+ VP +LA TY ++ N G
Sbjct: 943 FGWVMFTSTHGHAVDFWLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTNTDAG 1002
Query: 209 GLIVCCAPLLYRWYA 223
+I C LL W A
Sbjct: 1003 AIIAGCPMLLQLWAA 1017
>UniRef100_Q7F9T8 OSJNBb0015N08.1 protein [Oryza sativa]
Length = 775
Score = 44.7 bits (104), Expect = 0.006
Identities = 51/195 (26%), Positives = 87/195 (44%), Gaps = 28/195 (14%)
Query: 53 ILETLVQFYDPICHCFTFPDYQLVPTLEEYSFWVGLPVSEGE--PFNGLEPSPKNAT--- 107
+L LV + P H F P ++ PTL++ S+ +GLP++ P +G+ ++ T
Sbjct: 129 LLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGVFGWKEDITARF 188
Query: 108 --IAEALHLKPSDLVHPHFTI---KNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLI 162
+ HL P++ + P+ T+ K L TA L+ A D+ + A + L +
Sbjct: 189 EQVMRLPHLGPANTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEA----YLLWL 244
Query: 163 YGLFLFPN-----MDNFVDLNAIKIFLAK-NPVP------TLLANTYHSIHH--RNIREG 208
+G +F + +D ++ A I A+ VP +LA TY ++ N G
Sbjct: 245 FGWVMFTSTHGHAVDFWLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTNTDAG 304
Query: 209 GLIVCCAPLLYRWYA 223
+I C LL W A
Sbjct: 305 AIIAGCPMLLQLWAA 319
>UniRef100_Q9FW81 Mutator-like transposase [Oryza sativa]
Length = 1626
Score = 43.5 bits (101), Expect = 0.013
Identities = 58/242 (23%), Positives = 100/242 (40%), Gaps = 37/242 (15%)
Query: 6 RSIKKYTFKKPDLTRLRELASHVTNPRDFRERHGRLLDLLKVKVEDGILETLVQFYDPIC 65
R + Y + L L L + RD +R + +L LV + P
Sbjct: 923 RHVPWYALRAAGLLPLCRLVEAAADDRDPAKRWD---------ADRSLLAALVDRWRPET 973
Query: 66 HCFTFPDYQLVPTLEEYSFWVGLPVSEGE--PFNGLEPSPKNAT-----IAEALHLKPSD 118
H F P ++ PTL++ S+ +GLP++ P +G+ ++ T + HL P++
Sbjct: 974 HTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGVFGWKEDITARFEQVMRLPHLGPAN 1033
Query: 119 LVHPHFTI---KNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLIYGLFLFPN----- 170
+ P+ T+ K L TA L+ A D+ + A + L ++G +F +
Sbjct: 1034 TLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEA----YLLWLFGWVMFTSTHGHA 1089
Query: 171 MDNFVDLNAIKIFLAK-NPVP------TLLANTYHSIHHRNIR--EGGLIVCCAPLLYRW 221
+D ++ A I A+ VP +LA TY ++ + G +I C LL W
Sbjct: 1090 VDFWLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKTDAGAIIAGCPMLLQLW 1149
Query: 222 YA 223
A
Sbjct: 1150 AA 1151
>UniRef100_Q8LN97 Putative Mu transposable element [Oryza sativa]
Length = 1536
Score = 43.1 bits (100), Expect = 0.017
Identities = 48/195 (24%), Positives = 84/195 (42%), Gaps = 28/195 (14%)
Query: 53 ILETLVQFYDPICHCFTFPDYQLVPTLEEYSFWVGLPVSEGE--PFNGLEPSPKNAT--- 107
+L LV + P H F P ++ PTL++ S+ +GLP++ P +G+ ++ T
Sbjct: 959 LLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGVFGWKEDITARF 1018
Query: 108 --IAEALHLKPSDLVHPHFTI---KNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLI 162
+ HL P++ + P+ T+ K L TA L+ A D+ + A + L +
Sbjct: 1019 EQVMRLPHLGPANTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEA----YLLWL 1074
Query: 163 YGLFLFPNM-DNFVDLNAIKIFLA-----KNPVP------TLLANTYHSIHHRNIR--EG 208
+G +F + + VD + + VP +LA TY ++ + G
Sbjct: 1075 FGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKTDAG 1134
Query: 209 GLIVCCAPLLYRWYA 223
+I C LL W A
Sbjct: 1135 AIIAGCPMLLQLWAA 1149
>UniRef100_Q7XEH0 Putative mutator-like transposase [Oryza sativa]
Length = 1421
Score = 43.1 bits (100), Expect = 0.017
Identities = 48/195 (24%), Positives = 84/195 (42%), Gaps = 28/195 (14%)
Query: 53 ILETLVQFYDPICHCFTFPDYQLVPTLEEYSFWVGLPVSEGE--PFNGLEPSPKNAT--- 107
+L LV + P H F P ++ PTL++ S+ +GLP++ P +G+ ++ T
Sbjct: 783 LLAALVDRWRPETHMFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGVFGWKEDITARF 842
Query: 108 --IAEALHLKPSDLVHPHFTI---KNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLI 162
+ HL P++ + P+ T+ K L TA L+ A D+ + A + L +
Sbjct: 843 EQVMRLPHLGPANTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEA----YLLWL 898
Query: 163 YGLFLFPNM-DNFVDLNAIKIFLA-----KNPVP------TLLANTYHSIHHRNIR--EG 208
+G +F + + VD + + VP +LA TY ++ + G
Sbjct: 899 FGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKTDAG 958
Query: 209 GLIVCCAPLLYRWYA 223
+I C LL W A
Sbjct: 959 AIIAGCPMLLQLWAA 973
>UniRef100_Q947Y7 Putative mutator-like transposase [Oryza sativa]
Length = 2421
Score = 42.7 bits (99), Expect = 0.023
Identities = 48/195 (24%), Positives = 83/195 (41%), Gaps = 28/195 (14%)
Query: 53 ILETLVQFYDPICHCFTFPDYQLVPTLEEYSFWVGLPVSEGE--PFNGLEPSPKNAT--- 107
+L LV + P H F P ++ PTL++ S+ +GLP++ P +G+ ++ T
Sbjct: 865 LLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPIDGVFGWKEDITARF 924
Query: 108 --IAEALHLKPSDLVHPHFTI---KNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLI 162
+ HL P+ + P+ T+ K L TA L+ A D+ + A + L +
Sbjct: 925 EQVMRLPHLGPTTTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEA----YLLWL 980
Query: 163 YGLFLFPNM-DNFVDLNAIKIFLA-----KNPVP------TLLANTYHSIHHRNIR--EG 208
+G +F + + VD + + VP +LA TY ++ + G
Sbjct: 981 FGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKTDAG 1040
Query: 209 GLIVCCAPLLYRWYA 223
+I C LL W A
Sbjct: 1041 AIIAGCPMLLQLWAA 1055
>UniRef100_Q6L487 Putative mutator transposase [Oryza sativa]
Length = 1385
Score = 42.7 bits (99), Expect = 0.023
Identities = 47/196 (23%), Positives = 83/196 (41%), Gaps = 30/196 (15%)
Query: 53 ILETLVQFYDPICHCFTFPDYQLVPTLEEYSFWVGLPVSEGEPFNGLEP--------SPK 104
+L LV + P H F P ++ PTL++ S+ +GLP++ G P ++ + +
Sbjct: 959 LLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLA-GAPVGSVDGVVGWKEDITAR 1017
Query: 105 NATIAEALHLKPSDLVHPHFTI---KNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLL 161
+ HL P++ + P+ T+ K L TA L+ A D+ + A + L
Sbjct: 1018 FEQVMRLPHLGPANTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEA----YLLW 1073
Query: 162 IYGLFLFPNM-DNFVDLNAIKIFLA-----KNPVP------TLLANTYHSIHHRNIR--E 207
++G +F + + VD + + VP +LA TY ++ +
Sbjct: 1074 LFGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKTDA 1133
Query: 208 GGLIVCCAPLLYRWYA 223
G +I C LL W A
Sbjct: 1134 GAIIAGCPMLLQLWAA 1149
>UniRef100_Q6F2W5 Putative MuDR family transposase [Oryza sativa]
Length = 1043
Score = 42.7 bits (99), Expect = 0.023
Identities = 48/195 (24%), Positives = 83/195 (41%), Gaps = 28/195 (14%)
Query: 53 ILETLVQFYDPICHCFTFPDYQLVPTLEEYSFWVGLPVSEGE--PFNGLEPSPKNAT--- 107
+L LV + P H F P ++ PTL++ S+ +GLP++ P +G ++ T
Sbjct: 737 LLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLTGAPVGPVDGFFGWKEDITARF 796
Query: 108 --IAEALHLKPSDLVHPHFTI---KNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLI 162
+ HL P++ + P+ T+ K L TA L+ A D+ + A + L +
Sbjct: 797 EQVIRLPHLGPTNTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEA----YLLWL 852
Query: 163 YGLFLFPNM-DNFVDLNAIKIFLA-----KNPVP------TLLANTYHSIHHRNIR--EG 208
+G +F + + VD + + VP +LA TY ++ + G
Sbjct: 853 FGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKTDAG 912
Query: 209 GLIVCCAPLLYRWYA 223
+I C LL W A
Sbjct: 913 AIIAGCPMLLQLWAA 927
>UniRef100_Q9LDW9 EST C28952(C62945) corresponds to a region of the predicted gene
[Oryza sativa]
Length = 1591
Score = 42.4 bits (98), Expect = 0.029
Identities = 48/195 (24%), Positives = 83/195 (41%), Gaps = 28/195 (14%)
Query: 53 ILETLVQFYDPICHCFTFPDYQLVPTLEEYSFWVGLPVSEGE--PFNGLEPSPKNAT--- 107
+L LV + P H F P ++ PTL++ S+ +GLP++ P +G+ ++ T
Sbjct: 948 LLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAAVGPVDGVFGWKEDITARF 1007
Query: 108 --IAEALHLKPSDLVHPHFTI---KNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLI 162
+ HL P+ + P+ T+ K L TA L+ A D+ + A + L +
Sbjct: 1008 EQVMRLPHLGPTTTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEA----YLLWL 1063
Query: 163 YGLFLFPNM-DNFVDLNAIKIFLA-----KNPVP------TLLANTYHSIHHRNIR--EG 208
+G +F + + VD + + VP +LA TY ++ + G
Sbjct: 1064 FGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKTDAG 1123
Query: 209 GLIVCCAPLLYRWYA 223
+I C LL W A
Sbjct: 1124 AIIAGCPMLLQLWAA 1138
>UniRef100_Q8SB37 Putative transposon protein [Oryza sativa]
Length = 1656
Score = 42.4 bits (98), Expect = 0.029
Identities = 48/195 (24%), Positives = 83/195 (41%), Gaps = 28/195 (14%)
Query: 53 ILETLVQFYDPICHCFTFPDYQLVPTLEEYSFWVGLPVSEGE--PFNGLEPSPKNAT--- 107
+L LV + P H F P ++ PTL++ S+ +GLP++ P +G+ ++ T
Sbjct: 959 LLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGVFGWKEDITARF 1018
Query: 108 --IAEALHLKPSDLVHPHFTI---KNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLI 162
+ HL P+ + P+ T+ K L TA L+ A D+ + A + L +
Sbjct: 1019 EQVMRLPHLGPTTTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEA----YLLWL 1074
Query: 163 YGLFLFPNM-DNFVDLNAIKIFLA-----KNPVP------TLLANTYHSIHHRNIR--EG 208
+G +F + + VD + + VP +LA TY ++ + G
Sbjct: 1075 FGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKTDAG 1134
Query: 209 GLIVCCAPLLYRWYA 223
+I C LL W A
Sbjct: 1135 AIIAGCPMLLQLWAA 1149
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.138 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 850,387,654
Number of Sequences: 2790947
Number of extensions: 38136366
Number of successful extensions: 100834
Number of sequences better than 10.0: 155
Number of HSP's better than 10.0 without gapping: 63
Number of HSP's successfully gapped in prelim test: 92
Number of HSP's that attempted gapping in prelim test: 100530
Number of HSP's gapped (non-prelim): 351
length of query: 475
length of database: 848,049,833
effective HSP length: 131
effective length of query: 344
effective length of database: 482,435,776
effective search space: 165957906944
effective search space used: 165957906944
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC149496.4


