

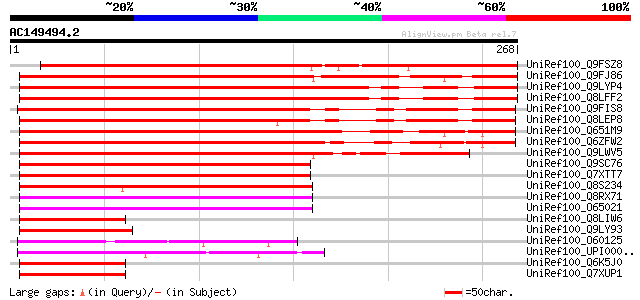
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149494.2 - phase: 0
(268 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FSZ8 Hypothetical protein [Cicer arietinum] 322 8e-87
UniRef100_Q9FJ86 Similarity to unknown protein [Arabidopsis thal... 263 3e-69
UniRef100_Q9LYP4 Hypothetical protein T28J14_160 [Arabidopsis th... 250 3e-65
UniRef100_Q8LFF2 Hypothetical protein [Arabidopsis thaliana] 250 3e-65
UniRef100_Q9FIS8 Similarity to unknown protein [Arabidopsis thal... 234 1e-60
UniRef100_Q8LEP8 Hypothetical protein [Arabidopsis thaliana] 228 2e-58
UniRef100_Q651M9 Hypothetical protein OSJNBa0047P18.22 [Oryza sa... 221 2e-56
UniRef100_Q6ZFW2 Putative BAG domain containing protein [Oryza s... 220 3e-56
UniRef100_Q9LWV5 BAG domain containing protein-like [Oryza sativa] 201 1e-50
UniRef100_Q9SC76 Hypothetical protein l1332.2 [Oryza sativa] 154 2e-36
UniRef100_Q7XTT7 OSJNBa0058K23.16 protein [Oryza sativa] 153 4e-36
UniRef100_Q8S234 P0446G04.18 protein [Oryza sativa] 122 1e-26
UniRef100_Q8RX71 At3g51780/ORF3 [Arabidopsis thaliana] 108 1e-22
UniRef100_O65021 Hypothetical protein [Arabidopsis thaliana] 108 1e-22
UniRef100_Q8LIW6 P0497A05.5 protein [Oryza sativa] 59 1e-07
UniRef100_Q9LY93 Hypothetical protein F18O22_150 [Arabidopsis th... 55 1e-06
UniRef100_O60125 BAG-family molecular chaperone regulator-1A [Sc... 52 1e-05
UniRef100_UPI000001534F UPI000001534F UniRef100 entry 51 3e-05
UniRef100_Q6K5J0 Ubiquitin-like protein [Oryza sativa] 51 3e-05
UniRef100_Q7XUP1 OSJNBb0011N17.22 protein [Oryza sativa] 51 3e-05
>UniRef100_Q9FSZ8 Hypothetical protein [Cicer arietinum]
Length = 262
Score = 322 bits (824), Expect = 8e-87
Identities = 175/264 (66%), Positives = 211/264 (79%), Gaps = 14/264 (5%)
Query: 17 GLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEMRKNIKMERAAK 76
GLHHQDQK+FYK+KERDSK FLDIVGVKDKSKLV++EDPI+QEKR LE+RKN KME+AAK
Sbjct: 1 GLHHQDQKLFYKDKERDSKVFLDIVGVKDKSKLVLVEDPISQEKRVLEIRKNAKMEKAAK 60
Query: 77 SISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLDGIVADGDVKLQ 136
SIS+ISLEVDRLAG+VSA E+IISKGGKVVETD+L LIE LMNQLLKLD I+ADGDVKLQ
Sbjct: 61 SISQISLEVDRLAGRVSAFESIISKGGKVVETDMLGLIELLMNQLLKLDSIIADGDVKLQ 120
Query: 137 RKMQVKRVQKYVETLDMLKIKN---SNGGHVPKKKPQQK----VKLPPIDEQLEGMSIGN 189
RKMQVKRVQKYVET+DMLK+K+ NG H P +PQQK +L I+EQ + + N
Sbjct: 121 RKMQVKRVQKYVETMDMLKVKSYMIGNGVHAP-IQPQQKNSNGKRLETIEEQPQQI-YSN 178
Query: 190 HKLQPSLEQQSQRNSNGNSQ----VFQQLQHKPSTNSTSEVVVTTKWETFDSLPPLIPVT 245
+ P EQQ ++ + NS+ +F++ +++PS NSTS VVVTTKWETFDS+PPLI +
Sbjct: 179 GSIVPIQEQQQEQQARNNSKELSLIFEEHENQPSNNSTSGVVVTTKWETFDSMPPLITLP 238
Query: 246 SASSSSSST-NNSVHPKFKWEHFE 268
S S++ S T NNS PKF WE F+
Sbjct: 239 STSTNHSMTNNNSTPPKFNWEFFD 262
>UniRef100_Q9FJ86 Similarity to unknown protein [Arabidopsis thaliana]
Length = 326
Score = 263 bits (672), Expect = 3e-69
Identities = 141/267 (52%), Positives = 187/267 (69%), Gaps = 17/267 (6%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELKKMLTGPTG+HHQDQK+ YK+KERDSKAFLD+ GVKDKSK+V++EDP++QEKR+LEM
Sbjct: 73 GELKKMLTGPTGIHHQDQKLMYKDKERDSKAFLDVSGVKDKSKMVLIEDPLSQEKRFLEM 132
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
RK K E+A+K+IS+ISLEVDRL G+VSA E + KGGK+ E D++++IE LMN+L+KLD
Sbjct: 133 RKIAKTEKASKAISDISLEVDRLGGRVSAFEMVTKKGGKIAEKDLVTVIELLMNELIKLD 192
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNS--NGGHVPKKKPQQKVKLPPIDEQLE 183
IVA+GDVKLQRKMQVKRVQ YVETLD LK+KNS NG +K+ +L PI E
Sbjct: 193 AIVAEGDVKLQRKMQVKRVQNYVETLDALKVKNSMANG---QQKQSSTAQRLAPIQEHNN 249
Query: 184 GMSIGNHKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSEVVV--TTKWETFDSLPPL 241
+Q ++ Q Q+++ +P + V+ + KWETFD P
Sbjct: 250 EERQEQKPIQSLMDMPIQYKEKK-----QEIEEEPRNSGEGPFVLDSSAKWETFDHHP-- 302
Query: 242 IPVTSASSSSSSTNNSVHPKFKWEHFE 268
+ SS+++ NN++ P+F WE F+
Sbjct: 303 ---VTPLSSTTAKNNAIPPRFNWEFFD 326
>UniRef100_Q9LYP4 Hypothetical protein T28J14_160 [Arabidopsis thaliana]
Length = 303
Score = 250 bits (638), Expect = 3e-65
Identities = 139/263 (52%), Positives = 178/263 (66%), Gaps = 26/263 (9%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELKKML+ GLHH+D K+ YK+KERDSK FLD+ GVKD+SKLVV EDPI+QEKR L
Sbjct: 67 GELKKMLSDQVGLHHEDMKVLYKDKERDSKMFLDLCGVKDRSKLVVKEDPISQEKRLLAK 126
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
RKN +E+A+KSIS+IS EVDRLAGQVSA ET+I+KGGKV E +++LIE LMNQLL+LD
Sbjct: 127 RKNAAIEKASKSISDISFEVDRLAGQVSAFETVINKGGKVEEKSLVNLIEMLMNQLLRLD 186
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLEGM 185
I+ADGDVKL RKMQV+RVQKYVE LD+LK+KNS K + K + EQ + +
Sbjct: 187 AIIADGDVKLMRKMQVQRVQKYVEALDLLKVKNSAKKVEVNKSVRHKPQTQTRFEQRDLL 246
Query: 186 SIGNHKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSEVVVTTKWETFDSLPPLIPVT 245
S E++ RNSN S++S + VV +KWE FDS
Sbjct: 247 SFVEE------EEEEPRNSNA------------SSSSGTPAVVASKWEMFDS-------- 280
Query: 246 SASSSSSSTNNSVHPKFKWEHFE 268
++++ ++ T V P+FKWE F+
Sbjct: 281 ASTAKAAETVKPVPPRFKWEFFD 303
>UniRef100_Q8LFF2 Hypothetical protein [Arabidopsis thaliana]
Length = 300
Score = 250 bits (638), Expect = 3e-65
Identities = 139/263 (52%), Positives = 178/263 (66%), Gaps = 26/263 (9%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELKKML+ GLHH+D K+ YK+KERDSK FLD+ GVKD+SKLVV EDPI+QEKR L
Sbjct: 64 GELKKMLSDQVGLHHEDMKVLYKDKERDSKMFLDLCGVKDRSKLVVKEDPISQEKRLLAK 123
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
RKN +E+A+KSIS+IS EVDRLAGQVSA ET+I+KGGKV E +++LIE LMNQLL+LD
Sbjct: 124 RKNAAIEKASKSISDISFEVDRLAGQVSAFETVINKGGKVEEKSLVNLIEMLMNQLLRLD 183
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLEGM 185
I+ADGDVKL RKMQV+RVQKYVE LD+LK+KNS K + K + EQ + +
Sbjct: 184 AIIADGDVKLMRKMQVQRVQKYVEALDLLKVKNSAKKVEVNKSVRHKPQTQTRFEQRDLL 243
Query: 186 SIGNHKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSEVVVTTKWETFDSLPPLIPVT 245
S E++ RNSN S++S + VV +KWE FDS
Sbjct: 244 SFVEE------EEEEPRNSNA------------SSSSGTPAVVASKWEMFDS-------- 277
Query: 246 SASSSSSSTNNSVHPKFKWEHFE 268
++++ ++ T V P+FKWE F+
Sbjct: 278 ASTAKAAETVKPVPPRFKWEFFD 300
>UniRef100_Q9FIS8 Similarity to unknown protein [Arabidopsis thaliana]
Length = 302
Score = 234 bits (598), Expect = 1e-60
Identities = 129/264 (48%), Positives = 179/264 (66%), Gaps = 40/264 (15%)
Query: 5 IGELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLE 64
I ELKK+L+G TG+HHQD +I YK+KERDSK FLD+ GVKD+SKL+++EDPI+QEKR LE
Sbjct: 77 IRELKKILSGATGVHHQDMQIIYKDKERDSKMFLDLSGVKDRSKLILIEDPISQEKRLLE 136
Query: 65 MRKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKL 124
+RK E+++K+IS+IS +V+RLAGQ+SA +T+I KGGKV E ++ +L+E LMNQL+KL
Sbjct: 137 LRKIATKEKSSKAISDISFQVERLAGQLSAFDTVIGKGGKVEEKNLENLMEMLMNQLVKL 196
Query: 125 DGIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLEG 184
D I DGDVKL++KMQ +R+ KYVE LD+LKIKNS ++PQ K K
Sbjct: 197 DAISGDGDVKLKKKMQEERLHKYVEALDLLKIKNS-------RQPQTKPK---------- 239
Query: 185 MSIGNHKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTS-EVVVTTKWETFDSLPPLIP 243
P +++ +++ KP+ +S+S V++TT+WETFDS
Sbjct: 240 ---------PQYKEREMLT------FYEEASRKPTASSSSPPVIITTRWETFDS------ 278
Query: 244 VTSASSSSSSTNNSVHPKFKWEHF 267
+SAS+++ VHPKFKWE F
Sbjct: 279 -SSASTATLQPVRPVHPKFKWELF 301
>UniRef100_Q8LEP8 Hypothetical protein [Arabidopsis thaliana]
Length = 293
Score = 228 bits (580), Expect = 2e-58
Identities = 129/274 (47%), Positives = 179/274 (65%), Gaps = 51/274 (18%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELKK+L+G TG+HHQD +I YK+KERDSK FLD+ GVKD+SKL+++EDPI+QEKR LE+
Sbjct: 58 GELKKILSGATGVHHQDMQIIYKDKERDSKMFLDLSGVKDRSKLILIEDPISQEKRLLEL 117
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
RK E+++K+IS+IS +V+RLAGQ+SA +T+I KGGKV E ++ +L+E LMNQL+KLD
Sbjct: 118 RKIATKEKSSKAISDISFQVERLAGQLSAFDTVIGKGGKVEEKNLENLMEMLMNQLVKLD 177
Query: 126 GIVADGDVKLQRKMQ-----------VKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVK 174
I DGDVKL++KMQ +R+ KYVE LD+LKIKNS ++PQ K K
Sbjct: 178 AISGDGDVKLKKKMQNLMIRFTNCWKEERLHKYVEALDLLKIKNS-------RQPQTKPK 230
Query: 175 LPPIDEQLEGMSIGNHKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSE-VVVTTKWE 233
P +++ +++ KP+ +S+S V++TT+WE
Sbjct: 231 -------------------PQYKEREMLT------FYEEASRKPTASSSSPPVIITTRWE 265
Query: 234 TFDSLPPLIPVTSASSSSSSTNNSVHPKFKWEHF 267
TFDS +SAS+++ VHPKFKWE F
Sbjct: 266 TFDS-------SSASTATLQPVRPVHPKFKWELF 292
>UniRef100_Q651M9 Hypothetical protein OSJNBa0047P18.22 [Oryza sativa]
Length = 334
Score = 221 bits (562), Expect = 2e-56
Identities = 128/272 (47%), Positives = 173/272 (63%), Gaps = 33/272 (12%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELKK L+ PTGLH +DQKI YK+KERDSKAFLD+ GVKD+SK+V++EDP AQ KR LE
Sbjct: 86 GELKKQLSAPTGLHPEDQKIVYKDKERDSKAFLDMAGVKDRSKMVLLEDPTAQAKRLLEQ 145
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
R+ K ERAAKSIS ISL+VD+LA +V+ALE I+ KGG+VV+ DV++L E LMN+L+KLD
Sbjct: 146 RRTDKAERAAKSISRISLDVDKLATKVTALEAIVGKGGRVVDADVVTLTEALMNELVKLD 205
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLEGM 185
I A+G+VK+QR+MQ KRVQKYVE+LD ++ KN+ + K + P
Sbjct: 206 AIAAEGEVKVQRRMQEKRVQKYVESLDAIRAKNAASHNKASGNGHAKPRAP--------- 256
Query: 186 SIGNHKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSEVVV---TTKWETFDSLPPLI 242
L P + Q+ Q + + P+T T+ T WE+FD L +
Sbjct: 257 -----HLPPPVSQRRQFQAPPPA--------APTTTKTAAAPAPPPTASWESFDLLSSM- 302
Query: 243 PVTSAS-------SSSSSTNNSVHPKFKWEHF 267
P TS+S +++++T S P+F WE F
Sbjct: 303 PSTSSSTVTTTMAAATTTTTTSPIPRFDWELF 334
>UniRef100_Q6ZFW2 Putative BAG domain containing protein [Oryza sativa]
Length = 316
Score = 220 bits (560), Expect = 3e-56
Identities = 126/269 (46%), Positives = 171/269 (62%), Gaps = 34/269 (12%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELKK+L+ TGLH DQK+ Y++KERDSKAFLDI GVKD+SK++++EDP AQ KR LE
Sbjct: 75 GELKKLLSEKTGLHPDDQKVVYRDKERDSKAFLDIAGVKDRSKMLLLEDPTAQAKRLLEE 134
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
R++ K ERAAKS+S ++L+VD+LA +VSALE I+SKGG+VV+ DV++L E LMN+L+KLD
Sbjct: 135 RRHCKAERAAKSVSRVALDVDKLASKVSALEAIVSKGGRVVDADVVALTEALMNELVKLD 194
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLEGM 185
I ADG+VK QR++Q KRVQKYVE LD ++ K P K + LPP
Sbjct: 195 SIAADGEVKEQRRVQEKRVQKYVEALDAIRAKTKKAAAAPPK--ARPPHLPP-------- 244
Query: 186 SIGNHKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSEV--VVTTKWETFDSLPPLIP 243
+P QQ QR +Q Q +T+ T WE+FD L +P
Sbjct: 245 -------RPPPAQQQQR---------RQFQPPAPATATAPAPQTATASWESFDLLSS-VP 287
Query: 244 VTSAS-----SSSSSTNNSVHPKFKWEHF 267
TS++ + +++T S P+F+WE F
Sbjct: 288 STSSAPVTTMAPATTTTTSPSPRFEWELF 316
>UniRef100_Q9LWV5 BAG domain containing protein-like [Oryza sativa]
Length = 321
Score = 201 bits (512), Expect = 1e-50
Identities = 114/243 (46%), Positives = 156/243 (63%), Gaps = 18/243 (7%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELKKM+ TGLH DQK+ YK+KERDSKAFLD+ GVKD+SKLVV+EDP A+ +R +E
Sbjct: 82 GELKKMVAARTGLHPDDQKVMYKDKERDSKAFLDMAGVKDRSKLVVVEDPEARARRLIEE 141
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
R+N +E+AAK+++ ++ EVD+LA +V+AL+ + KG KV E DV+ + E LMN+LLKLD
Sbjct: 142 RRNGHLEKAAKAVAAVTAEVDKLAPKVAALDASVRKGEKVAENDVVQVTELLMNELLKLD 201
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNS-----NGGHVPKKKPQQKVKLPPIDE 180
+VADGDVK QR++QVKRVQKYVETLD + KN+ +G + K+ PP +
Sbjct: 202 AVVADGDVKAQRRLQVKRVQKYVETLDAVMAKNAAIVRKSGEKLTSKQHHH----PPARQ 257
Query: 181 QLEGMSIGNHKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSEVVVTTKWETFDSLPP 240
Q + H QP+ Q ++F L PST+S S + + + PP
Sbjct: 258 QQQ--QAHQHHQQPAAGQT-------RWEMFDLLSSLPSTSSASSTTTVSSTASSGAPPP 308
Query: 241 LIP 243
L P
Sbjct: 309 LPP 311
>UniRef100_Q9SC76 Hypothetical protein l1332.2 [Oryza sativa]
Length = 268
Score = 154 bits (390), Expect = 2e-36
Identities = 79/154 (51%), Positives = 110/154 (71%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELKK+L TGL DQ++ Y+ KER + +LD+ GVK+KSKL + EDP + E+RY+E
Sbjct: 89 GELKKLLAARTGLPAADQRLTYRGKERGNADYLDVCGVKNKSKLYLAEDPTSVERRYIER 148
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
+K+ K+E A ++I I+LEVD+LA QV ++E I++G KV E + +LIE LM +KLD
Sbjct: 149 QKSAKIETANRAIGAIALEVDKLADQVRSIEKSITRGSKVAEVQITTLIELLMRLAVKLD 208
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNS 159
I A+GD Q+ +Q KRVQK VETLD+LKI N+
Sbjct: 209 SIHAEGDSSSQKNIQAKRVQKCVETLDVLKISNA 242
>UniRef100_Q7XTT7 OSJNBa0058K23.16 protein [Oryza sativa]
Length = 272
Score = 153 bits (387), Expect = 4e-36
Identities = 78/154 (50%), Positives = 110/154 (70%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELKK+L TGL DQ++ Y+ KER + +LD+ GVK++SKL + EDP + E+RY+E
Sbjct: 93 GELKKLLAARTGLPAADQRLTYRGKERGNADYLDVCGVKNRSKLYLAEDPTSVERRYIER 152
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
+K+ K+E A ++I I+LEVD+LA QV ++E I++G KV E + +LIE LM +KLD
Sbjct: 153 QKSAKIETANRAIGAIALEVDKLADQVRSIEKSITRGSKVAEVQITTLIELLMRLAVKLD 212
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNS 159
I A+GD Q+ +Q KRVQK VETLD+LKI N+
Sbjct: 213 SIHAEGDSSSQKNIQAKRVQKCVETLDVLKISNA 246
>UniRef100_Q8S234 P0446G04.18 protein [Oryza sativa]
Length = 280
Score = 122 bits (305), Expect = 1e-26
Identities = 66/157 (42%), Positives = 100/157 (63%), Gaps = 2/157 (1%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQ--EKRYL 63
GELK +LT TG+ + Q++F++ KE+ FL GVKD +KL+++E P E+R
Sbjct: 79 GELKGVLTQATGVEPERQRLFFRGKEKSDNEFLHTAGVKDGAKLLLLEKPAPANVEQRAE 138
Query: 64 EMRKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLK 123
+ + M +A +++ + EVDRL+ +V LE + G K+ + D + L E LM +LLK
Sbjct: 139 PVIMDESMMKACEAVGRVRAEVDRLSAKVCDLEKSVFAGRKIEDKDFVVLTELLMMELLK 198
Query: 124 LDGIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSN 160
LDGI A+G+ + QRK +V+RVQ VETLD LK +N+N
Sbjct: 199 LDGIEAEGEARAQRKAEVRRVQGLVETLDKLKARNAN 235
>UniRef100_Q8RX71 At3g51780/ORF3 [Arabidopsis thaliana]
Length = 269
Score = 108 bits (270), Expect = 1e-22
Identities = 62/155 (40%), Positives = 91/155 (58%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
G++KK L TGL + KI ++ ERD L GVKD SKLVV+ + +
Sbjct: 70 GDVKKALVQKTGLEASELKILFRGVERDDAEQLQAAGVKDASKLVVVVEDTNKRVEQQPP 129
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
+ME+A +++ ++ EVD+L+ +V ALE ++ G +V + E LM QLLKLD
Sbjct: 130 VVTKEMEKAIAAVNAVTGEVDKLSDRVVALEVAVNGGTQVAVREFDMAAELLMRQLLKLD 189
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSN 160
GI A+GD K+QRK +V+R+Q E +D LK + SN
Sbjct: 190 GIEAEGDAKVQRKAEVRRIQNLQEAVDKLKARCSN 224
>UniRef100_O65021 Hypothetical protein [Arabidopsis thaliana]
Length = 269
Score = 108 bits (270), Expect = 1e-22
Identities = 62/155 (40%), Positives = 91/155 (58%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
G++KK L TGL + KI ++ ERD L GVKD SKLVV+ + +
Sbjct: 70 GDVKKALVQKTGLEASELKILFRGVERDDAEQLQAAGVKDASKLVVVVEDTNKRVEQQPP 129
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
+ME+A +++ ++ EVD+L+ +V ALE ++ G +V + E LM QLLKLD
Sbjct: 130 VVTKEMEKAIAAVNAVTGEVDKLSDRVVALEVAVNGGTQVAVREFDMAAELLMRQLLKLD 189
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSN 160
GI A+GD K+QRK +V+R+Q E +D LK + SN
Sbjct: 190 GIEAEGDAKVQRKAEVRRIQNLQEAVDKLKARCSN 224
>UniRef100_Q8LIW6 P0497A05.5 protein [Oryza sativa]
Length = 172
Score = 58.9 bits (141), Expect = 1e-07
Identities = 27/56 (48%), Positives = 41/56 (73%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKR 61
G+LK ML+ TGL +DQ++ YK KERD L +VGV+DK K++++EDP +E++
Sbjct: 97 GDLKVMLSLVTGLWPRDQRLLYKGKERDDGDHLHMVGVQDKDKVLLLEDPAVKERK 152
>UniRef100_Q9LY93 Hypothetical protein F18O22_150 [Arabidopsis thaliana]
Length = 163
Score = 55.5 bits (132), Expect = 1e-06
Identities = 25/60 (41%), Positives = 43/60 (71%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELK +L+ TGL + Q++ +K KER+ +L +VGV DK K++++EDP ++K+ L++
Sbjct: 92 GELKMVLSLLTGLEPKQQRLVFKGKEREDHEYLHMVGVGDKDKVLLLEDPGFKDKKLLDL 151
>UniRef100_O60125 BAG-family molecular chaperone regulator-1A [Schizosaccharomyces
pombe]
Length = 195
Score = 52.4 bits (124), Expect = 1e-05
Identities = 46/166 (27%), Positives = 79/166 (46%), Gaps = 23/166 (13%)
Query: 5 IGELKKMLTGPTGLHHQDQKIFYKNKE-RDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYL 63
+ EL L T + + K+FY K +D KA L +G+K+ SK++ I K+
Sbjct: 27 LSELIDDLLETTEISEKKVKLFYAGKRLKDKKASLSKLGLKNHSKILC----IRPHKQQR 82
Query: 64 EMRKNIKMERAAKSISEISLEVDRLAGQVSALETIISK---------------GGKVVET 108
++ +E A K+ +E + R++G++ A++ + K K
Sbjct: 83 GSKEKDTVEPAPKAEAENPV-FSRISGEIKAIDQYVDKELSPMYDNYVNKPSNDPKQKNK 141
Query: 109 DVLSLIEKLMNQLLKLDGIVADGDVKL--QRKMQVKRVQKYVETLD 152
L + E L+ QLLKLDG+ G KL +RK V ++QK ++ +D
Sbjct: 142 QKLMISELLLQQLLKLDGVDVLGSEKLRFERKQLVSKIQKMLDHVD 187
>UniRef100_UPI000001534F UPI000001534F UniRef100 entry
Length = 201
Score = 51.2 bits (121), Expect = 3e-05
Identities = 48/171 (28%), Positives = 86/171 (50%), Gaps = 12/171 (7%)
Query: 5 IGELKKMLTGPTGLHHQDQKIFYKNKE-RDSKAFLDIVGVKDKSKLVVM-EDPIAQEKRY 62
+ +L LT TG+ QKI +K K +D + L GVK+ KL+++ + +E+
Sbjct: 24 VKDLSDALTAATGVPQTSQKIIFKGKSLKDMEERLTSYGVKEGCKLMMIGKRNSPEEEAE 83
Query: 63 LEMRKNIK--MERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQ 120
L+ K+I+ +E+ AK + ++ E+ L A + + GK ++ V + E+LM
Sbjct: 84 LKKLKDIEKSVEQTAKKLEKVGGELTGLKNGFLAKDLQVEALGK-LDHRVKTAAEQLMKT 142
Query: 121 LLKLDGIVAD-----GDVKLQRKMQVKRVQKYVETLDMLKIKNSNGGHVPK 166
L ++D +V D +L++K VK VQ ++ D KI+ H+ K
Sbjct: 143 LEQIDALVIHVPENFSDCRLKKKGLVKTVQGFLAECD--KIEACISDHLSK 191
>UniRef100_Q6K5J0 Ubiquitin-like protein [Oryza sativa]
Length = 168
Score = 51.2 bits (121), Expect = 3e-05
Identities = 22/56 (39%), Positives = 39/56 (69%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKR 61
GELK L+ TGL ++Q++ ++ KER+ L +VGV+DK K++++EDP ++ +
Sbjct: 92 GELKVRLSMVTGLEPREQRLLFRGKEREDTDHLHMVGVRDKDKVLLLEDPALKDMK 147
>UniRef100_Q7XUP1 OSJNBb0011N17.22 protein [Oryza sativa]
Length = 167
Score = 51.2 bits (121), Expect = 3e-05
Identities = 21/56 (37%), Positives = 40/56 (70%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKR 61
GELK +++ TGL ++Q++ ++ KER+ L +VGV+DK K++++EDP ++ +
Sbjct: 91 GELKTVVSIVTGLEPREQRLLFRGKEREDSDHLHMVGVRDKDKVLLLEDPALKDMK 146
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.311 0.129 0.350
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 418,651,187
Number of Sequences: 2790947
Number of extensions: 16220002
Number of successful extensions: 61188
Number of sequences better than 10.0: 267
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 239
Number of HSP's that attempted gapping in prelim test: 60984
Number of HSP's gapped (non-prelim): 390
length of query: 268
length of database: 848,049,833
effective HSP length: 125
effective length of query: 143
effective length of database: 499,181,458
effective search space: 71382948494
effective search space used: 71382948494
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 74 (33.1 bits)
Medicago: description of AC149494.2


