

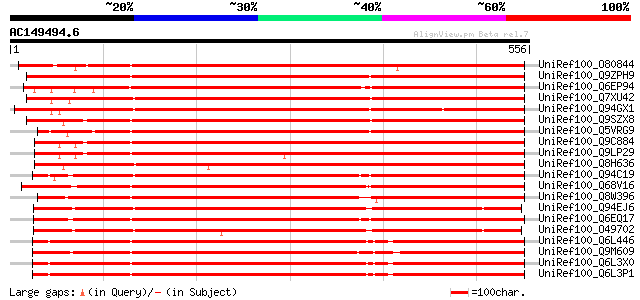
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149494.6 + phase: 0 /pseudo
(556 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O80844 Hypothetical protein At2g45750 [Arabidopsis tha... 716 0.0
UniRef100_Q9ZPH9 F15P23.1 protein [Arabidopsis thaliana] 712 0.0
UniRef100_Q6EP94 Dehydration-responsive family protein-like [Ory... 708 0.0
UniRef100_Q7XU42 OSJNBa0088I22.11 protein [Oryza sativa] 706 0.0
UniRef100_Q94GX1 Hypothetical protein OSJNBa0005K07.2 [Oryza sat... 691 0.0
UniRef100_Q9SZX8 Hypothetical protein F7L13.20 [Arabidopsis thal... 665 0.0
UniRef100_Q5VRG9 Hypothetical protein [Oryza sativa] 665 0.0
UniRef100_Q9C884 Hypothetical protein T16O9.7 [Arabidopsis thali... 651 0.0
UniRef100_Q9LP29 T9L6.6 protein [Arabidopsis thaliana] 641 0.0
UniRef100_Q8H636 Dehydration-responsive protein-like [Oryza sativa] 630 e-179
UniRef100_Q94C19 At1g26850/T2P11_4 [Arabidopsis thaliana] 615 e-174
UniRef100_Q68V16 Ankyrin protein kinase-like [Poa pratensis] 609 e-173
UniRef100_Q8W396 Hypothetical protein OSJNBa0013O08.23 [Oryza sa... 602 e-171
UniRef100_Q94EJ6 AT4g18030/T6K21_210 [Arabidopsis thaliana] 600 e-170
UniRef100_Q6EQ17 Dehydration-responsive protein-like [Oryza sativa] 596 e-169
UniRef100_O49702 Hypothetical protein AT4g18030 [Arabidopsis tha... 592 e-168
UniRef100_Q6L446 Putative methyltransferase [Solanum demissum] 592 e-167
UniRef100_Q9M609 Hypothetical protein [Malus domestica] 590 e-167
UniRef100_Q6L3X0 Putative methyltransferase family protein [Sola... 590 e-167
UniRef100_Q6L3P1 Putative methyltransferase [Solanum demissum] 590 e-167
>UniRef100_O80844 Hypothetical protein At2g45750 [Arabidopsis thaliana]
Length = 631
Score = 716 bits (1849), Expect = 0.0
Identities = 338/553 (61%), Positives = 426/553 (76%), Gaps = 16/553 (2%)
Query: 10 NKFPRPSSM-RKPHLYF--LIAFLCAAFYLLGAYQQRASFTSLTKKAIITSPSCTIQQVN 66
N F R SS +K +LY+ L+A LC A YLLG +Q A + A S +
Sbjct: 2 NLFTRISSRTKKANLYYVTLVALLCIASYLLGIWQNTAVNP---RAAFDDSDGTPCEGFT 58
Query: 67 KPT----LDFQSHHNSSDTIIALSSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYR 122
+P LDF +HHN D ++ +FP C +E+TPCED RSL++ R R+ YR
Sbjct: 59 RPNSTKDLDFDAHHNIQDPP-PVTETAVSFPSCAAALSEHTPCEDAKRSLKFSRERLEYR 117
Query: 123 ERHCPVKGEEDLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGD 182
+RHCP + EE LKCR+P P+GYKTPF WPASRDVAW+ANVPH ELTVEK QNW+RY+ D
Sbjct: 118 QRHCPER-EEILKCRIPAPYGYKTPFRWPASRDVAWFANVPHTELTVEKKNQNWVRYEND 176
Query: 183 RFFFPGGGTMFPNGAGAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSL 242
RF+FPGGGTMFP GA AYIDDIG+LI+L DGSIRTA+DTGCGVAS+GAYL SRNI T+S
Sbjct: 177 RFWFPGGGTMFPRGADAYIDDIGRLIDLSDGSIRTAIDTGCGVASFGAYLLSRNITTMSF 236
Query: 243 APRDTHEAQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEV 302
APRDTHEAQVQFALERGVPA+IG++A+ RLP+PSRAFD++HCSRCLIPW + DG +L EV
Sbjct: 237 APRDTHEAQVQFALERGVPAMIGIMATIRLPYPSRAFDLAHCSRCLIPWGQNDGAYLMEV 296
Query: 303 DRVLRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAI 362
DRVLRPGGYWILSGPPINW K +GW+RT DLN EQT+IE+VA+SLCW K++++DD+AI
Sbjct: 297 DRVLRPGGYWILSGPPINWQKRWKGWERTMDDLNAEQTQIEQVARSLCWKKVVQRDDLAI 356
Query: 363 WQKPINHLDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEE---TAGGV 419
WQKP NH+DC+ R++ + FC ++PD AWYT + +CL P+P+V + E+ AGG
Sbjct: 357 WQKPFNHIDCKKTREVLKNPEFCRHDQDPDMAWYTKMDSCLTPLPEVDDAEDLKTVAGGK 416
Query: 420 LKNWPQRLESVPPRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLG-TKRYRNLVD 478
++ WP RL ++PPR++ G +E +T E + ++ +LWK+R+ +YKK++ QLG T RYRNLVD
Sbjct: 417 VEKWPARLNAIPPRVNKGALEEITPEAFLENTKLWKQRVSYYKKLDYQLGETGRYRNLVD 476
Query: 479 MNANLGGFASALVKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDL 538
MNA LGGFA+AL +PVWVMNVVPV+AK++TLG IYERGLIGTY +WCEAMSTYPRTYD
Sbjct: 477 MNAYLGGFAAALADDPVWVMNVVPVEAKLNTLGVIYERGLIGTYQNWCEAMSTYPRTYDF 536
Query: 539 IHADSLFSLYNGR 551
IHADS+F+LY G+
Sbjct: 537 IHADSVFTLYQGQ 549
>UniRef100_Q9ZPH9 F15P23.1 protein [Arabidopsis thaliana]
Length = 633
Score = 712 bits (1837), Expect = 0.0
Identities = 336/541 (62%), Positives = 413/541 (76%), Gaps = 10/541 (1%)
Query: 19 RKPHLY--FLIAFLCAAFYLLGAYQQ--RASFTSLTKKAIITSPSCTIQQVNKPTLDFQS 74
++ +LY LIA LC FY +G +Q R S +TS CT P L+F S
Sbjct: 18 KQTNLYRVILIAILCVTFYFVGVWQHSGRGISRSSISNHELTSVPCTFPHQTTPILNFAS 77
Query: 75 HHNSSDTIIALS-SETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEED 133
H + D ++ + P CGV F+EYTPCE RSL + R R+IYRERHCP K E
Sbjct: 78 RHTAPDLPPTITDARVVQIPSCGVEFSEYTPCEFVNRSLNFPRERLIYRERHCPEK-HEI 136
Query: 134 LKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMF 193
++CR+P P+GY PF WP SRDVAW+ANVPH ELTVEK QNW+RY+ DRF FPGGGTMF
Sbjct: 137 VRCRIPAPYGYSLPFRWPESRDVAWFANVPHTELTVEKKNQNWVRYEKDRFLFPGGGTMF 196
Query: 194 PNGAGAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQ 253
P GA AYID+IG+LINLKDGSIRTA+DTGCGVAS+GAYL SRNI+T+S APRDTHEAQVQ
Sbjct: 197 PRGADAYIDEIGRLINLKDGSIRTAIDTGCGVASFGAYLMSRNIVTMSFAPRDTHEAQVQ 256
Query: 254 FALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWI 313
FALERGVPA+IGVLAS RLPFP+RAFDI+HCSRCLIPW +Y+G +L EVDRVLRPGGYWI
Sbjct: 257 FALERGVPAIIGVLASIRLPFPARAFDIAHCSRCLIPWGQYNGTYLIEVDRVLRPGGYWI 316
Query: 314 LSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCR 373
LSGPPINW +H +GW+RT+ DLN EQ++IE+VA+SLCW KL++++D+A+WQKP NH+ C+
Sbjct: 317 LSGPPINWQRHWKGWERTRDDLNSEQSQIERVARSLCWRKLVQREDLAVWQKPTNHVHCK 376
Query: 374 SARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKE--ETAGGVLKNWPQRLESVP 431
R PFC + P++ WYT L+TCL P+P+V+ E E AGG L WP+RL ++P
Sbjct: 377 RNRIALGRPPFC-HRTLPNQGWYTKLETCLTPLPEVTGSEIKEVAGGQLARWPERLNALP 435
Query: 432 PRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLG-TKRYRNLVDMNANLGGFASAL 490
PRI G++EG+T + + + E W++R+ +YKK + QL T RYRN +DMNA+LGGFASAL
Sbjct: 436 PRIKSGSLEGITEDEFVSNTEKWQRRVSYYKKYDQQLAETGRYRNFLDMNAHLGGFASAL 495
Query: 491 VKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNG 550
V +PVWVMNVVPV+A V+TLG IYERGLIGTY +WCEAMSTYPRTYD IHADS+FSLY
Sbjct: 496 VDDPVWVMNVVPVEASVNTLGVIYERGLIGTYQNWCEAMSTYPRTYDFIHADSVFSLYKD 555
Query: 551 R 551
R
Sbjct: 556 R 556
>UniRef100_Q6EP94 Dehydration-responsive family protein-like [Oryza sativa]
Length = 646
Score = 708 bits (1828), Expect = 0.0
Identities = 342/565 (60%), Positives = 415/565 (72%), Gaps = 33/565 (5%)
Query: 15 PSSMRKPHLYF---LIAFLCAAFYLLGAYQQR---------ASFTSLTKKAIITSPSCTI 62
P++ R+P + +A LC+A Y LGA+Q +S + T A T+ + T
Sbjct: 14 PAAARRPSSFLPIATVALLCSASYFLGAWQHGGFSSPSASPSSVSVATAVACTTTTTATT 73
Query: 63 QQVNKP----------TLDFQSHHNSSDTIIALSSE-----TFNFPRCGVNFTEYTPCED 107
+ +P LDF +HH ++ LSS T + C ++EYTPCED
Sbjct: 74 RSATRPRKRTPAGQGQALDFSAHHAAAADGAVLSSSGDSAATRRYQACPARYSEYTPCED 133
Query: 108 PTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHGYKTPFTWPASRDVAWYANVPHREL 167
RSLRY R R++YRERHCP G E L+C VP P GY+ PF WPASRDVAW+ANVPH+EL
Sbjct: 134 VKRSLRYPRERLVYRERHCPT-GRERLRCLVPAPSGYRNPFPWPASRDVAWFANVPHKEL 192
Query: 168 TVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIGKLINLKDGSIRTALDTGCGVAS 227
TVEKAVQNWIR DGD+F FPGGGTMFP+GA AYIDDIGKLI L DGS+RTALDTGCGVAS
Sbjct: 193 TVEKAVQNWIRVDGDKFRFPGGGTMFPHGADAYIDDIGKLIPLHDGSVRTALDTGCGVAS 252
Query: 228 WGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRC 287
WGAYL SR+I+ +S APRD+HEAQVQFALERGVPA+IGVLAS RL +P+RAFD++HCSRC
Sbjct: 253 WGAYLLSRDILAMSFAPRDSHEAQVQFALERGVPAMIGVLASNRLTYPARAFDMAHCSRC 312
Query: 288 LIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAK 347
LIPW YDG++L EVDRVLRPGGYWILSGPPINW K+ +GW+RTK+DLN EQ IE VA+
Sbjct: 313 LIPWHLYDGLYLIEVDRVLRPGGYWILSGPPINWKKYWKGWERTKEDLNAEQQAIEAVAR 372
Query: 348 SLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVP 407
SLCW K+ E DIA+WQKP NH C+++RK PFC +NPD AWY ++ C+ P+P
Sbjct: 373 SLCWKKIKEAGDIAVWQKPANHASCKASRK---SPPFCS-HKNPDAAWYDKMEACVTPLP 428
Query: 408 QVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQ 467
+VS+ E AGG LK WPQRL +VPPRI G+I+GVTS+ + +D ELW+KRI HYK V NQ
Sbjct: 429 EVSDASEVAGGALKKWPQRLTAVPPRISRGSIKGVTSKAFVQDTELWRKRIQHYKGVINQ 488
Query: 468 LGTK-RYRNLVDMNANLGGFASALVKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWC 526
K RYRN++DMNA LGGFA+AL +P+WVMN+VP TLG +YERGLIG+Y DWC
Sbjct: 489 FEQKGRYRNVLDMNAGLGGFAAALASDPLWVMNMVPTVGNSSTLGVVYERGLIGSYQDWC 548
Query: 527 EAMSTYPRTYDLIHADSLFSLYNGR 551
E MSTYPRTYDLIHADS+F+LY R
Sbjct: 549 EGMSTYPRTYDLIHADSVFTLYKNR 573
Score = 34.7 bits (78), Expect = 7.5
Identities = 28/105 (26%), Positives = 45/105 (42%), Gaps = 5/105 (4%)
Query: 213 GSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLASKRL 272
G R LD G+ + A L S + +++ P + + + ERG+
Sbjct: 493 GRYRNVLDMNAGLGGFAAALASDPLWVMNMVPTVGNSSTLGVVYERGLIGSYQDWCEGMS 552
Query: 273 PFPSRAFDISHCSRCLIPW---AEYDGIFLNEVDRVLRPGGYWIL 314
+P R +D+ H + E D I L E+DR+LRP G I+
Sbjct: 553 TYP-RTYDLIHADSVFTLYKNRCEMD-IILLEMDRILRPEGTVII 595
>UniRef100_Q7XU42 OSJNBa0088I22.11 protein [Oryza sativa]
Length = 646
Score = 706 bits (1821), Expect = 0.0
Identities = 339/558 (60%), Positives = 414/558 (73%), Gaps = 27/558 (4%)
Query: 19 RKPHLYFL--IAFLCAAFYLLGAYQQR----------ASFTSLTKKAIIT--SPSCTI-- 62
R+P L+ L +A LC YL+G + AS S+ A ++ SP+ T+
Sbjct: 18 RRPSLFHLAAVAVLCTVSYLIGIWHHGGFSASPAGGVASSVSIATTASVSCVSPTPTLLG 77
Query: 63 ------QQVNKPTLDFQSHHNSSDTIIALSSETFNFPRCGVNFTEYTPCEDPTRSLRYKR 116
+ LDF +HH + +A + C ++EYTPCED RSLR+ R
Sbjct: 78 GGGGGGDSSSSAPLDFAAHHTAEGMEVASGQVHRTYEACPAKYSEYTPCEDVERSLRFPR 137
Query: 117 SRMIYRERHCPVKGEEDLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNW 176
R++YRERHCP +GE L+C VP P GY+ PF WP SRDVAW+ANVPH+ELTVEKAVQNW
Sbjct: 138 DRLVYRERHCPSEGER-LRCLVPAPQGYRNPFPWPTSRDVAWFANVPHKELTVEKAVQNW 196
Query: 177 IRYDGDRFFFPGGGTMFPNGAGAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRN 236
IR +G++F FPGGGTMFP+GAGAYIDDIGK+I L DGSIRTALDTGCGVASWGAYL SRN
Sbjct: 197 IRVEGEKFRFPGGGTMFPHGAGAYIDDIGKIIPLHDGSIRTALDTGCGVASWGAYLLSRN 256
Query: 237 IITLSLAPRDTHEAQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDG 296
I+ +S APRD+HEAQVQFALERGVPA+IGVL+S RL +P+RAFD++HCSRCLIPW YDG
Sbjct: 257 ILAMSFAPRDSHEAQVQFALERGVPAMIGVLSSNRLTYPARAFDMAHCSRCLIPWQLYDG 316
Query: 297 IFLNEVDRVLRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIE 356
++L EVDR+LRPGGYWILSGPPINW KH +GWQRTK+DLN EQ IE VAKSLCW K+
Sbjct: 317 LYLAEVDRILRPGGYWILSGPPINWKKHWKGWQRTKEDLNAEQQAIEAVAKSLCWKKITL 376
Query: 357 KD--DIAIWQKPINHLDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEE 414
K+ DIAIWQKP NH+ C+++RK+ PFC +NPD AWY ++ C+ P+P+VS+ +E
Sbjct: 377 KEVGDIAIWQKPTNHIHCKASRKVVKSPPFCS-NKNPDAAWYDKMEACITPLPEVSDIKE 435
Query: 415 TAGGVLKNWPQRLESVPPRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLGTK-RY 473
AGG LK WP+RL +VPPRI G+IEGVT E + +D +LW+KR+ HYK V +Q G K RY
Sbjct: 436 IAGGQLKKWPERLTAVPPRIASGSIEGVTDEMFVEDTKLWQKRVGHYKSVISQFGQKGRY 495
Query: 474 RNLVDMNANLGGFASALVKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYP 533
RNL+DMNA GGFA+ALV +PVWVMN+VP TLG IYERGLIG+Y DWCE MSTYP
Sbjct: 496 RNLLDMNARFGGFAAALVDDPVWVMNMVPTVGNSTTLGVIYERGLIGSYQDWCEGMSTYP 555
Query: 534 RTYDLIHADSLFSLYNGR 551
RTYDLIHADS+F+LY R
Sbjct: 556 RTYDLIHADSVFTLYKDR 573
>UniRef100_Q94GX1 Hypothetical protein OSJNBa0005K07.2 [Oryza sativa]
Length = 634
Score = 691 bits (1783), Expect = 0.0
Identities = 322/559 (57%), Positives = 415/559 (73%), Gaps = 16/559 (2%)
Query: 6 YSRNNKFPRPSSMRKPHLYFL--IAFLCAAFYLLGAYQQR-----ASFTSLTK------K 52
Y + K + +K L ++ ++ LC AFY+LGA+Q A+ +++TK
Sbjct: 5 YPASPKAQQLQESKKQRLTYILVVSALCVAFYVLGAWQNTTVPKPAASSAITKVGCDPAA 64
Query: 53 AIITSPSCTIQQVNKPTLDFQSHHNSSDTIIALSSETFNFPRCGVNFTEYTPCEDPTRSL 112
A +S + ++ +LDF++HH S + FP C +NF+EYTPCED R
Sbjct: 65 AGQSSAVPSFGSASQESLDFEAHHQLSLDDTGAEAAVQPFPACPLNFSEYTPCEDRKRGR 124
Query: 113 RYKRSRMIYRERHCPVKGEEDLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKA 172
R++R+ ++YRERHCP K EE ++C +P P Y+TPF WP SRD AW+ N+PH+EL++EKA
Sbjct: 125 RFERAMLVYRERHCPGKDEE-IRCLIPAPPKYRTPFKWPQSRDFAWFNNIPHKELSIEKA 183
Query: 173 VQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYL 232
VQNWI+ DG RF FPGGGTMFP GA AYIDDIGKLI+L DG IRTA+DTGCGVASWGAYL
Sbjct: 184 VQNWIQVDGQRFRFPGGGTMFPRGADAYIDDIGKLISLTDGKIRTAIDTGCGVASWGAYL 243
Query: 233 QSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWA 292
RNI+ +S APRDTHEAQVQFALERGVPA+IGV+ +RLP+PSR+FD++HCSRCLIPW
Sbjct: 244 LKRNILAMSFAPRDTHEAQVQFALERGVPAIIGVMGKQRLPYPSRSFDMAHCSRCLIPWH 303
Query: 293 EYDGIFLNEVDRVLRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWN 352
E+DGI+L EVDR+LRPGGYWILSGPPINW H++GW+RTK+DL +EQ IE VA+SLCWN
Sbjct: 304 EFDGIYLAEVDRILRPGGYWILSGPPINWKTHYKGWERTKEDLKEEQDNIEDVARSLCWN 363
Query: 353 KLIEKDDIAIWQKPINHLDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNK 412
K++EK D++IWQKP NHL+C + +K C +NPD AWY ++ C+ P+P+VSN+
Sbjct: 364 KVVEKGDLSIWQKPKNHLECANIKKKYKTPHIC-KSDNPDAAWYKQMEACVTPLPEVSNQ 422
Query: 413 EETAGGVLKNWPQRLESVPPRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKR 472
E AGG L+ WPQR +VPPR+ G I G+ + + +D +LW+KR+ +YK+ + R
Sbjct: 423 GEIAGGALERWPQRAFAVPPRVKRGMIPGIDASKFEEDKKLWEKRVAYYKR-TLPIADGR 481
Query: 473 YRNLVDMNANLGGFASALVKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTY 532
YRN++DMNANLGGFA++LVK PVWVMNVVPV + DTLGAIYERG IGTY DWCEA STY
Sbjct: 482 YRNVMDMNANLGGFAASLVKYPVWVMNVVPVNSDRDTLGAIYERGFIGTYQDWCEAFSTY 541
Query: 533 PRTYDLIHADSLFSLYNGR 551
PRTYDL+HAD+LFS+Y R
Sbjct: 542 PRTYDLLHADNLFSIYQDR 560
Score = 34.3 bits (77), Expect = 9.7
Identities = 31/157 (19%), Positives = 63/157 (39%), Gaps = 22/157 (14%)
Query: 206 KLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIG 265
+ + + DG R +D + + A L + +++ P ++ + ERG
Sbjct: 473 RTLPIADGRYRNVMDMNANLGGFAASLVKYPVWVMNVVPVNSDRDTLGAIYERGFIGTYQ 532
Query: 266 VLASKRLPFPSRAFDISHCSRCLIPWAEYDGI--FLNEVDRVLRPGGYWILSGPPINWNK 323
+P R +D+ H + + I L E+DR+LRP G I+
Sbjct: 533 DWCEAFSTYP-RTYDLLHADNLFSIYQDRCDITNILLEMDRILRPEGTAII--------- 582
Query: 324 HHRGWQRTKKDLNQEQTKIEKVAKSLCW-NKLIEKDD 359
+D TK++ +AK + W +++++ +D
Sbjct: 583 ---------RDTVDVLTKVQAIAKRMRWESRILDHED 610
>UniRef100_Q9SZX8 Hypothetical protein F7L13.20 [Arabidopsis thaliana]
Length = 633
Score = 665 bits (1717), Expect = 0.0
Identities = 313/539 (58%), Positives = 401/539 (74%), Gaps = 11/539 (2%)
Query: 19 RKPHLYFLIAFLCAAFYLLGAYQQRASFTSLTKKAIIT-----SPSCTIQQVNKPTLDFQ 73
+K L ++ LC FY+LGA+Q +S++K T S S + LDF+
Sbjct: 17 KKLTLILGVSGLCILFYVLGAWQANTVPSSISKLGCETQSNPSSSSSSSSSSESAELDFK 76
Query: 74 SHHNSSDTIIALSSETFN-FPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEE 132
SH+ + +++T F C ++ +EYTPCED R R+ R+ M YRERHCPVK +E
Sbjct: 77 SHNQIE---LKETNQTIKYFEPCELSLSEYTPCEDRQRGRRFDRNMMKYRERHCPVK-DE 132
Query: 133 DLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTM 192
L C +PPP YK PF WP SRD AWY N+PH+EL+VEKAVQNWI+ +GDRF FPGGGTM
Sbjct: 133 LLYCLIPPPPNYKIPFKWPQSRDYAWYDNIPHKELSVEKAVQNWIQVEGDRFRFPGGGTM 192
Query: 193 FPNGAGAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQV 252
FP GA AYIDDI +LI L DG IRTA+DTGCGVAS+GAYL R+I+ +S APRDTHEAQV
Sbjct: 193 FPRGADAYIDDIARLIPLTDGGIRTAIDTGCGVASFGAYLLKRDIMAVSFAPRDTHEAQV 252
Query: 253 QFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYW 312
QFALERGVPA+IG++ S+RLP+P+RAFD++HCSRCLIPW + DG++L EVDRVLRPGGYW
Sbjct: 253 QFALERGVPAIIGIMGSRRLPYPARAFDLAHCSRCLIPWFKNDGLYLMEVDRVLRPGGYW 312
Query: 313 ILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDC 372
ILSGPPINW ++ RGW+RT++DL +EQ IE VAKSLCW K+ EK D++IWQKP+NH++C
Sbjct: 313 ILSGPPINWKQYWRGWERTEEDLKKEQDSIEDVAKSLCWKKVTEKGDLSIWQKPLNHIEC 372
Query: 373 RSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPP 432
+ ++ P C +N D AWY DL+TC+ P+P+ +N +++AGG L++WP R +VPP
Sbjct: 373 KKLKQNNKSPPICS-SDNADSAWYKDLETCITPLPETNNPDDSAGGALEDWPDRAFAVPP 431
Query: 433 RIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVK 492
RI GTI + +E + +DNE+WK+RI HYKK+ +L R+RN++DMNA LGGFA++++K
Sbjct: 432 RIIRGTIPEMNAEKFREDNEVWKERIAHYKKIVPELSHGRFRNIMDMNAFLGGFAASMLK 491
Query: 493 NPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
P WVMNVVPV A+ TLG IYERGLIGTY DWCE STYPRTYD+IHA LFSLY R
Sbjct: 492 YPSWVMNVVPVDAEKQTLGVIYERGLIGTYQDWCEGFSTYPRTYDMIHAGGLFSLYEHR 550
>UniRef100_Q5VRG9 Hypothetical protein [Oryza sativa]
Length = 631
Score = 665 bits (1715), Expect = 0.0
Identities = 308/533 (57%), Positives = 400/533 (74%), Gaps = 16/533 (3%)
Query: 30 LCAAFYLLGAYQQRASFTSLTKKAIITSPSC-----------TIQQVNKPTLDFQSHHNS 78
LC A Y+LGA+Q S TS+ I T C + + + LDFQ+HH
Sbjct: 30 LCVASYILGAWQG-TSTTSIHPSIIYTKSQCGESILRTSSNSSGRSSSDARLDFQAHHQV 88
Query: 79 SDTIIALSSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRV 138
S +L +E FP C + ++EYTPC+DP R+ ++ ++ M YRERHCP K EE +C +
Sbjct: 89 SFNESSLVAE--KFPPCQLKYSEYTPCQDPRRARKFPKTMMQYRERHCPRK-EELFRCLI 145
Query: 139 PPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAG 198
P P YK PF WP RD AWY N+PHREL++EKAVQNWI+ +G RF FPGGGTMFP+GA
Sbjct: 146 PAPPKYKNPFKWPQCRDFAWYDNIPHRELSIEKAVQNWIQVEGKRFRFPGGGTMFPHGAD 205
Query: 199 AYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALER 258
AYIDDI LI+L DG+IRTALDTGCGVASWGAYL RNIIT+S APRD+HEAQVQFALER
Sbjct: 206 AYIDDINALISLTDGNIRTALDTGCGVASWGAYLIKRNIITMSFAPRDSHEAQVQFALER 265
Query: 259 GVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPP 318
GVPA+IGV++++R+P+P+R+FD++HCSRCLIPW ++DGI+L EVDRV+RPGGYWILSGPP
Sbjct: 266 GVPAMIGVISTERIPYPARSFDMAHCSRCLIPWNKFDGIYLIEVDRVIRPGGYWILSGPP 325
Query: 319 INWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKL 378
I+W K+ +GW+RT++DL QEQ +IE +AK LCW K++EKDD+AIWQKPINH++C ++RK+
Sbjct: 326 IHWKKYFKGWERTEEDLKQEQDEIEDLAKRLCWKKVVEKDDLAIWQKPINHIECVNSRKI 385
Query: 379 ATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGT 438
C + D AWY ++TC+ P+P V++++E AGG L+ WP+R +VPPRI G+
Sbjct: 386 YETPQIC-KSNDVDSAWYKKMETCISPLPDVNSEDEVAGGALEKWPKRAFAVPPRISRGS 444
Query: 439 IEGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVM 498
+ G+T+E + +DN++W +R +YKK+ L RYRN++DMNA +GGFA+AL+K P+WVM
Sbjct: 445 VSGLTTEKFQEDNKVWAERADYYKKLIPPLTKGRYRNVMDMNAGMGGFAAALMKYPLWVM 504
Query: 499 NVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
NVVP + DTLG IYERG IGTY DWCEA STYPRTYD IHAD +FS Y R
Sbjct: 505 NVVPSGSAHDTLGIIYERGFIGTYQDWCEAFSTYPRTYDFIHADKIFSFYQDR 557
>UniRef100_Q9C884 Hypothetical protein T16O9.7 [Arabidopsis thaliana]
Length = 639
Score = 651 bits (1680), Expect = 0.0
Identities = 302/549 (55%), Positives = 398/549 (72%), Gaps = 28/549 (5%)
Query: 27 IAFLCAAFYLLGAYQQRASFTSLTK--------KAIITSPSCTIQQVNKPT--------- 69
++ LC Y+LG++Q TS ++ + T+ + T Q P+
Sbjct: 24 VSGLCILSYVLGSWQTNTVPTSSSEAYSRMGCDETSTTTRAQTTQTQTNPSSDDTSSSLS 83
Query: 70 ------LDFQSHHNSSDTIIALSSETFN-FPRCGVNFTEYTPCEDPTRSLRYKRSRMIYR 122
LDF+SHH + ++++T F C ++ +EYTPCED R R+ R+ M YR
Sbjct: 84 SSEPVELDFESHHKLE---LKITNQTVKYFEPCDMSLSEYTPCEDRERGRRFDRNMMKYR 140
Query: 123 ERHCPVKGEEDLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGD 182
ERHCP K +E L C +PPP YK PF WP SRD AWY N+PH+EL++EKA+QNWI+ +G+
Sbjct: 141 ERHCPSK-DELLYCLIPPPPNYKIPFKWPQSRDYAWYDNIPHKELSIEKAIQNWIQVEGE 199
Query: 183 RFFFPGGGTMFPNGAGAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSL 242
RF FPGGGTMFP GA AYIDDI +LI L DG+IRTA+DTGCGVAS+GAYL R+I+ +S
Sbjct: 200 RFRFPGGGTMFPRGADAYIDDIARLIPLTDGAIRTAIDTGCGVASFGAYLLKRDIVAMSF 259
Query: 243 APRDTHEAQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEV 302
APRDTHEAQVQFALERGVPA+IG++ S+RLP+P+RAFD++HCSRCLIPW + DG++L EV
Sbjct: 260 APRDTHEAQVQFALERGVPAIIGIMGSRRLPYPARAFDLAHCSRCLIPWFQNDGLYLTEV 319
Query: 303 DRVLRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAI 362
DRVLRPGGYWILSGPPINW K+ +GW+R+++DL QEQ IE A+SLCW K+ EK D++I
Sbjct: 320 DRVLRPGGYWILSGPPINWKKYWKGWERSQEDLKQEQDSIEDAARSLCWKKVTEKGDLSI 379
Query: 363 WQKPINHLDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKN 422
WQKPINH++C +++ P C + PD AWY DL++C+ P+P+ ++ +E AGG L++
Sbjct: 380 WQKPINHVECNKLKRVHKTPPLCSKSDLPDFAWYKDLESCVTPLPEANSSDEFAGGALED 439
Query: 423 WPQRLESVPPRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNAN 482
WP R +VPPRI GTI + +E + +DNE+WK+RI +YK++ +L R+RN++DMNA
Sbjct: 440 WPNRAFAVPPRIIGGTIPDINAEKFREDNEVWKERISYYKQIMPELSRGRFRNIMDMNAY 499
Query: 483 LGGFASALVKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHAD 542
LGGFA+A++K P WVMNVVPV A+ TLG I+ERG IGTY DWCE STYPRTYDLIHA
Sbjct: 500 LGGFAAAMMKYPSWVMNVVPVDAEKQTLGVIFERGFIGTYQDWCEGFSTYPRTYDLIHAG 559
Query: 543 SLFSLYNGR 551
LFS+Y R
Sbjct: 560 GLFSIYENR 568
>UniRef100_Q9LP29 T9L6.6 protein [Arabidopsis thaliana]
Length = 656
Score = 641 bits (1653), Expect = 0.0
Identities = 302/566 (53%), Positives = 398/566 (69%), Gaps = 45/566 (7%)
Query: 27 IAFLCAAFYLLGAYQQRASFTSLTK--------KAIITSPSCTIQQVNKPT--------- 69
++ LC Y+LG++Q TS ++ + T+ + T Q P+
Sbjct: 24 VSGLCILSYVLGSWQTNTVPTSSSEAYSRMGCDETSTTTRAQTTQTQTNPSSDDTSSSLS 83
Query: 70 ------LDFQSHHNSSDTIIALSSETFN-FPRCGVNFTEYTPCEDPTRSLRYKRSRMIYR 122
LDF+SHH + ++++T F C ++ +EYTPCED R R+ R+ M YR
Sbjct: 84 SSEPVELDFESHHKLE---LKITNQTVKYFEPCDMSLSEYTPCEDRERGRRFDRNMMKYR 140
Query: 123 ERHCPVKGEEDLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGD 182
ERHCP K +E L C +PPP YK PF WP SRD AWY N+PH+EL++EKA+QNWI+ +G+
Sbjct: 141 ERHCPSK-DELLYCLIPPPPNYKIPFKWPQSRDYAWYDNIPHKELSIEKAIQNWIQVEGE 199
Query: 183 RFFFPGGGTMFPNGAGAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSL 242
RF FPGGGTMFP GA AYIDDI +LI L DG+IRTA+DTGCGVAS+GAYL R+I+ +S
Sbjct: 200 RFRFPGGGTMFPRGADAYIDDIARLIPLTDGAIRTAIDTGCGVASFGAYLLKRDIVAMSF 259
Query: 243 APRDTHEAQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAE--------- 293
APRDTHEAQVQFALERGVPA+IG++ S+RLP+P+RAFD++HCSRCLIPW +
Sbjct: 260 APRDTHEAQVQFALERGVPAIIGIMGSRRLPYPARAFDLAHCSRCLIPWFQNGFLIGVAN 319
Query: 294 --------YDGIFLNEVDRVLRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKV 345
DG++L EVDRVLRPGGYWILSGPPINW K+ +GW+R+++DL QEQ IE
Sbjct: 320 NQKKNWMCVDGLYLTEVDRVLRPGGYWILSGPPINWKKYWKGWERSQEDLKQEQDSIEDA 379
Query: 346 AKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMP 405
A+SLCW K+ EK D++IWQKPINH++C +++ P C + PD AWY DL++C+ P
Sbjct: 380 ARSLCWKKVTEKGDLSIWQKPINHVECNKLKRVHKTPPLCSKSDLPDFAWYKDLESCVTP 439
Query: 406 VPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVN 465
+P+ ++ +E AGG L++WP R +VPPRI GTI + +E + +DNE+WK+RI +YK++
Sbjct: 440 LPEANSSDEFAGGALEDWPNRAFAVPPRIIGGTIPDINAEKFREDNEVWKERISYYKQIM 499
Query: 466 NQLGTKRYRNLVDMNANLGGFASALVKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDW 525
+L R+RN++DMNA LGGFA+A++K P WVMNVVPV A+ TLG I+ERG IGTY DW
Sbjct: 500 PELSRGRFRNIMDMNAYLGGFAAAMMKYPSWVMNVVPVDAEKQTLGVIFERGFIGTYQDW 559
Query: 526 CEAMSTYPRTYDLIHADSLFSLYNGR 551
CE STYPRTYDLIHA LFS+Y R
Sbjct: 560 CEGFSTYPRTYDLIHAGGLFSIYENR 585
>UniRef100_Q8H636 Dehydration-responsive protein-like [Oryza sativa]
Length = 618
Score = 630 bits (1625), Expect = e-179
Identities = 297/536 (55%), Positives = 376/536 (69%), Gaps = 12/536 (2%)
Query: 27 IAFLCAAFYLLGAYQQRASFTSLTKKAIIT----SPSCTIQQVNKPTLDFQSHHNSS--D 80
+ LCAA YLL + A + A + P N LDF HH +S D
Sbjct: 16 VVALCAASYLLAVWTHPAPPLPASSLAAVPCNTRQPPAPAASKNDTALDFSIHHGASEED 75
Query: 81 TIIALSSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPP 140
A + + P C ++E+TPCE SLR R R YRERHCP E +C VP
Sbjct: 76 AAEAGAPPSRRVPACDAGYSEHTPCEGQRWSLRQPRRRFAYRERHCPPPAERR-RCLVPA 134
Query: 141 PHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAY 200
P GY+ P WP SRD AWYAN PH EL EK VQNWIR DGD FPGGGTMFP+GA Y
Sbjct: 135 PRGYRAPLRWPRSRDAAWYANAPHEELVTEKGVQNWIRRDGDVLRFPGGGTMFPHGADRY 194
Query: 201 IDDIGKLINLK---DGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALE 257
IDDI + G++RTALDTGCGVASWGAYL SR+++T+S AP+DTHEAQV FALE
Sbjct: 195 IDDIAAAAGITLGGGGAVRTALDTGCGVASWGAYLLSRDVLTMSFAPKDTHEAQVLFALE 254
Query: 258 RGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGP 317
RGVPA++G++A+KRLP+P+RAFD++HCSRCLIPW++Y+G+++ EVDRVLRPGGYW+LSGP
Sbjct: 255 RGVPAMLGIMATKRLPYPARAFDMAHCSRCLIPWSKYNGLYMIEVDRVLRPGGYWVLSGP 314
Query: 318 PINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARK 377
P+NW +H +GW+RT +DL+ EQ+ IE +AKSLCW K+ + DIA+WQK INH+ C+++R
Sbjct: 315 PVNWERHFKGWKRTPEDLSSEQSAIEAIAKSLCWTKVQQMGDIAVWQKQINHVSCKASRN 374
Query: 378 LATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMG 437
FC ++PD WY +++ C+ P+P+VS + AGG +K WP+RL S PPRI G
Sbjct: 375 ELGGLGFCNSNQDPDAGWYVNMEECITPLPEVSGPGDVAGGEVKRWPERLTSPPPRIAGG 434
Query: 438 TI-EGVTSEGYSKDNELWKKRIPHYKKVNNQLGTK-RYRNLVDMNANLGGFASALVKNPV 495
++ VT + + KD+E+W++R+ YK V+ L K RYRNL+DMNA LGGFA+ALV +PV
Sbjct: 435 SLGSSVTVDTFIKDSEMWRRRVDRYKGVSGGLAEKGRYRNLLDMNAGLGGFAAALVDDPV 494
Query: 496 WVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
WVMNVVP A +TLG IYERGLIGTY DWCEAMSTYPRTYDLIHA SLF++Y R
Sbjct: 495 WVMNVVPTAAVANTLGVIYERGLIGTYQDWCEAMSTYPRTYDLIHAYSLFTMYKDR 550
>UniRef100_Q94C19 At1g26850/T2P11_4 [Arabidopsis thaliana]
Length = 616
Score = 615 bits (1585), Expect = e-174
Identities = 292/532 (54%), Positives = 378/532 (70%), Gaps = 13/532 (2%)
Query: 25 FLIAFLCAAFYLLGAYQQRASF---TSLTKKAIITSPSCTIQQVNKPTLDFQSHHNSSDT 81
F++ LC FY+LGA+Q R+ F S+ + + C I P+L+F++HH +
Sbjct: 19 FIVFSLCCFFYILGAWQ-RSGFGKGDSIALEMTNSGADCNIV----PSLNFETHHAGESS 73
Query: 82 IIALS--SETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVP 139
++ S ++ F C +T+YTPC+D R++ + R MIYRERHC + E+ L C +P
Sbjct: 74 LVGASEAAKVKAFEPCDGRYTDYTPCQDQRRAMTFPRDSMIYRERHCAPENEK-LHCLIP 132
Query: 140 PPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGA 199
P GY TPF+WP SRD YAN P++ LTVEKA+QNWI+Y+GD F FPGGGT FP GA
Sbjct: 133 APKGYVTPFSWPKSRDYVPYANAPYKALTVEKAIQNWIQYEGDVFRFPGGGTQFPQGADK 192
Query: 200 YIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERG 259
YID + +I +++G++RTALDTGCGVASWGAYL SRN+ +S APRD+HEAQVQFALERG
Sbjct: 193 YIDQLASVIPMENGTVRTALDTGCGVASWGAYLWSRNVRAMSFAPRDSHEAQVQFALERG 252
Query: 260 VPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPI 319
VPA+IGVL + +LP+P+RAFD++HCSRCLIPW DG++L EVDRVLRPGGYWILSGPPI
Sbjct: 253 VPAVIGVLGTIKLPYPTRAFDMAHCSRCLIPWGANDGMYLMEVDRVLRPGGYWILSGPPI 312
Query: 320 NWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLA 379
NW +++ WQR K+DL +EQ KIE+ AK LCW K E +IAIWQK +N CRS R+
Sbjct: 313 NWKVNYKAWQRPKEDLQEEQRKIEEAAKLLCWEKKYEHGEIAIWQKRVNDEACRS-RQDD 371
Query: 380 TDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTI 439
FC ++ D WY ++ C+ P P+ S+ +E AGG L+ +P RL +VPPRI G+I
Sbjct: 372 PRANFC-KTDDTDDVWYKKMEACITPYPETSSSDEVAGGELQAFPDRLNAVPPRISSGSI 430
Query: 440 EGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVMN 499
GVT + Y DN WKK + YK++N+ L T RYRN++DMNA GGFA+AL +WVMN
Sbjct: 431 SGVTVDAYEDDNRQWKKHVKAYKRINSLLDTGRYRNIMDMNAGFGGFAAALESQKLWVMN 490
Query: 500 VVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
VVP A+ + LG +YERGLIG YHDWCEA STYPRTYDLIHA+ LFSLY +
Sbjct: 491 VVPTIAEKNRLGVVYERGLIGIYHDWCEAFSTYPRTYDLIHANHLFSLYKNK 542
Score = 36.6 bits (83), Expect = 2.0
Identities = 27/108 (25%), Positives = 48/108 (44%), Gaps = 5/108 (4%)
Query: 210 LKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLAS 269
L G R +D G + A L+S+ + +++ P + ++ ERG+ +
Sbjct: 459 LDTGRYRNIMDMNAGFGGFAAALESQKLWVMNVVPTIAEKNRLGVVYERGLIGIYHDWCE 518
Query: 270 KRLPFPSRAFDISHCSRCLIPW---AEYDGIFLNEVDRVLRPGGYWIL 314
+P R +D+ H + + D I L E+DR+LRP G I+
Sbjct: 519 AFSTYP-RTYDLIHANHLFSLYKNKCNADDILL-EMDRILRPEGAVII 564
>UniRef100_Q68V16 Ankyrin protein kinase-like [Poa pratensis]
Length = 613
Score = 609 bits (1571), Expect = e-173
Identities = 287/540 (53%), Positives = 372/540 (68%), Gaps = 9/540 (1%)
Query: 13 PRPSSMRKPHLYFLIAFLCAAFYLLGAYQQRASFTSLTKKAIITSPS-CTIQQVNKPTLD 71
P S R ++ +C FY+LGA+Q+ + IT + CTI ++
Sbjct: 7 PAESRTRSTVSICIVVGMCVFFYILGAWQKSGFGKGDSIALEITKRTDCTILPIS----- 61
Query: 72 FQSHHNSSDTIIALSSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGE 131
+ +HH+ + L S F C FT+YTPC+D R++++ R M YRERHCP++ +
Sbjct: 62 YDTHHSKKGSSGDLVSPVKKFKPCPDRFTDYTPCQDQNRAMKFPRENMNYRERHCPLQ-K 120
Query: 132 EDLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGT 191
E L C VPPP GY PF WP SRD +AN P++ LTVEKA+QNW++Y+G+ F FPGGGT
Sbjct: 121 EKLHCLVPPPKGYVAPFPWPKSRDYVPFANCPYKSLTVEKAIQNWVQYEGNVFRFPGGGT 180
Query: 192 MFPNGAGAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQ 251
FP GA YID + +I + +G++RTALDTGCGVASWGAYL RN++ + APRD+HEAQ
Sbjct: 181 QFPQGADKYIDQLAAVIPIANGTVRTALDTGCGVASWGAYLLKRNVLAMPFAPRDSHEAQ 240
Query: 252 VQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGY 311
VQFALERGVPA+IGVL + +LP+PSRAFD++HCSRCLIPW DG+++ EVDRVLRPGGY
Sbjct: 241 VQFALERGVPAVIGVLGTIKLPYPSRAFDMAHCSRCLIPWGLNDGLYMMEVDRVLRPGGY 300
Query: 312 WILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLD 371
W+LSGPPINW +++GWQRTKKDL EQ KIE++A+ LCW K+ EK + AIW+K +N
Sbjct: 301 WVLSGPPINWKVNYKGWQRTKKDLEAEQNKIEEIAELLCWEKVSEKGETAIWRKRVNTES 360
Query: 372 CRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVP 431
C S + +T + C N D WY +K C+ P+P V N E AGG +K +P RL ++P
Sbjct: 361 CPSRHEESTVQ-MC-KSTNADDVWYKTMKACVTPLPDVENPSEVAGGAIKPFPSRLNAIP 418
Query: 432 PRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALV 491
PRI G I GV+S+ Y KDN++WKK + Y VN L T RYRN++DMNA GGFA+A+
Sbjct: 419 PRIANGLIPGVSSQAYEKDNKMWKKHVKAYSNVNKYLLTGRYRNIMDMNAGFGGFAAAIE 478
Query: 492 KNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
WVMNVVP K+ TLG++Y RGLIG YHDWCEA STYPRTYDLIHA LF+LY +
Sbjct: 479 SPKSWVMNVVPTIGKIATLGSVYGRGLIGIYHDWCEAFSTYPRTYDLIHASGLFTLYKNK 538
Score = 34.3 bits (77), Expect = 9.7
Identities = 34/153 (22%), Positives = 61/153 (39%), Gaps = 22/153 (14%)
Query: 210 LKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLAS 269
L G R +D G + A ++S +++ P A + RG+ +
Sbjct: 455 LLTGRYRNIMDMNAGFGGFAAAIESPKSWVMNVVPTIGKIATLGSVYGRGLIGIYHDWCE 514
Query: 270 KRLPFPSRAFDISHCSRCLIPWAEYDGI--FLNEVDRVLRPGGYWILSGPPINWNKHHRG 327
+P R +D+ H S + + L E+DR+LRP G I+
Sbjct: 515 AFSTYP-RTYDLIHASGLFTLYKNKCSLEDILLEMDRILRPEGAVIM------------- 560
Query: 328 WQRTKKDLNQEQTKIEKVAKSLCWN-KLIEKDD 359
+D TK++K A+ + WN +L++ +D
Sbjct: 561 -----RDDVDILTKVDKFARGMRWNTRLVDHED 588
>UniRef100_Q8W396 Hypothetical protein OSJNBa0013O08.23 [Oryza sativa]
Length = 611
Score = 602 bits (1553), Expect = e-171
Identities = 289/533 (54%), Positives = 364/533 (68%), Gaps = 27/533 (5%)
Query: 30 LCAAFYLLGAYQQRASFTSLTKKAIITSPSCTIQQVNKPTLDFQSHHNSSDTIIAL-SSE 88
LC+ FYLLG +Q+ + A++ + + V P L+F++HH++SD S+E
Sbjct: 22 LCSFFYLLGVWQRSGFGRGDSIAAVVNEQT---KCVVLPNLNFETHHSASDLPNDTGSTE 78
Query: 89 TFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHGYKTPF 148
F C +T+YTPCE+ R++ + R MIYRERHCP + ++ L C VP P GY PF
Sbjct: 79 VKTFEPCDAQYTDYTPCEEQKRAMTFPRDNMIYRERHCPPE-KDKLYCLVPAPKGYAAPF 137
Query: 149 TWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIGKLI 208
WP SRD YAN+PH+ LTVEKA+QNW+ Y+G F FPGGGT FP GA YID + +I
Sbjct: 138 HWPKSRDYVHYANIPHKSLTVEKAIQNWVHYEGKVFRFPGGGTQFPQGADKYIDHLASVI 197
Query: 209 NLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLA 268
+ +G +RTALDTGCGVAS GAYL +N++T+S APRD HEAQVQFALERGVPA IGVL
Sbjct: 198 PIANGKVRTALDTGCGVASLGAYLLKKNVLTMSFAPRDNHEAQVQFALERGVPAYIGVLG 257
Query: 269 SKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHHRGW 328
S +L FPSR FD++HCSRCLIPW+ DG+++ EVDRVLRPGGYW+LSGPPI W H++GW
Sbjct: 258 SMKLSFPSRVFDMAHCSRCLIPWSGNDGMYMMEVDRVLRPGGYWVLSGPPIGWKIHYKGW 317
Query: 329 QRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPFCGPQ 388
QRTK DL EQ +IE+ A+ LCWNK+ EKD IAIW+K IN C Q
Sbjct: 318 QRTKDDLQSEQRRIEQFAELLCWNKISEKDGIAIWRKRINDKSCPM------------KQ 365
Query: 389 ENP----------DKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGT 438
ENP + WY ++ C+ P+P+V E AGG L+ +PQRL +VPPRI G
Sbjct: 366 ENPKVDKCELAYDNDVWYKKMEVCVTPLPEVKTMTEVAGGQLEPFPQRLNAVPPRITHGF 425
Query: 439 IEGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVM 498
+ G + + Y DN+LW+K I YKK+NN L T RYRN++DMNA LG FA+AL +WVM
Sbjct: 426 VPGFSVQSYQDDNKLWQKHINAYKKINNLLDTGRYRNIMDMNAGLGSFAAALESTKLWVM 485
Query: 499 NVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
NVVP A TLG IYERGLIG YHDWCE STYPRTYDLIHA+++FSLY +
Sbjct: 486 NVVPTIADTSTLGVIYERGLIGMYHDWCEGFSTYPRTYDLIHANAVFSLYENK 538
Score = 42.4 bits (98), Expect = 0.036
Identities = 39/156 (25%), Positives = 70/156 (44%), Gaps = 24/156 (15%)
Query: 200 YIDDIGKLINLKD-GSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALER 258
+I+ K+ NL D G R +D G+ S+ A L+S + +++ P + + ER
Sbjct: 444 HINAYKKINNLLDTGRYRNIMDMNAGLGSFAAALESTKLWVMNVVPTIADTSTLGVIYER 503
Query: 259 GVPALIGVLASKRLPFPSRAFDISHCSRCLIPW---AEYDGIFLNEVDRVLRPGGYWILS 315
G+ + +P R +D+ H + + +++ I L E+DR+LRP G I+
Sbjct: 504 GLIGMYHDWCEGFSTYP-RTYDLIHANAVFSLYENKCKFEDILL-EMDRILRPEGAVII- 560
Query: 316 GPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCW 351
R K D+ K+EK+A ++ W
Sbjct: 561 --------------RDKVDV---LVKVEKIANAMRW 579
>UniRef100_Q94EJ6 AT4g18030/T6K21_210 [Arabidopsis thaliana]
Length = 621
Score = 600 bits (1546), Expect = e-170
Identities = 286/523 (54%), Positives = 363/523 (68%), Gaps = 9/523 (1%)
Query: 26 LIAFLCAAFYLLGAYQQRASFTSLTKKAIITSPSCTIQQVNKPTLDFQSHHNSSDTIIAL 85
++ LC FYLLGA+Q+ + IT + V LDF+ HHN+
Sbjct: 21 VVVGLCCFFYLLGAWQKSGFGKGDSIAMEITKQAQCTDIVTD--LDFEPHHNTVKIPHKA 78
Query: 86 SSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHGYK 145
+ +F C V +YTPC++ R++++ R MIYRERHCP E+ L+C VP P GY
Sbjct: 79 DPKPVSFKPCDVKLKDYTPCQEQDRAMKFPRENMIYRERHCPPDNEK-LRCLVPAPKGYM 137
Query: 146 TPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIG 205
TPF WP SRD YAN P + LTVEKA QNW+++ G+ F FPGGGTMFP GA AYI+++
Sbjct: 138 TPFPWPKSRDYVHYANAPFKSLTVEKAGQNWVQFQGNVFKFPGGGTMFPQGADAYIEELA 197
Query: 206 KLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIG 265
+I +KDGS+RTALDTGCGVASWGAY+ RN++T+S APRD HEAQVQFALERGVPA+I
Sbjct: 198 SVIPIKDGSVRTALDTGCGVASWGAYMLKRNVLTMSFAPRDNHEAQVQFALERGVPAIIA 257
Query: 266 VLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHH 325
VL S LP+P+RAFD++ CSRCLIPW +G +L EVDRVLRPGGYW+LSGPPINW H
Sbjct: 258 VLGSILLPYPARAFDMAQCSRCLIPWTANEGTYLMEVDRVLRPGGYWVLSGPPINWKTWH 317
Query: 326 RGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPFC 385
+ W RTK +LN EQ +IE +A+SLCW K EK DIAI++K IN C + + T +
Sbjct: 318 KTWNRTKAELNAEQKRIEGIAESLCWEKKYEKGDIAIFRKKINDRSCDRSTPVDTCK--- 374
Query: 386 GPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVTSE 445
+++ D WY +++TC+ P P+VSN+EE AGG LK +P+RL +VPP I G I GV E
Sbjct: 375 --RKDTDDVWYKEIETCVTPFPKVSNEEEVAGGKLKKFPERLFAVPPSISKGLINGVDEE 432
Query: 446 GYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVMNVVPVQA 505
Y +D LWKKR+ YK++N +G+ RYRN++DMNA LGGFA+AL WVMNV+P
Sbjct: 433 SYQEDINLWKKRVTGYKRINRLIGSTRYRNVMDMNAGLGGFAAALESPKSWVMNVIPTIN 492
Query: 506 KVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLY 548
K +TL +YERGLIG YHDWCE STYPRTYD IHA +FSLY
Sbjct: 493 K-NTLSVVYERGLIGIYHDWCEGFSTYPRTYDFIHASGVFSLY 534
>UniRef100_Q6EQ17 Dehydration-responsive protein-like [Oryza sativa]
Length = 616
Score = 596 bits (1537), Expect = e-169
Identities = 283/527 (53%), Positives = 364/527 (68%), Gaps = 8/527 (1%)
Query: 26 LIAFLCAAFYLLGAYQQRASFTSLTKKAIITSPS-CTIQQVNKPTLDFQSHHNSSDTIIA 84
++ +C FY+LGA+Q+ + IT + CTI P L F +H
Sbjct: 20 IVIGMCCFFYILGAWQKSGFGKGDSIALEITKRTDCTIL----PNLSFDTHLAKQARPRD 75
Query: 85 LSSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHGY 144
L S F C +T+YTPC+D R++++ R M YRERHCP + +E L C +PPP GY
Sbjct: 76 LVSPAKKFKPCPDRYTDYTPCQDQNRAMKFPRENMNYRERHCPPQ-KEKLHCLIPPPKGY 134
Query: 145 KTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDI 204
PF WP SRD +AN P++ LTVEKA+QNW++++G+ F FPGGGT FP GA YID +
Sbjct: 135 VAPFPWPKSRDYVPFANCPYKSLTVEKAIQNWVQFEGNVFRFPGGGTQFPQGADKYIDQL 194
Query: 205 GKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALI 264
++ + +G++RTALDTGCGVASWGAYL RN++ +S APRD+HEAQVQFALERGVPA+I
Sbjct: 195 ASVVPIANGTVRTALDTGCGVASWGAYLLKRNVLAMSFAPRDSHEAQVQFALERGVPAVI 254
Query: 265 GVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKH 324
GVL + +LP+PSRAFD++HCSRCLIPW GI++ EVDRVLRPGGYW+LSGPPINW +
Sbjct: 255 GVLGTIKLPYPSRAFDMAHCSRCLIPWGANGGIYMMEVDRVLRPGGYWVLSGPPINWKVN 314
Query: 325 HRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPF 384
++GWQRTKKDL EQ KIE++A LCW K+ E ++AIW+K +N C S R+ +
Sbjct: 315 YKGWQRTKKDLEAEQNKIEEIADLLCWEKVKEIGEMAIWRKRLNTESCPS-RQDESSVQM 373
Query: 385 CGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVTS 444
C N D WY +K C+ P+P V++ E AGG +K +P RL +VPPRI G I GV+S
Sbjct: 374 C-DSTNADDVWYKKMKPCVTPIPDVNDPSEVAGGAIKPFPSRLNAVPPRIANGLIPGVSS 432
Query: 445 EGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVMNVVPVQ 504
+ Y KD ++WKK + Y VN L T RYRN++DMNA GGFA+A+ WVMN VP
Sbjct: 433 QAYQKDIKMWKKHVKAYSSVNKYLLTGRYRNIMDMNAGFGGFAAAIESPKSWVMNAVPTI 492
Query: 505 AKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
+K+ TLGAIYERGLIG YHDWCEA STYPRTYDLIHA LF+LY +
Sbjct: 493 SKMSTLGAIYERGLIGIYHDWCEAFSTYPRTYDLIHASGLFTLYKNK 539
>UniRef100_O49702 Hypothetical protein AT4g18030 [Arabidopsis thaliana]
Length = 629
Score = 592 bits (1527), Expect = e-168
Identities = 286/531 (53%), Positives = 363/531 (67%), Gaps = 17/531 (3%)
Query: 26 LIAFLCAAFYLLGAYQQRASFTSLTKKAIITSPSCTIQQVNKPTLDFQSHHNSSDTIIAL 85
++ LC FYLLGA+Q+ + IT + V LDF+ HHN+
Sbjct: 21 VVVGLCCFFYLLGAWQKSGFGKGDSIAMEITKQAQCTDIVTD--LDFEPHHNTVKIPHKA 78
Query: 86 SSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHGYK 145
+ +F C V +YTPC++ R++++ R MIYRERHCP E+ L+C VP P GY
Sbjct: 79 DPKPVSFKPCDVKLKDYTPCQEQDRAMKFPRENMIYRERHCPPDNEK-LRCLVPAPKGYM 137
Query: 146 TPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIG 205
TPF WP SRD YAN P + LTVEKA QNW+++ G+ F FPGGGTMFP GA AYI+++
Sbjct: 138 TPFPWPKSRDYVHYANAPFKSLTVEKAGQNWVQFQGNVFKFPGGGTMFPQGADAYIEELA 197
Query: 206 KLINLKDGSIRTALDTGCGV--------ASWGAYLQSRNIITLSLAPRDTHEAQVQFALE 257
+I +KDGS+RTALDTGCGV ASWGAY+ RN++T+S APRD HEAQVQFALE
Sbjct: 198 SVIPIKDGSVRTALDTGCGVSRFLFDLVASWGAYMLKRNVLTMSFAPRDNHEAQVQFALE 257
Query: 258 RGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGP 317
RGVPA+I VL S LP+P+RAFD++ CSRCLIPW +G +L EVDRVLRPGGYW+LSGP
Sbjct: 258 RGVPAIIAVLGSILLPYPARAFDMAQCSRCLIPWTANEGTYLMEVDRVLRPGGYWVLSGP 317
Query: 318 PINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARK 377
PINW H+ W RTK +LN EQ +IE +A+SLCW K EK DIAI++K IN C +
Sbjct: 318 PINWKTWHKTWNRTKAELNAEQKRIEGIAESLCWEKKYEKGDIAIFRKKINDRSCDRSTP 377
Query: 378 LATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMG 437
+ T + +++ D WY +++TC+ P P+VSN+EE AGG LK +P+RL +VPP I G
Sbjct: 378 VDTCK-----RKDTDDVWYKEIETCVTPFPKVSNEEEVAGGKLKKFPERLFAVPPSISKG 432
Query: 438 TIEGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWV 497
I GV E Y +D LWKKR+ YK++N +G+ RYRN++DMNA LGGFA+AL WV
Sbjct: 433 LINGVDEESYQEDINLWKKRVTGYKRINRLIGSTRYRNVMDMNAGLGGFAAALESPKSWV 492
Query: 498 MNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLY 548
MNV+P K +TL +YERGLIG YHDWCE STYPRTYD IHA +FSLY
Sbjct: 493 MNVIPTINK-NTLSVVYERGLIGIYHDWCEGFSTYPRTYDFIHASGVFSLY 542
>UniRef100_Q6L446 Putative methyltransferase [Solanum demissum]
Length = 612
Score = 592 bits (1525), Expect = e-167
Identities = 284/527 (53%), Positives = 362/527 (67%), Gaps = 9/527 (1%)
Query: 25 FLIAFLCAAFYLLGAYQQRASFTSLTKKAIITSPSCTIQQVNKPTLDFQSHHNSSDTIIA 84
F++A LC FYLLGA+Q R+ F A+ + + P L+F++ H
Sbjct: 18 FIVAGLCCFFYLLGAWQ-RSGFGKGDSIAVAVTKTAGENCDILPNLNFETRHAGEAGGTD 76
Query: 85 LSSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHGY 144
S E C +T+YTPC+D R++ + R M YRERHCP + EE L C +P P GY
Sbjct: 77 ESEEVEELKPCDPQYTDYTPCQDQKRAMTFPRENMNYRERHCPPQ-EEKLHCLIPAPKGY 135
Query: 145 KTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDI 204
TPF WP SRD YAN P++ LTVEKA+QNW++Y+G+ F FPGGGT FP GA YID +
Sbjct: 136 VTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWVQYEGNMFRFPGGGTQFPQGADKYIDQL 195
Query: 205 GKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALI 264
++ +++G++RTALDTGCGVASWGAYL RN+I +S APRD+HEAQVQFALERGVPA+I
Sbjct: 196 ASVVPIENGTVRTALDTGCGVASWGAYLWKRNVIAMSFAPRDSHEAQVQFALERGVPAVI 255
Query: 265 GVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKH 324
GVL + ++P+PS+AFD++HCSRCLIPW DGI + EVDRVLRPGGYW+LSGPPINW +
Sbjct: 256 GVLGTIKMPYPSKAFDMAHCSRCLIPWGAADGILMMEVDRVLRPGGYWVLSGPPINWKVN 315
Query: 325 HRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPF 384
+ WQR K+DL +EQ KIE+ AK LCW K+ EK + AIWQK + CRSA++ + R
Sbjct: 316 FKAWQRPKEDLEEEQRKIEEAAKLLCWEKISEKGETAIWQKRKDSASCRSAQENSAAR-V 374
Query: 385 CGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVTS 444
C P + PD WY ++ C+ P N LK +P+RL +VPPRI G + GV+
Sbjct: 375 CKPSD-PDSVWYNKMEMCITP-----NNGNGGDESLKPFPERLYAVPPRIANGLVSGVSV 428
Query: 445 EGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVMNVVPVQ 504
Y +D++ WKK I YKK+N L T RYRN++DMNA LGGFA+AL WVMNV+P
Sbjct: 429 AKYQEDSKKWKKHISAYKKINKLLDTGRYRNIMDMNAGLGGFAAALHSPKFWVMNVMPTI 488
Query: 505 AKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
A+ +TLG I+ERGLIG YHDWCEA STYPRTYDLIHA LFSLY +
Sbjct: 489 AEKNTLGVIFERGLIGIYHDWCEAFSTYPRTYDLIHASGLFSLYKDK 535
Score = 38.9 bits (89), Expect = 0.40
Identities = 37/154 (24%), Positives = 63/154 (40%), Gaps = 24/154 (15%)
Query: 210 LKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLAS 269
L G R +D G+ + A L S +++ P + + ERG+ +
Sbjct: 452 LDTGRYRNIMDMNAGLGGFAAALHSPKFWVMNVMPTIAEKNTLGVIFERGLIGIYHDWCE 511
Query: 270 KRLPFPSRAFDISHCSRCLIPW---AEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHHR 326
+P R +D+ H S + E++ I L E+DR+LRP G IL
Sbjct: 512 AFSTYP-RTYDLIHASGLFSLYKDKCEFEDILL-EMDRILRPEGAVIL------------ 557
Query: 327 GWQRTKKDLNQEQTKIEKVAKSLCWN-KLIEKDD 359
+D K++K+ + WN KL++ +D
Sbjct: 558 ------RDNVDVLIKVKKIIGGMRWNFKLMDHED 585
>UniRef100_Q9M609 Hypothetical protein [Malus domestica]
Length = 608
Score = 590 bits (1522), Expect = e-167
Identities = 286/528 (54%), Positives = 365/528 (68%), Gaps = 12/528 (2%)
Query: 25 FLIAFLCAAFYLLGAYQQRASFTSLTKKAIITSPSCTIQQVNKPTLDFQSHHNSS-DTII 83
F+ A LC FY+LGA+Q+ + IT + P+L F S H I
Sbjct: 18 FIAAGLCCFFYILGAWQRSGFGKGDSIALAITKNEADCNII--PSLSFDSQHAGEVGNID 75
Query: 84 ALSSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHG 143
S+ F C +T+YTPC+D R++ + R M YRERHCP + EE L C +P P G
Sbjct: 76 ESESKPKVFEPCHHRYTDYTPCQDQKRAMTFPREDMNYRERHCPPE-EEKLHCLIPAPKG 134
Query: 144 YKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDD 203
Y TPF WP SRD YAN P++ LTVEKAVQNWI+Y+G+ F FPGGGT FP GA YID
Sbjct: 135 YVTPFPWPKSRDYVPYANAPYKSLTVEKAVQNWIQYEGNVFRFPGGGTQFPQGADKYIDQ 194
Query: 204 IGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPAL 263
+ +I +K+G++RTALDTGCGVASWGAYL SRN++ +S APRD+HEAQVQFALERGVPA+
Sbjct: 195 LAAVIPIKNGTVRTALDTGCGVASWGAYLLSRNVLAMSFAPRDSHEAQVQFALERGVPAV 254
Query: 264 IGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNK 323
IGVL + +LP+PSRAFD++HCSRCLIPW DG +L EVDRVLRPGGYW+LSGPPINW
Sbjct: 255 IGVLGTIKLPYPSRAFDMAHCSRCLIPWGINDGKYLKEVDRVLRPGGYWVLSGPPINWKN 314
Query: 324 HHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRP 383
+++ WQR K+DL +EQ +IE+ AK LCW K EK + AIWQK ++ C R+ +
Sbjct: 315 NYQAWQRPKEDLQEEQRQIEEAAKLLCWEKKSEKGETAIWQKRVDSDSC-GDRQDDSRAN 373
Query: 384 FCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVT 443
FC E D WY ++ C+ P P+VS+ G LK +P+RL +VPPRI G++ GV+
Sbjct: 374 FCKADE-ADSVWYKKMEGCITPYPKVSS------GELKPFPKRLYAVPPRISSGSVPGVS 426
Query: 444 SEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVMNVVPV 503
E Y +DN WKK + YK++N + T RYRN++DMNA LGGFA+A+ +WVMNV+P
Sbjct: 427 VEDYEEDNNKWKKHVNAYKRINKLIDTGRYRNIMDMNAGLGGFAAAIESPKLWVMNVMPT 486
Query: 504 QAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
A+ +TLG +YERGLIG YHDWCE STYPRTYDLIHA +FS+YNG+
Sbjct: 487 IAEKNTLGVVYERGLIGIYHDWCEGFSTYPRTYDLIHAHGVFSMYNGK 534
>UniRef100_Q6L3X0 Putative methyltransferase family protein [Solanum demissum]
Length = 612
Score = 590 bits (1522), Expect = e-167
Identities = 284/527 (53%), Positives = 362/527 (67%), Gaps = 9/527 (1%)
Query: 25 FLIAFLCAAFYLLGAYQQRASFTSLTKKAIITSPSCTIQQVNKPTLDFQSHHNSSDTIIA 84
F+IA LC FYLLGA+Q R+ F A+ + + P L+F++ H
Sbjct: 18 FIIAGLCCFFYLLGAWQ-RSGFGKGDSIAVAVTKTAGENCDILPNLNFETRHAGEAGGTD 76
Query: 85 LSSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHGY 144
S E C +T+YTPC+D R++ + R M YRERHCP + EE L C +P P GY
Sbjct: 77 ESEEVEELKPCDPQYTDYTPCQDQKRAMTFPRENMNYRERHCPPQ-EEKLHCLIPAPKGY 135
Query: 145 KTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDI 204
TPF WP SRD YAN P++ LTVEKA+QNW++Y+G+ F FPGGGT FP GA YID +
Sbjct: 136 VTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWVQYEGNFFRFPGGGTQFPQGADKYIDQL 195
Query: 205 GKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALI 264
++ +++G++RTALDTGCGVASWGAYL RN+I +S APRD+HEAQVQFALERGVPA+I
Sbjct: 196 ASVVPIENGTVRTALDTGCGVASWGAYLWKRNVIAMSFAPRDSHEAQVQFALERGVPAVI 255
Query: 265 GVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKH 324
GVL + ++P+PS+AFD++HCSRCLIPW DGI + EVDRVLRPGGYW+LSGPPINW +
Sbjct: 256 GVLGTIKMPYPSKAFDMAHCSRCLIPWGAADGILMMEVDRVLRPGGYWVLSGPPINWKVN 315
Query: 325 HRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPF 384
+ WQR K+DL +EQ KIE+ AK LCW K+ EK + AIWQK + CRSA++ + R
Sbjct: 316 FKAWQRPKEDLEEEQRKIEEAAKLLCWEKISEKGETAIWQKRKDSASCRSAQENSAAR-V 374
Query: 385 CGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVTS 444
C P + PD WY ++ C+ P N LK +P+RL +VPPRI G + GV+
Sbjct: 375 CKPSD-PDSVWYNKMEMCITP-----NTGNGGDESLKPFPERLYAVPPRIANGLVSGVSV 428
Query: 445 EGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVMNVVPVQ 504
Y +D++ WKK + YKK+N L T RYRN++DMNA LGGFA+AL WVMNV+P
Sbjct: 429 AKYQEDSKKWKKHVSPYKKINKLLDTGRYRNIMDMNAGLGGFAAALHSPKFWVMNVMPTI 488
Query: 505 AKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
A+ +TLG I+ERGLIG YHDWCEA STYPRTYDLIHA LFSLY +
Sbjct: 489 AEKNTLGVIFERGLIGIYHDWCEAFSTYPRTYDLIHASGLFSLYKDK 535
Score = 38.9 bits (89), Expect = 0.40
Identities = 37/154 (24%), Positives = 63/154 (40%), Gaps = 24/154 (15%)
Query: 210 LKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLAS 269
L G R +D G+ + A L S +++ P + + ERG+ +
Sbjct: 452 LDTGRYRNIMDMNAGLGGFAAALHSPKFWVMNVMPTIAEKNTLGVIFERGLIGIYHDWCE 511
Query: 270 KRLPFPSRAFDISHCSRCLIPW---AEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHHR 326
+P R +D+ H S + E++ I L E+DR+LRP G IL
Sbjct: 512 AFSTYP-RTYDLIHASGLFSLYKDKCEFEDILL-EMDRILRPEGAVIL------------ 557
Query: 327 GWQRTKKDLNQEQTKIEKVAKSLCWN-KLIEKDD 359
+D K++K+ + WN KL++ +D
Sbjct: 558 ------RDNVDVLIKVKKIIGGMRWNFKLMDHED 585
>UniRef100_Q6L3P1 Putative methyltransferase [Solanum demissum]
Length = 612
Score = 590 bits (1522), Expect = e-167
Identities = 284/527 (53%), Positives = 362/527 (67%), Gaps = 9/527 (1%)
Query: 25 FLIAFLCAAFYLLGAYQQRASFTSLTKKAIITSPSCTIQQVNKPTLDFQSHHNSSDTIIA 84
F+IA LC FYLLGA+Q R+ F A+ + + P L+F++ H
Sbjct: 18 FIIAGLCCFFYLLGAWQ-RSGFGKGDSIAVAITKTAGENCDILPNLNFETRHAGEAGGTD 76
Query: 85 LSSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHGY 144
S E C +T+YTPC+D R++ + R M YRERHCP + EE L C +P P GY
Sbjct: 77 ESEEVEELKPCDPQYTDYTPCQDQKRAMTFPRENMNYRERHCPPQ-EEKLHCLIPAPKGY 135
Query: 145 KTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDI 204
TPF WP SRD YAN P++ LTVEKA+QNW++Y+G+ F FPGGGT FP GA YID +
Sbjct: 136 VTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWVQYEGNVFRFPGGGTQFPQGADKYIDQL 195
Query: 205 GKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALI 264
++ +++G++RTALDTGCGVASWGAYL RN+I +S APRD+HEAQVQFALERGVPA+I
Sbjct: 196 ASVVPIENGTVRTALDTGCGVASWGAYLWKRNVIAMSFAPRDSHEAQVQFALERGVPAVI 255
Query: 265 GVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKH 324
GVL + ++P+PS+AFD++HCSRCLIPW DGI + EVDRVLRPGGYW+LSGPPINW +
Sbjct: 256 GVLGTIKMPYPSKAFDMAHCSRCLIPWGAADGILMMEVDRVLRPGGYWVLSGPPINWKVN 315
Query: 325 HRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPF 384
+ WQR K+DL +EQ KIE+ AK LCW K+ EK + AIWQK + CRSA++ + R
Sbjct: 316 FKAWQRPKEDLEEEQRKIEEAAKLLCWEKISEKGETAIWQKRKDSASCRSAQENSAAR-V 374
Query: 385 CGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVTS 444
C P + PD WY ++ C+ P N LK +P+RL +VPPRI G + GV+
Sbjct: 375 CKPSD-PDSVWYNKMEMCITP-----NNGNGGDESLKPFPERLYAVPPRIANGLVSGVSV 428
Query: 445 EGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVMNVVPVQ 504
Y +D++ WKK + YKK+N L T RYRN++DMNA LGGFA+AL WVMNV+P
Sbjct: 429 AKYQEDSKKWKKHVSAYKKINKLLDTGRYRNIMDMNAGLGGFAAALHNPKFWVMNVMPTI 488
Query: 505 AKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
A+ +TLG I+ERGLIG YHDWCEA STYPRTYDLIHA LFSLY +
Sbjct: 489 AEKNTLGVIFERGLIGIYHDWCEAFSTYPRTYDLIHASGLFSLYKDK 535
Score = 37.7 bits (86), Expect = 0.88
Identities = 36/154 (23%), Positives = 63/154 (40%), Gaps = 24/154 (15%)
Query: 210 LKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLAS 269
L G R +D G+ + A L + +++ P + + ERG+ +
Sbjct: 452 LDTGRYRNIMDMNAGLGGFAAALHNPKFWVMNVMPTIAEKNTLGVIFERGLIGIYHDWCE 511
Query: 270 KRLPFPSRAFDISHCSRCLIPW---AEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHHR 326
+P R +D+ H S + E++ I L E+DR+LRP G IL
Sbjct: 512 AFSTYP-RTYDLIHASGLFSLYKDKCEFEDILL-EMDRILRPEGAVIL------------ 557
Query: 327 GWQRTKKDLNQEQTKIEKVAKSLCWN-KLIEKDD 359
+D K++K+ + WN KL++ +D
Sbjct: 558 ------RDNVDVLIKVKKIIGGMRWNFKLMDHED 585
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.137 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,041,520,772
Number of Sequences: 2790947
Number of extensions: 46948274
Number of successful extensions: 90492
Number of sequences better than 10.0: 213
Number of HSP's better than 10.0 without gapping: 88
Number of HSP's successfully gapped in prelim test: 125
Number of HSP's that attempted gapping in prelim test: 89826
Number of HSP's gapped (non-prelim): 308
length of query: 556
length of database: 848,049,833
effective HSP length: 133
effective length of query: 423
effective length of database: 476,853,882
effective search space: 201709192086
effective search space used: 201709192086
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC149494.6


