

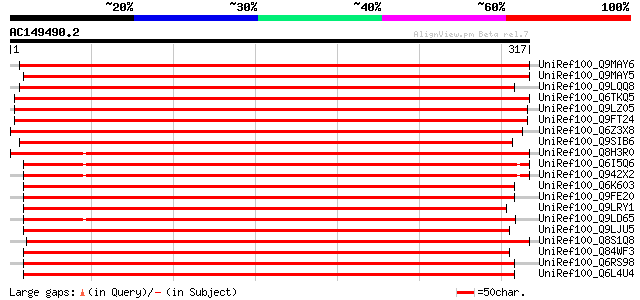
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149490.2 - phase: 1
(317 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9MAY6 Protein kinase 1 [Populus nigra] 523 e-147
UniRef100_Q9MAY5 Protein kinase 2 [Populus nigra] 515 e-145
UniRef100_Q9LQQ8 Putative serine/threonine-protein kinase RLCKVI... 496 e-139
UniRef100_Q6TKQ5 Protein kinase-like protein [Vitis aestivalis] 495 e-139
UniRef100_Q9LZ05 Protein kinase-like [Arabidopsis thaliana] 489 e-137
UniRef100_Q9FT24 Protein serine/threonine kinase BNK1 [Brassica ... 486 e-136
UniRef100_Q6Z3X8 Hypothetical protein P0627E10.22 [Oryza sativa] 484 e-135
UniRef100_Q9SIB6 Hypothetical protein At2g28590 [Arabidopsis tha... 484 e-135
UniRef100_Q8H3R0 Hypothetical protein P0625E02.115 [Oryza sativa] 470 e-131
UniRef100_Q6I5Q6 Putative serine/threonine protein kinase [Oryza... 466 e-130
UniRef100_Q942X2 Putative serine/threonine kinase PBS1 protein [... 466 e-130
UniRef100_Q6K603 Hypothetical protein OJ1789_D08.20 [Oryza sativa] 446 e-124
UniRef100_Q9FE20 Serine/threonine-protein kinase PBS1 [Arabidops... 439 e-122
UniRef100_Q9LRY1 Receptor protein kinase-like protein [Arabidops... 410 e-113
UniRef100_Q9LD65 Hypothetical protein P0462H08.22 (EST C22619(S1... 409 e-113
UniRef100_Q9LJU5 Receptor protein kinase-like protein [Arabidops... 408 e-113
UniRef100_Q8S1Q8 Protein kinase-like [Oryza sativa] 408 e-113
UniRef100_Q84WF3 Hypothetical protein At3g20530 [Arabidopsis tha... 406 e-112
UniRef100_Q6RS98 Protein kinase [Triticum turgidum] 402 e-111
UniRef100_Q6L4U4 Putative receptor-like protein kinase [Oryza sa... 402 e-111
>UniRef100_Q9MAY6 Protein kinase 1 [Populus nigra]
Length = 405
Score = 523 bits (1348), Expect = e-147
Identities = 249/311 (80%), Positives = 279/311 (89%)
Query: 7 KVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREF 66
K A+ FTF+ELAAAT +FR DCF+GEGGFGKVYKGY+ KINQ VAIKQLD NG+QG REF
Sbjct: 81 KQAQTFTFEELAAATSNFRSDCFLGEGGFGKVYKGYLDKINQAVAIKQLDRNGVQGIREF 140
Query: 67 AVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRM 126
VEV+TLSLA+HPNLVKL+GFCAEG+QRLLVYEYMPLGSLENHLHD+PP ++PLDWNTRM
Sbjct: 141 VVEVVTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPLGSLENHLHDIPPNRQPLDWNTRM 200
Query: 127 RIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVST 186
+IAAG AKGLEYLH+EMKPPVIYRDLKCSNILLG+ YHPKLSDFGLAKVGP GD THVST
Sbjct: 201 KIAAGAAKGLEYLHNEMKPPVIYRDLKCSNILLGEGYHPKLSDFGLAKVGPSGDKTHVST 260
Query: 187 RVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPL 246
RVMGTYGYCAPDYAMTGQLT KSDIYS GV LLELITGRKA D K EQNLVAWA P+
Sbjct: 261 RVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDQRKERGEQNLVAWARPM 320
Query: 247 FKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKY 306
FK++R FS MVDPLL+GQYP RGLYQALA+AAMCV+EQ +MRP ++D+V AL+++ASHKY
Sbjct: 321 FKDRRNFSCMVDPLLQGQYPIRGLYQALAIAAMCVQEQPNMRPAVSDLVMALNYLASHKY 380
Query: 307 DPQVHPIQSSR 317
DPQVH +Q SR
Sbjct: 381 DPQVHSVQDSR 391
>UniRef100_Q9MAY5 Protein kinase 2 [Populus nigra]
Length = 406
Score = 515 bits (1326), Expect = e-145
Identities = 242/309 (78%), Positives = 275/309 (88%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A+ FTF+EL AAT +FR DCF+GEGGFGKVYKGY++KINQ VAIKQLD NGLQG REF V
Sbjct: 83 AQTFTFEELVAATDNFRSDCFLGEGGFGKVYKGYLEKINQVVAIKQLDQNGLQGIREFVV 142
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EVLTLSLA++PNLVKL+GFCAEG+QRLLVYEYMPLGSLENHLHD+PP ++PLDWN RM+I
Sbjct: 143 EVLTLSLADNPNLVKLIGFCAEGDQRLLVYEYMPLGSLENHLHDIPPNRQPLDWNARMKI 202
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
AAG AKGLEYLH+EM PPVIYRDLKCSNILLG+ YHPKLSDFGLAKVGP GD THVSTRV
Sbjct: 203 AAGAAKGLEYLHNEMAPPVIYRDLKCSNILLGEGYHPKLSDFGLAKVGPSGDHTHVSTRV 262
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
MGTYGYCAPDYAMTGQLT KSD+YS GV LLELITGRKA D +K EQNLVAWA P+FK
Sbjct: 263 MGTYGYCAPDYAMTGQLTFKSDVYSFGVVLLELITGRKAIDQTKERSEQNLVAWARPMFK 322
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYDP 308
++R FS MVDP L+GQYP +GLYQALA+AAMCV+EQ +MRP ++DVV AL+++ASHKYDP
Sbjct: 323 DRRNFSGMVDPFLQGQYPIKGLYQALAIAAMCVQEQPNMRPAVSDVVLALNYLASHKYDP 382
Query: 309 QVHPIQSSR 317
Q+HP + R
Sbjct: 383 QIHPFKDPR 391
>UniRef100_Q9LQQ8 Putative serine/threonine-protein kinase RLCKVII [Arabidopsis
thaliana]
Length = 423
Score = 496 bits (1276), Expect = e-139
Identities = 239/302 (79%), Positives = 267/302 (88%)
Query: 7 KVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREF 66
K A+ FTF ELA AT +FR DCF+GEGGFGKV+KG I+K++Q VAIKQLD NG+QG REF
Sbjct: 86 KKAQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREF 145
Query: 67 AVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRM 126
VEVLTLSLA+HPNLVKL+GFCAEG+QRLLVYEYMP GSLE+HLH LP GKKPLDWNTRM
Sbjct: 146 VVEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRM 205
Query: 127 RIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVST 186
+IAAG A+GLEYLHD M PPVIYRDLKCSNILLG+DY PKLSDFGLAKVGP GD THVST
Sbjct: 206 KIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVST 265
Query: 187 RVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPL 246
RVMGTYGYCAPDYAMTGQLT KSDIYS GV LLELITGRKA D +K K+QNLV WA PL
Sbjct: 266 RVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPL 325
Query: 247 FKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKY 306
FK++R F KMVDPLL+GQYP RGLYQALA++AMCV+EQ +MRPV++DVV AL+F+AS KY
Sbjct: 326 FKDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLALNFLASSKY 385
Query: 307 DP 308
DP
Sbjct: 386 DP 387
>UniRef100_Q6TKQ5 Protein kinase-like protein [Vitis aestivalis]
Length = 376
Score = 495 bits (1275), Expect = e-139
Identities = 237/314 (75%), Positives = 269/314 (85%)
Query: 4 TDEKVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGT 63
+D A FTF ELAAATK+FR +C +GEGGFG+VYKG ++ N+ VAIKQLD NGLQG
Sbjct: 51 SDHIAAHTFTFRELAAATKNFRAECLLGEGGFGRVYKGRLESTNKIVAIKQLDRNGLQGN 110
Query: 64 REFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWN 123
REF VEVL LSL HPNLV L+G+CA+G+QRLLVYEYM LGSLE+HLHDLPP KK LDWN
Sbjct: 111 REFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEYMALGSLEDHLHDLPPDKKRLDWN 170
Query: 124 TRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTH 183
TRM+IAAG AKGLEYLHD+ PPVIYRDLKCSNILLG+ YHPKLSDFGLAK+GP+GD TH
Sbjct: 171 TRMKIAAGAAKGLEYLHDKASPPVIYRDLKCSNILLGEGYHPKLSDFGLAKLGPVGDKTH 230
Query: 184 VSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWA 243
VSTRVMGTYGYCAP+YAMTGQLT KSD+YS GV LLE+ITGRKA D SK A E NLVAWA
Sbjct: 231 VSTRVMGTYGYCAPEYAMTGQLTLKSDVYSFGVVLLEIITGRKAIDNSKAAGEHNLVAWA 290
Query: 244 YPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIAS 303
PLFK++RKFS+M DP+L GQYP RGLYQALAVAAMCV+EQ +MRP+IADVV AL ++AS
Sbjct: 291 RPLFKDRRKFSQMADPMLHGQYPLRGLYQALAVAAMCVQEQPNMRPLIADVVTALTYLAS 350
Query: 304 HKYDPQVHPIQSSR 317
KYDP+ P+QSSR
Sbjct: 351 QKYDPETQPVQSSR 364
>UniRef100_Q9LZ05 Protein kinase-like [Arabidopsis thaliana]
Length = 395
Score = 489 bits (1258), Expect = e-137
Identities = 230/313 (73%), Positives = 268/313 (85%)
Query: 4 TDEKVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGT 63
+D VA+ FTF ELA AT++FR +C +GEGGFG+VYKGY+ +Q AIKQLD NGLQG
Sbjct: 53 SDHIVAQTFTFSELATATRNFRKECLIGEGGFGRVYKGYLASTSQTAAIKQLDHNGLQGN 112
Query: 64 REFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWN 123
REF VEVL LSL HPNLV L+G+CA+G+QRLLVYEYMPLGSLE+HLHD+ PGK+PLDWN
Sbjct: 113 REFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLHDISPGKQPLDWN 172
Query: 124 TRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTH 183
TRM+IAAG AKGLEYLHD+ PPVIYRDLKCSNILL DDY PKLSDFGLAK+GP+GD +H
Sbjct: 173 TRMKIAAGAAKGLEYLHDKTMPPVIYRDLKCSNILLDDDYFPKLSDFGLAKLGPVGDKSH 232
Query: 184 VSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWA 243
VSTRVMGTYGYCAP+YAMTGQLT KSD+YS GV LLE+ITGRKA D S+ EQNLVAWA
Sbjct: 233 VSTRVMGTYGYCAPEYAMTGQLTLKSDVYSFGVVLLEIITGRKAIDSSRSTGEQNLVAWA 292
Query: 244 YPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIAS 303
PLFK++RKFS+M DP+L+GQYP RGLYQALAVAAMCV+EQ ++RP+IADVV AL ++AS
Sbjct: 293 RPLFKDRRKFSQMADPMLQGQYPPRGLYQALAVAAMCVQEQPNLRPLIADVVTALSYLAS 352
Query: 304 HKYDPQVHPIQSS 316
K+DP P+Q S
Sbjct: 353 QKFDPLAQPVQGS 365
>UniRef100_Q9FT24 Protein serine/threonine kinase BNK1 [Brassica napus]
Length = 376
Score = 486 bits (1252), Expect = e-136
Identities = 229/313 (73%), Positives = 267/313 (85%)
Query: 4 TDEKVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGT 63
+D VA+ FTF ELA AT++FR +C +GEGGFG+VYKGY+ Q AIKQLD NGLQG
Sbjct: 50 SDNIVAQTFTFSELATATRNFRKECLIGEGGFGRVYKGYLASTGQTAAIKQLDHNGLQGN 109
Query: 64 REFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWN 123
REF VEVL LSL HPNLV L+G+CA+G+QRLLVYEYMPLGSLE+HLHD+ P K+PLDWN
Sbjct: 110 REFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLHDISPSKQPLDWN 169
Query: 124 TRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTH 183
TRM+IAAG AKGLEYLHD+ PPVIYRDLKCSNILLGDDY PKLSDFGLAK+GP+GD +H
Sbjct: 170 TRMKIAAGAAKGLEYLHDKTMPPVIYRDLKCSNILLGDDYFPKLSDFGLAKLGPVGDKSH 229
Query: 184 VSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWA 243
VSTRVMGTYGYCAP+YAMTGQLT KSD+YS GV LLE+ITGRKA D S+ EQNLVAWA
Sbjct: 230 VSTRVMGTYGYCAPEYAMTGQLTLKSDVYSFGVVLLEIITGRKAIDNSRCTGEQNLVAWA 289
Query: 244 YPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIAS 303
PLFK++RKFS+M DP+++GQYP RGLYQALAVAAMCV+EQ ++RPVIADVV AL ++AS
Sbjct: 290 RPLFKDRRKFSQMADPMIQGQYPPRGLYQALAVAAMCVQEQPNLRPVIADVVTALTYLAS 349
Query: 304 HKYDPQVHPIQSS 316
++DP P+Q S
Sbjct: 350 QRFDPMSQPVQGS 362
>UniRef100_Q6Z3X8 Hypothetical protein P0627E10.22 [Oryza sativa]
Length = 390
Score = 484 bits (1246), Expect = e-135
Identities = 230/314 (73%), Positives = 269/314 (85%), Gaps = 1/314 (0%)
Query: 1 GVSTDEKVA-KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNG 59
G S + ++A + FTF ELAAAT +FRVDC +GEGGFG+VYKGY++ ++Q VAIKQLD NG
Sbjct: 63 GSSENRRIAARTFTFRELAAATSNFRVDCLLGEGGFGRVYKGYLETVDQVVAIKQLDRNG 122
Query: 60 LQGTREFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKP 119
LQG REF VEVL LS+ HPNLV L+G+CA+G+QRLLVYEYMPLGSLE+HLHD PPGK
Sbjct: 123 LQGNREFLVEVLMLSMLHHPNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLHDPPPGKSR 182
Query: 120 LDWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIG 179
LDWNTRM+IAAG AKGLEYLHD+ PPVIYRDLKCSNILLG+ YHPKLSDFGLAK+GPIG
Sbjct: 183 LDWNTRMKIAAGAAKGLEYLHDKANPPVIYRDLKCSNILLGEGYHPKLSDFGLAKLGPIG 242
Query: 180 DMTHVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNL 239
D +HVSTRVMGTYGYCAP+YAMTGQLT KSD+YS GV LLE+ITGR+A D ++ A EQNL
Sbjct: 243 DKSHVSTRVMGTYGYCAPEYAMTGQLTLKSDVYSFGVVLLEIITGRRAIDNTRAAGEQNL 302
Query: 240 VAWAYPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALD 299
VAWA PLFK++RKF +M DP L GQYP+RGLYQALAVAAMCV+EQ +MRP+I DVV AL
Sbjct: 303 VAWARPLFKDRRKFPQMADPALHGQYPSRGLYQALAVAAMCVQEQPTMRPLIGDVVTALA 362
Query: 300 FIASHKYDPQVHPI 313
++AS YDP+ H +
Sbjct: 363 YLASQTYDPEAHGV 376
>UniRef100_Q9SIB6 Hypothetical protein At2g28590 [Arabidopsis thaliana]
Length = 424
Score = 484 bits (1245), Expect = e-135
Identities = 233/301 (77%), Positives = 261/301 (86%)
Query: 7 KVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREF 66
K A+ FTF+EL+ +T +F+ DCF+GEGGFGKVYKG+I+KINQ VAIKQLD NG QG REF
Sbjct: 81 KKAQTFTFEELSVSTGNFKSDCFLGEGGFGKVYKGFIEKINQVVAIKQLDRNGAQGIREF 140
Query: 67 AVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRM 126
VEVLTLSLA+HPNLVKL+GFCAEG QRLLVYEYMPLGSL+NHLHDLP GK PL WNTRM
Sbjct: 141 VVEVLTLSLADHPNLVKLIGFCAEGVQRLLVYEYMPLGSLDNHLHDLPSGKNPLAWNTRM 200
Query: 127 RIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVST 186
+IAAG A+GLEYLHD MKPPVIYRDLKCSNIL+ + YH KLSDFGLAKVGP G THVST
Sbjct: 201 KIAAGAARGLEYLHDTMKPPVIYRDLKCSNILIDEGYHAKLSDFGLAKVGPRGSETHVST 260
Query: 187 RVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPL 246
RVMGTYGYCAPDYA+TGQLT KSD+YS GV LLELITGRKA+D ++ Q+LV WA PL
Sbjct: 261 RVMGTYGYCAPDYALTGQLTFKSDVYSFGVVLLELITGRKAYDNTRTRNHQSLVEWANPL 320
Query: 247 FKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKY 306
FK+++ F KMVDPLLEG YP RGLYQALA+AAMCV+EQ SMRPVIADVV ALD +AS KY
Sbjct: 321 FKDRKNFKKMVDPLLEGDYPVRGLYQALAIAAMCVQEQPSMRPVIADVVMALDHLASSKY 380
Query: 307 D 307
D
Sbjct: 381 D 381
>UniRef100_Q8H3R0 Hypothetical protein P0625E02.115 [Oryza sativa]
Length = 479
Score = 470 bits (1210), Expect = e-131
Identities = 225/317 (70%), Positives = 265/317 (82%), Gaps = 1/317 (0%)
Query: 1 GVSTDEKVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGL 60
G S ++ AKIFTF ELA ATK+FR DC +GEGGFG+VYKG ++ Q +A+KQLD NGL
Sbjct: 56 GSSANDGPAKIFTFRELAVATKNFRKDCLLGEGGFGRVYKGQMEN-GQVIAVKQLDRNGL 114
Query: 61 QGTREFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPL 120
QG REF VEVL LSL HPNLV+L+G+CA+G+QRLLVYEYM LGSLENHLHD PPGKKPL
Sbjct: 115 QGNREFLVEVLMLSLLHHPNLVRLIGYCADGDQRLLVYEYMLLGSLENHLHDRPPGKKPL 174
Query: 121 DWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGD 180
DWN RM+IA G AKGLEYLHD+ PPVIYRD K SNILLG+DY+PKLSDFGLAK+GP+GD
Sbjct: 175 DWNARMKIAVGAAKGLEYLHDKANPPVIYRDFKSSNILLGEDYYPKLSDFGLAKLGPVGD 234
Query: 181 MTHVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLV 240
THVSTRVMGTYGYCAP+YAMTGQLT KSD+YS GV LELITGRKA D ++PA EQNLV
Sbjct: 235 KTHVSTRVMGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDHTQPAGEQNLV 294
Query: 241 AWAYPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDF 300
AWA PLF+++RKF +M DP L+G YP RGLYQALAVA+MC++E ++ RP+IAD+V AL +
Sbjct: 295 AWARPLFRDRRKFCQMADPSLQGCYPKRGLYQALAVASMCLQENATSRPLIADIVTALSY 354
Query: 301 IASHKYDPQVHPIQSSR 317
+AS+ YDP +SSR
Sbjct: 355 LASNHYDPNAPSAKSSR 371
>UniRef100_Q6I5Q6 Putative serine/threonine protein kinase [Oryza sativa]
Length = 491
Score = 466 bits (1200), Expect = e-130
Identities = 225/309 (72%), Positives = 260/309 (83%), Gaps = 2/309 (0%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A FTF ELAAATK+FR DC +GEGGFG+VYKG+++ Q VA+KQLD NGLQG REF V
Sbjct: 65 AHTFTFRELAAATKNFRQDCLLGEGGFGRVYKGHLEN-GQAVAVKQLDRNGLQGNREFLV 123
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EVL LSL H NLV L+G+CA+G+QRLLVYE+MPLGSLE+HLHD+PP K+PLDWNTRM+I
Sbjct: 124 EVLMLSLLHHDNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDIPPDKEPLDWNTRMKI 183
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
AAG AKGLE+LHD+ PPVIYRD K SNILLG+ YHPKLSDFGLAK+GP+GD THVSTRV
Sbjct: 184 AAGAAKGLEFLHDKANPPVIYRDFKSSNILLGEGYHPKLSDFGLAKLGPVGDKTHVSTRV 243
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
MGTYGYCAP+YAMTGQLT KSD+YS GV LELITGRKA D +KP EQNLVAWA PLFK
Sbjct: 244 MGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDNTKPLGEQNLVAWARPLFK 303
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYDP 308
++RKF KM DPLL G++P RGLYQALAVAAMC++EQ++ RP I DVV AL ++AS YDP
Sbjct: 304 DRRKFPKMADPLLAGRFPMRGLYQALAVAAMCLQEQAATRPFIGDVVTALSYLASQTYDP 363
Query: 309 QVHPIQSSR 317
P+Q SR
Sbjct: 364 NT-PVQHSR 371
>UniRef100_Q942X2 Putative serine/threonine kinase PBS1 protein [Oryza sativa]
Length = 497
Score = 466 bits (1198), Expect = e-130
Identities = 225/309 (72%), Positives = 261/309 (83%), Gaps = 2/309 (0%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A+ FTF ELAAATK+FR DC +GEGGFG+VYKG ++ Q VA+KQLD NGLQG REF V
Sbjct: 74 AQTFTFRELAAATKNFRQDCLLGEGGFGRVYKGRLET-GQAVAVKQLDRNGLQGNREFLV 132
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EVL LSL H NLV L+G+CA+G+QRLLVYE+MPLGSLE+HLHDLPP K+PLDWNTRM+I
Sbjct: 133 EVLMLSLLHHTNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEPLDWNTRMKI 192
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
AAG AKGLEYLHD+ PPVIYRD K SNILLG+ +HPKLSDFGLAK+GP+GD THVSTRV
Sbjct: 193 AAGAAKGLEYLHDKASPPVIYRDFKSSNILLGEGFHPKLSDFGLAKLGPVGDKTHVSTRV 252
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
MGTYGYCAP+YAMTGQLT KSD+YS GV LELITGRKA D +KP EQNLVAWA PLFK
Sbjct: 253 MGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDNTKPQGEQNLVAWARPLFK 312
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYDP 308
++RKF KM DP+L+G++P RGLYQALAVAAMC++EQ++ RP I DVV AL ++AS YDP
Sbjct: 313 DRRKFPKMADPMLQGRFPMRGLYQALAVAAMCLQEQATTRPHIGDVVTALSYLASQTYDP 372
Query: 309 QVHPIQSSR 317
P+Q SR
Sbjct: 373 NA-PVQHSR 380
>UniRef100_Q6K603 Hypothetical protein OJ1789_D08.20 [Oryza sativa]
Length = 526
Score = 446 bits (1146), Expect = e-124
Identities = 210/300 (70%), Positives = 251/300 (83%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A+ FTF ELA AT++FR +CF+GEGGFG+VYKG ++ Q VAIKQL+ +GLQG REF V
Sbjct: 107 AQTFTFRELATATRNFRPECFLGEGGFGRVYKGRLESTGQVVAIKQLNRDGLQGNREFLV 166
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EVL LSL H NLV L+G+CA+G+QRLLVYEYM GSLE+HLHDLPP K+ LDWNTRM+I
Sbjct: 167 EVLMLSLLHHQNLVNLIGYCADGDQRLLVYEYMHFGSLEDHLHDLPPDKEALDWNTRMKI 226
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
AAG AKGLEYLHD+ PPVIYRD K SNILL + +HPKLSDFGLAK+GP+GD +HVSTRV
Sbjct: 227 AAGAAKGLEYLHDKANPPVIYRDFKSSNILLDESFHPKLSDFGLAKLGPVGDKSHVSTRV 286
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
MGTYGYCAP+YAMTGQLT KSD+YS GV LLELITGR+A D ++P EQNLV+WA PLF
Sbjct: 287 MGTYGYCAPEYAMTGQLTVKSDVYSFGVVLLELITGRRAIDSTRPHGEQNLVSWARPLFN 346
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYDP 308
++RK KM DP LEG+YP RGLYQALAVA+MC++ +++ RP+IADVV AL ++AS YDP
Sbjct: 347 DRRKLPKMADPRLEGRYPMRGLYQALAVASMCIQSEAASRPLIADVVTALSYLASQSYDP 406
>UniRef100_Q9FE20 Serine/threonine-protein kinase PBS1 [Arabidopsis thaliana]
Length = 456
Score = 439 bits (1129), Expect = e-122
Identities = 209/300 (69%), Positives = 245/300 (81%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A F F ELAAAT +F D F+GEGGFG+VYKG + Q VA+KQLD NGLQG REF V
Sbjct: 71 AHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLV 130
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EVL LSL HPNLV L+G+CA+G+QRLLVYE+MPLGSLE+HLHDLPP K+ LDWN RM+I
Sbjct: 131 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKI 190
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
AAG AKGLE+LHD+ PPVIYRD K SNILL + +HPKLSDFGLAK+GP GD +HVSTRV
Sbjct: 191 AAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRV 250
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
MGTYGYCAP+YAMTGQLT KSD+YS GV LELITGRKA D P EQNLVAWA PLF
Sbjct: 251 MGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFN 310
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYDP 308
++RKF K+ DP L+G++P R LYQALAVA+MC++EQ++ RP+IADVV AL ++A+ YDP
Sbjct: 311 DRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTALSYLANQAYDP 370
>UniRef100_Q9LRY1 Receptor protein kinase-like protein [Arabidopsis thaliana]
Length = 381
Score = 410 bits (1053), Expect = e-113
Identities = 192/295 (65%), Positives = 239/295 (80%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A+IFTF ELA ATK+FR +C +GEGGFG+VYKG ++ Q VA+KQLD NGLQG REF V
Sbjct: 50 ARIFTFRELATATKNFRQECLIGEGGFGRVYKGKLENPAQVVAVKQLDRNGLQGQREFLV 109
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EVL LSL H NLV L+G+CA+G+QRLLVYEYMPLGSLE+HL DL PG+KPLDWNTR++I
Sbjct: 110 EVLMLSLLHHRNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLLDLEPGQKPLDWNTRIKI 169
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
A G AKG+EYLHDE PPVIYRDLK SNILL +Y KLSDFGLAK+GP+GD HVS+RV
Sbjct: 170 ALGAAKGIEYLHDEADPPVIYRDLKSSNILLDPEYVAKLSDFGLAKLGPVGDTLHVSSRV 229
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
MGTYGYCAP+Y TG LT+KSD+YS GV LLELI+GR+ D +P+ EQNLV WA P+F+
Sbjct: 230 MGTYGYCAPEYQRTGYLTNKSDVYSFGVVLLELISGRRVIDTMRPSHEQNLVTWALPIFR 289
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIAS 303
+ ++ ++ DPLL G YP + L QA+AVAAMC+ E+ ++RP+++DV+ AL F+ +
Sbjct: 290 DPTRYWQLADPLLRGDYPEKSLNQAIAVAAMCLHEEPTVRPLMSDVITALSFLGA 344
>UniRef100_Q9LD65 Hypothetical protein P0462H08.22 (EST C22619(S11214) corresponds to
a region of the predicted gene) [Oryza sativa]
Length = 407
Score = 409 bits (1050), Expect = e-113
Identities = 197/301 (65%), Positives = 237/301 (78%), Gaps = 1/301 (0%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A IFT ELA AT +F +C +G GGFG VYK ++ Q VA+KQLD NGLQG REF V
Sbjct: 13 ATIFTLRELADATNNFSTECLLGRGGFGSVYKAFLND-RQVVAVKQLDLNGLQGNREFLV 71
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EVL LSL HPNLVKL G+C +G+QRLL+YEYMPLGSLE+ LHDL PG++PLDW TRM+I
Sbjct: 72 EVLMLSLLHHPNLVKLFGYCVDGDQRLLIYEYMPLGSLEDRLHDLRPGQEPLDWTTRMKI 131
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
AA A GLEYLHDE P VIYRD+K SNILLG+ Y+ KLSDFGLAK+GP+GD THV+TRV
Sbjct: 132 AADAAAGLEYLHDEAIPAVIYRDIKPSNILLGEGYNAKLSDFGLAKLGPVGDKTHVTTRV 191
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
MGT+GYCAP+Y TG+LT KSDIYS GV LELITGR+A D ++P EQ+LVAWA PLFK
Sbjct: 192 MGTHGYCAPEYLSTGKLTIKSDIYSFGVVFLELITGRRALDSNRPPDEQDLVAWARPLFK 251
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYDP 308
+QRKF KM DP L G +P RGL+QALA+AAMC++E++ RP I +V AL ++AS ++
Sbjct: 252 DQRKFPKMADPSLHGHFPKRGLFQALAIAAMCLQEKAKNRPSIREVAVALSYLASQTHES 311
Query: 309 Q 309
Q
Sbjct: 312 Q 312
>UniRef100_Q9LJU5 Receptor protein kinase-like protein [Arabidopsis thaliana]
Length = 377
Score = 408 bits (1049), Expect = e-113
Identities = 197/298 (66%), Positives = 242/298 (81%), Gaps = 1/298 (0%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A IFTF EL ATK+F D +GEGGFG+VYKG I+ Q VA+KQLD NG QG REF V
Sbjct: 58 AHIFTFRELCVATKNFNPDNQLGEGGFGRVYKGQIETPEQVVAVKQLDRNGYQGNREFLV 117
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKK-PLDWNTRMR 127
EV+ LSL H NLV L+G+CA+G+QR+LVYEYM GSLE+HL +L KK PLDW+TRM+
Sbjct: 118 EVMMLSLLHHQNLVNLVGYCADGDQRILVYEYMQNGSLEDHLLELARNKKKPLDWDTRMK 177
Query: 128 IAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTR 187
+AAG A+GLEYLH+ PPVIYRD K SNILL ++++PKLSDFGLAKVGP G THVSTR
Sbjct: 178 VAAGAARGLEYLHETADPPVIYRDFKASNILLDEEFNPKLSDFGLAKVGPTGGETHVSTR 237
Query: 188 VMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLF 247
VMGTYGYCAP+YA+TGQLT KSD+YS GV LE+ITGR+ D +KP +EQNLV WA PLF
Sbjct: 238 VMGTYGYCAPEYALTGQLTVKSDVYSFGVVFLEMITGRRVIDTTKPTEEQNLVTWASPLF 297
Query: 248 KEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHK 305
K++RKF+ M DPLLEG+YP +GLYQALAVAAMC++E+++ RP+++DVV AL+++A K
Sbjct: 298 KDRRKFTLMADPLLEGKYPIKGLYQALAVAAMCLQEEAATRPMMSDVVTALEYLAVTK 355
>UniRef100_Q8S1Q8 Protein kinase-like [Oryza sativa]
Length = 467
Score = 408 bits (1049), Expect = e-113
Identities = 194/307 (63%), Positives = 246/307 (79%)
Query: 11 IFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVEV 70
+ T+ +L AT SF + +GEGGFG+VY+G++++IN+ VA+KQLD +G QG REF VEV
Sbjct: 132 VLTYRQLCNATDSFSPNNLLGEGGFGRVYRGHLEEINEIVAVKQLDKDGFQGNREFLVEV 191
Query: 71 LTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIAA 130
L LSL HPNLVKLLG+C + +QR+LVYE M GSLE+HL DLPP KPL W TRM+IA
Sbjct: 192 LMLSLLHHPNLVKLLGYCTDMDQRILVYECMRNGSLEDHLLDLPPKAKPLPWQTRMKIAV 251
Query: 131 GVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVMG 190
G AKG+EYLH+ PPVIYRDLK SNILL +D++ KLSDFGLAK+GP+GD +HVSTRVMG
Sbjct: 252 GAAKGIEYLHEVANPPVIYRDLKTSNILLDEDFNSKLSDFGLAKLGPVGDKSHVSTRVMG 311
Query: 191 TYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFKEQ 250
TYGYCAP+YAMTG+LT SDIYS GV LLE+ITGR+A D S+P EQ LV WA PL K++
Sbjct: 312 TYGYCAPEYAMTGKLTKTSDIYSFGVVLLEIITGRRAIDTSRPTHEQVLVQWAAPLVKDK 371
Query: 251 RKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYDPQV 310
++F ++ DPLLE ++P +GLYQALA+A+MC++E +S RP+I+DVVAAL F+A KY PQ
Sbjct: 372 KRFVRLADPLLEEKFPLKGLYQALAIASMCLQEDASNRPMISDVVAALSFLAEQKYHPQD 431
Query: 311 HPIQSSR 317
P Q++R
Sbjct: 432 GPDQAAR 438
>UniRef100_Q84WF3 Hypothetical protein At3g20530 [Arabidopsis thaliana]
Length = 386
Score = 406 bits (1043), Expect = e-112
Identities = 196/298 (65%), Positives = 241/298 (80%), Gaps = 1/298 (0%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A IFTF EL ATK+F D +GEGGFG+VYKG I+ Q VA+KQLD NG QG REF V
Sbjct: 67 AHIFTFRELCVATKNFNPDNQLGEGGFGRVYKGQIETPEQVVAVKQLDRNGYQGNREFLV 126
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKK-PLDWNTRMR 127
EV+ LSL H NLV L+G+CA+G+QR+LVYEYM GSLE+HL +L KK PLDW+TRM+
Sbjct: 127 EVMMLSLLHHQNLVNLVGYCADGDQRILVYEYMQNGSLEDHLLELARNKKKPLDWDTRMK 186
Query: 128 IAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTR 187
+AAG A+GLEYLH+ PPVIYRD K SNILL ++++PKLSDFGLAKVGP G HVSTR
Sbjct: 187 VAAGAARGLEYLHETADPPVIYRDFKASNILLDEEFNPKLSDFGLAKVGPTGGEIHVSTR 246
Query: 188 VMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLF 247
VMGTYGYCAP+YA+TGQLT KSD+YS GV LE+ITGR+ D +KP +EQNLV WA PLF
Sbjct: 247 VMGTYGYCAPEYALTGQLTVKSDVYSFGVVFLEMITGRRVIDTTKPTEEQNLVTWASPLF 306
Query: 248 KEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHK 305
K++RKF+ M DPLLEG+YP +GLYQALAVAAMC++E+++ RP+++DVV AL+++A K
Sbjct: 307 KDRRKFTLMADPLLEGKYPIKGLYQALAVAAMCLQEEAATRPMMSDVVTALEYLAVTK 364
>UniRef100_Q6RS98 Protein kinase [Triticum turgidum]
Length = 568
Score = 402 bits (1033), Expect = e-111
Identities = 186/300 (62%), Positives = 241/300 (80%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
+++F + +L+ AT SF + +GEGGFG+VYKGYI + + +A+KQLD +GLQG REF V
Sbjct: 231 SRVFAYSQLSDATNSFSQENLLGEGGFGRVYKGYISETMEVIAVKQLDKDGLQGNREFLV 290
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EVL LSL HP+LV LLG+C E +Q++LVYEYMPLGSL++HL DL P +PL WNTRM+I
Sbjct: 291 EVLMLSLLHHPHLVTLLGYCTECDQKILVYEYMPLGSLQDHLLDLTPKSQPLSWNTRMKI 350
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
A A+GLEYLH+ PPV+YRDLK SNILL ++ KL+DFGLAK+GP+GD THV+TRV
Sbjct: 351 AVDAARGLEYLHEVANPPVVYRDLKASNILLDGNFSAKLADFGLAKLGPVGDKTHVTTRV 410
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
MGTYGYCAP+YAM+G+LT SDIY GV LLELITGR+A D +KP +EQ LV WA PLFK
Sbjct: 411 MGTYGYCAPEYAMSGKLTKMSDIYCFGVVLLELITGRRAIDTTKPTREQILVHWAAPLFK 470
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYDP 308
+++KF+KM DP L+ +YP +GLYQALA+++MC++E++S RP+I+DVV AL F+A YDP
Sbjct: 471 DKKKFTKMADPKLDSKYPLKGLYQALAISSMCLQEEASSRPLISDVVTALTFLADPNYDP 530
>UniRef100_Q6L4U4 Putative receptor-like protein kinase [Oryza sativa]
Length = 484
Score = 402 bits (1033), Expect = e-111
Identities = 189/300 (63%), Positives = 241/300 (80%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
+++FTF +LA AT SF + +GEGGFG+VYKG+I + +A+KQLD +GLQG REF V
Sbjct: 147 SRVFTFRQLADATGSFSPENLLGEGGFGRVYKGFIPDTKEVIAVKQLDKDGLQGNREFLV 206
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EVL LSL HPNLV LLG+ E +QR+LVYEYMPLGSL++HL DL P PL W+TRM+I
Sbjct: 207 EVLMLSLLHHPNLVTLLGYSTECDQRILVYEYMPLGSLQDHLLDLTPNSSPLSWHTRMKI 266
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
A G A+G+EYLH+ PPVIYRDLK SNILL ++ KLSDFGLAK+GP+GD +HV+TRV
Sbjct: 267 AVGAARGMEYLHEIANPPVIYRDLKASNILLDGGFNAKLSDFGLAKLGPVGDKSHVTTRV 326
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
MGTYGYCAP+YAMTG+LT SDIYS GV LLE+ITGR+A D +KP +EQ LV WA PLF+
Sbjct: 327 MGTYGYCAPEYAMTGKLTKMSDIYSFGVVLLEIITGRRAIDTTKPTREQILVHWAAPLFR 386
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYDP 308
+++KF KM DPLL+ ++P +GLYQALA+++MC++E++S RP+I+DVV AL F+A YDP
Sbjct: 387 DKKKFVKMADPLLDMKFPLKGLYQALAISSMCLQEEASSRPLISDVVTALTFLADPNYDP 446
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 531,240,599
Number of Sequences: 2790947
Number of extensions: 21796856
Number of successful extensions: 90568
Number of sequences better than 10.0: 17635
Number of HSP's better than 10.0 without gapping: 6917
Number of HSP's successfully gapped in prelim test: 10718
Number of HSP's that attempted gapping in prelim test: 53777
Number of HSP's gapped (non-prelim): 19807
length of query: 317
length of database: 848,049,833
effective HSP length: 127
effective length of query: 190
effective length of database: 493,599,564
effective search space: 93783917160
effective search space used: 93783917160
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC149490.2


