

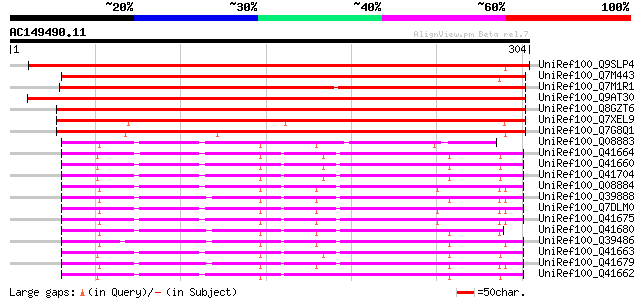
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149490.11 + phase: 0
(304 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SLP4 Chitinase 1 precursor [Tulipa bakeri] 380 e-104
UniRef100_Q7M443 Chitinase 2 [Tulipa bakeri] 374 e-102
UniRef100_Q7M1R1 Chitinase (EC 3.2.1.14) a [Gladiolus gandavensis] 338 8e-92
UniRef100_Q9AT30 Class III chitinase RCB4 [Oryza sativa] 335 1e-90
UniRef100_Q8GZT6 Chitinase [Oryza sativa] 332 7e-90
UniRef100_Q7XEL9 Putative class III chitinase [Oryza sativa] 286 4e-76
UniRef100_Q7G8Q1 Class III chitinase [Oryza sativa] 282 7e-75
UniRef100_Q08883 Narbonin [Vicia pannonica] 139 1e-31
UniRef100_Q41664 Nodulin homologous to narbonin [Vicia faba] 135 2e-30
UniRef100_Q41660 Putative narbonin-like 2S protein [Vicia faba] 132 9e-30
UniRef100_Q41704 Putative narbonin-like 2S protein [Vicia sativa] 131 2e-29
UniRef100_Q08884 Narbonin [Vicia narbonensis] 128 2e-28
UniRef100_Q39888 Putative narbonin [Glycine max] 128 2e-28
UniRef100_Q7DLM0 Narbonin [Vicia narbonensis] 128 2e-28
UniRef100_Q41675 Narbonin [Vicia narbonensis] 128 2e-28
UniRef100_Q41680 Narbonin [Vicia pannonica] 125 1e-27
UniRef100_Q39486 Putative narbonin [Canavalia ensiformis] 124 4e-27
UniRef100_Q41663 Nodulin homologous to narbonin [Vicia faba] 123 7e-27
UniRef100_Q41679 Narbonin [Vicia pannonica] 122 2e-26
UniRef100_Q41662 Putative narbonin-like 2S protein [Vicia faba] 121 3e-26
>UniRef100_Q9SLP4 Chitinase 1 precursor [Tulipa bakeri]
Length = 314
Score = 380 bits (977), Expect = e-104
Identities = 188/295 (63%), Positives = 228/295 (76%), Gaps = 2/295 (0%)
Query: 12 LLILTTSLFASTILAANSNLFREYIGAQFKGVKFSDVPINPNVNFHFILSFAIDYDTSSP 71
LL+L +L + + ++ +FREYIG+QF VKFSDVPINP+V+FHFIL+FAIDY + S
Sbjct: 9 LLLLFFALLSPLLPLTSALVFREYIGSQFNDVKFSDVPINPDVDFHFILAFAIDYTSGSS 68
Query: 72 PSPTNGNFNIFWDTQNLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVS 131
P+PTNGNF FWDT NLSPS+V+ +K + NVKV+LSLGGD+V G V F PSS+ SWV
Sbjct: 69 PTPTNGNFKPFWDTNNLSPSQVAAVKRTHSNVKVSLSLGGDSVGGKNVFFSPSSVSSWVE 128
Query: 132 NAVSSLTSIIKTYNLDGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISFASIAPFDDA 191
NAVSSLT IIK Y+LDGID+DYEHF GDPNTFAECIG+L+T LK N V+SF SIAPFDDA
Sbjct: 129 NAVSSLTRIIKQYHLDGIDIDYEHFKGDPNTFAECIGQLVTRLKKNEVVSFVSIAPFDDA 188
Query: 192 EVQNHYLALWKSYSNIIDYVNFQFYAYDKSTTVAQFIDYYNKQSSNYNSEKVLVSFVSDG 251
+VQ+HY ALW+ Y + IDYVNFQFYAY T+V QF+ Y+ +QSSNY+ KVLVSF +D
Sbjct: 189 QVQSHYQALWEKYGHQIDYVNFQFYAYSARTSVEQFLKYFEEQSSNYHGGKVLVSFSTDS 248
Query: 252 STGLGPGNGFLDGCRMLKSQQKLNGIFVYSADDSKADG--FRYEKEAQEVLASPN 304
S GL P NGF C +LK Q KL+GIFV+SADDS FRYE +AQ +LAS N
Sbjct: 249 SGGLKPDNGFFRACSILKKQGKLHGIFVWSADDSLMSNNVFRYEMQAQSMLASVN 303
>UniRef100_Q7M443 Chitinase 2 [Tulipa bakeri]
Length = 275
Score = 374 bits (960), Expect = e-102
Identities = 184/274 (67%), Positives = 215/274 (78%), Gaps = 2/274 (0%)
Query: 31 LFREYIGAQFKGVKFSDVPINPNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQNLSP 90
LFREYIGAQF VKFSDVPINPNV+FHFIL+FAIDY + S P+PTNGNFN FWDT NLSP
Sbjct: 2 LFREYIGAQFNDVKFSDVPINPNVDFHFILAFAIDYTSGSSPTPTNGNFNPFWDTNNLSP 61
Query: 91 SEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYNLDGID 150
S+V+ IK NVKV++SLGG++V G+ V F PSS+ SWV NAVSSLT IIK Y+LDGID
Sbjct: 62 SQVAAIKRTYNNVKVSVSLGGNSVGGERVFFNPSSVSSWVDNAVSSLTKIIKQYHLDGID 121
Query: 151 VDYEHFIGDPNTFAECIGRLITTLKNNNVISFASIAPFDDAEVQNHYLALWKSYSNIIDY 210
+DYEHF GDPNTFAECIG+L+T LK N V+SF SIAPFDDA+VQ+HY ALW+ Y + IDY
Sbjct: 122 IDYEHFKGDPNTFAECIGQLVTRLKKNGVVSFVSIAPFDDAQVQSHYQALWEKYGHQIDY 181
Query: 211 VNFQFYAYDKSTTVAQFIDYYNKQSSNYNSEKVLVSFVSDGSTGLGPGNGFLDGCRMLKS 270
VNFQFY Y +V QF+ Y+ Q SNY KVLVSF +D S GL P NGF D C +LK
Sbjct: 182 VNFQFYVYSSRMSVEQFLKYFEMQRSNYPGGKVLVSFSTDNSGGLKPRNGFFDACSILKK 241
Query: 271 QQKLNGIFVYSADDS--KADGFRYEKEAQEVLAS 302
Q KL+GIFV+SADDS D F+YE +AQ +LAS
Sbjct: 242 QGKLHGIFVWSADDSLMSNDVFKYEMQAQSLLAS 275
>UniRef100_Q7M1R1 Chitinase (EC 3.2.1.14) a [Gladiolus gandavensis]
Length = 274
Score = 338 bits (868), Expect = 8e-92
Identities = 171/276 (61%), Positives = 211/276 (75%), Gaps = 5/276 (1%)
Query: 30 NLFREYIGAQFKGVKFSDVPINPNV-NFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQNL 88
NLF EYIG+QF G+KF+DVPINP + +F F+LSFAIDY S+P +PTNG FN+FWD+ NL
Sbjct: 1 NLFVEYIGSQFTGIKFTDVPINPRITDFQFVLSFAIDYTVSTPHTPTNGKFNVFWDSSNL 60
Query: 89 SPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYNLDG 148
PS+V IK+ + NV+VA+SLGG TV G V F PSSIDSWV+NAVSSLT II+ YNLDG
Sbjct: 61 GPSQVDAIKSSHNNVRVAVSLGGATVGGKSVQFQPSSIDSWVNNAVSSLTQIIQRYNLDG 120
Query: 149 IDVDYEHFIG-DPNTFAECIGRLITTLKNNNVISFASIAPFDDAEVQNHYLALWKSYSNI 207
ID+DYE+F DP+TFAECIGRLITTLK N VI+FASIAPF D V+ +YLALW Y N+
Sbjct: 121 IDIDYENFQNTDPDTFAECIGRLITTLKRNRVINFASIAPFPD--VEEYYLALWNRYKNV 178
Query: 208 IDYVNFQFYAYDKSTTVAQFIDYYNKQSSNYNSEKVLVSFVSD-GSTGLGPGNGFLDGCR 266
I+++NFQFYAYD STTV QF+ YY+ S Y VL+SF ++ + GL GF D
Sbjct: 179 INHINFQFYAYDSSTTVRQFLKYYDTAVSKYRGGNVLISFSTERDAGGLTVDRGFFDAAS 238
Query: 267 MLKSQQKLNGIFVYSADDSKADGFRYEKEAQEVLAS 302
+LK Q KL+GI V+SAD SK++GFRY++EAQ L S
Sbjct: 239 ILKKQGKLHGIAVWSADTSKSNGFRYDEEAQSFLVS 274
>UniRef100_Q9AT30 Class III chitinase RCB4 [Oryza sativa]
Length = 307
Score = 335 bits (858), Expect = 1e-90
Identities = 166/293 (56%), Positives = 209/293 (70%), Gaps = 1/293 (0%)
Query: 11 ILLILTTSLFASTILAANSNLFREYIGAQFKGVKFSDVPINPNVNFHFILSFAIDYDT-S 69
+LL + A A NSNLFR+YIGA F GVKF+DVPINP V F FIL+F IDY T +
Sbjct: 10 VLLPALLAFQAPMATAVNSNLFRDYIGAIFNGVKFTDVPINPKVRFDFILAFIIDYTTET 69
Query: 70 SPPSPTNGNFNIFWDTQNLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSW 129
+PP+PTNG FNIFW L+PS V+ IK NPNV+VA+S+GG TV PV F +S+DSW
Sbjct: 70 NPPTPTNGKFNIFWQNTVLTPSAVASIKQSNPNVRVAVSMGGATVNDRPVFFNITSVDSW 129
Query: 130 VSNAVSSLTSIIKTYNLDGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISFASIAPFD 189
V+NAV SLT II+ NLDGID+DYE F DP+TF EC+GRLIT LK VI FASIAPF
Sbjct: 130 VNNAVESLTGIIQDNNLDGIDIDYEQFQVDPDTFTECVGRLITVLKAKGVIKFASIAPFG 189
Query: 190 DAEVQNHYLALWKSYSNIIDYVNFQFYAYDKSTTVAQFIDYYNKQSSNYNSEKVLVSFVS 249
+AEVQ HY+ALW Y +IDY+NFQFYAY STT AQ++D++N+Q NY +L SF +
Sbjct: 190 NAEVQRHYMALWAKYGAVIDYINFQFYAYGASTTEAQYVDFFNQQIVNYPGGNILASFTT 249
Query: 250 DGSTGLGPGNGFLDGCRMLKSQQKLNGIFVYSADDSKADGFRYEKEAQEVLAS 302
+T P L CR L+ + KL GIF+++AD S++ GF+YE E+Q +LA+
Sbjct: 250 AATTTSVPVETALSACRTLQKEGKLYGIFIWAADHSRSQGFKYETESQALLAN 302
>UniRef100_Q8GZT6 Chitinase [Oryza sativa]
Length = 282
Score = 332 bits (851), Expect = 7e-90
Identities = 162/276 (58%), Positives = 203/276 (72%), Gaps = 1/276 (0%)
Query: 28 NSNLFREYIGAQFKGVKFSDVPINPNVNFHFILSFAIDYDT-SSPPSPTNGNFNIFWDTQ 86
NSNLFR+YIGA F GVKF+DVPINP V F FIL+F IDY T ++PP+PTNG FNIFW
Sbjct: 2 NSNLFRDYIGAIFNGVKFTDVPINPKVRFDFILAFIIDYTTETNPPTPTNGKFNIFWQNT 61
Query: 87 NLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYNL 146
L+PS V+ IK NPNV+VA+S+GG TV PV F +S+DSWV+NAV SLT II+ NL
Sbjct: 62 VLTPSAVASIKQSNPNVRVAVSMGGATVNDRPVFFNITSVDSWVNNAVESLTGIIQDNNL 121
Query: 147 DGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISFASIAPFDDAEVQNHYLALWKSYSN 206
DGID+DYE F DP+TF EC+GRLIT LK VI FASIAPF +AEVQ HY+ALW Y
Sbjct: 122 DGIDIDYEQFQVDPDTFTECVGRLITVLKAKGVIKFASIAPFGNAEVQRHYMALWAKYGA 181
Query: 207 IIDYVNFQFYAYDKSTTVAQFIDYYNKQSSNYNSEKVLVSFVSDGSTGLGPGNGFLDGCR 266
+IDY+NFQFYAY STT AQ++D++N+Q NY +L SF + +T P L CR
Sbjct: 182 VIDYINFQFYAYGASTTEAQYVDFFNQQIVNYPGGNILASFTTAATTTSVPVETALSACR 241
Query: 267 MLKSQQKLNGIFVYSADDSKADGFRYEKEAQEVLAS 302
L+ + KL GIF+++AD S++ GF+YE E+Q +LA+
Sbjct: 242 TLQKEGKLYGIFIWAADHSRSQGFKYETESQALLAN 277
>UniRef100_Q7XEL9 Putative class III chitinase [Oryza sativa]
Length = 288
Score = 286 bits (733), Expect = 4e-76
Identities = 149/285 (52%), Positives = 199/285 (69%), Gaps = 10/285 (3%)
Query: 28 NSNLFREYIGAQFKGVKFSDVPINPNVNFHFILSFAIDYDT---SSPPSPTNGNFNIFWD 84
N LFREYIGAQF GV+FSDVPINPN++F+FILSFAIDY + + P+PTNG F+ +WD
Sbjct: 3 NGYLFREYIGAQFTGVRFSDVPINPNLSFNFILSFAIDYTSPAGGATPAPTNGVFSPYWD 62
Query: 85 TQNLSPSEVSFIKNQNPNVKVALSLGGDTVQGDP-VNFIPSSIDSWVSNAVSSLTSIIKT 143
T NLSP++V+ +K +PNV V + LGGD+VQ V F P+S+DSWV+NAV+S++ II
Sbjct: 63 TANLSPADVAAVKAAHPNVSVMVGLGGDSVQDTAKVFFSPTSVDSWVANAVASVSGIIDA 122
Query: 144 YNLDGIDVDYEHFIGDP----NTFAECIGRLITTLKNNNVISFASIAPFDDAEVQNHYLA 199
Y LDG+DVDYEHF D +TF ECIGRL+T LK + SIAPF+DA VQ +Y
Sbjct: 123 YGLDGVDVDYEHFNDDGGAGVDTFVECIGRLLTELKARHPNITTSIAPFEDAVVQRYYQP 182
Query: 200 LWKSYSNIIDYVNFQFYAYDKSTTVAQFIDYYNKQSSNYNSEKVLVSFVSDGSTGLGPGN 259
LW+ Y+ +ID VNFQFY Y +T V ++ +Y++Q++NY KVL SF + GL
Sbjct: 183 LWRRYAGVIDLVNFQFYGYGDNTDVPTYVMFYDEQAANYPGGKVLASFKTGDVAGLLWPE 242
Query: 260 GFLDGCRMLKSQQKLNGIFVYSADDSKAD--GFRYEKEAQEVLAS 302
+ G + L+ Q KL G+F++SAD SK GF YE +AQE++A+
Sbjct: 243 QGIAGAKELQRQGKLPGLFIWSADSSKVSSYGFEYEIKAQEIIAN 287
>UniRef100_Q7G8Q1 Class III chitinase [Oryza sativa]
Length = 286
Score = 282 bits (722), Expect = 7e-75
Identities = 145/283 (51%), Positives = 191/283 (67%), Gaps = 8/283 (2%)
Query: 28 NSNLFREYIGAQFKGVKFSDVPINPNVNFHFILSFAIDY---DTSSPPSPTNGNFNIFWD 84
N LFREYIGAQF GV+FSDVP+NP ++FHFIL+FAIDY SS P+P NG F +WD
Sbjct: 3 NGYLFREYIGAQFTGVRFSDVPVNPGLSFHFILAFAIDYFMATQSSKPAPANGVFAPYWD 62
Query: 85 TQNLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVN--FIP-SSIDSWVSNAVSSLTSII 141
T NLSP+ V+ K +PN+ V L+LGGDTVQ VN F P SS+D+WV NA S++ +I
Sbjct: 63 TANLSPAAVAAAKAAHPNLSVILALGGDTVQNTGVNATFAPTSSVDAWVRNAADSVSGLI 122
Query: 142 KTYNLDGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISFASIAPFDDAEVQNHYLALW 201
Y LDG+DVDYEHF +TF ECIGRL+T LK + SIAPF+ VQ +Y LW
Sbjct: 123 DAYGLDGVDVDYEHFAAGVDTFVECIGRLLTELKARHPNIATSIAPFEHPVVQRYYQPLW 182
Query: 202 KSYSNIIDYVNFQFYAYDKSTTVAQFIDYYNKQSSNYNSEKVLVSFVSDGSTGLGPGNGF 261
+ Y+ +IDYVNFQFY Y +T VA ++ +Y++Q++NY K+L SF + TGL
Sbjct: 183 RRYAGVIDYVNFQFYGYGANTDVATYVMFYDEQAANYPGSKLLASFKTGNVTGLLSPEQG 242
Query: 262 LDGCRMLKSQQKLNGIFVYSADDSKADG--FRYEKEAQEVLAS 302
+ G + L+ Q KL G+F++SAD S F YE +AQE++A+
Sbjct: 243 IAGAKELQRQGKLPGLFIWSADSSMVSSYKFEYETKAQEIVAN 285
>UniRef100_Q08883 Narbonin [Vicia pannonica]
Length = 292
Score = 139 bits (349), Expect = 1e-31
Identities = 96/274 (35%), Positives = 137/274 (49%), Gaps = 29/274 (10%)
Query: 31 LFREYIGAQFKGVKFSDVPIN----PNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQ 86
+FREYIG + D P N+ FHFIL FA + S S GNF WD +
Sbjct: 5 IFREYIGVKPNSETLHDFPHEIIDTENLEFHFILGFATESYYESGKS--TGNFEESWDVE 62
Query: 87 NLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN- 145
P V +K ++P VKV +S+ G D F P + WV AV SL IIK Y
Sbjct: 63 LFGPENVKNLKTKHPEVKVVISIRG---HDDKTPFDPDEENIWVWKAVKSLKQIIKKYRN 119
Query: 146 -----LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNN--VISFASIAPFDDAEVQNHYL 198
+DGID++YEH D F CIG++I LK ++ I SIAP ++ + N
Sbjct: 120 ESGNMIDGIDINYEHINSDDELFVNCIGQVIRELKKDDDLNIDVVSIAPSENNQSSNQ-- 177
Query: 199 ALWKSYSNIIDYVNFQFYAYDKS-TTVAQFIDYYNKQSSNYNSEKVLVSF------VSDG 251
L+ + ++ I++V++QF K TTV F+D YN +Y++ KVL F + D
Sbjct: 178 KLYNANTDYINWVDYQFSNQVKPVTTVDAFVDIYNSLVKDYDAGKVLPGFNTEPLDIKDT 237
Query: 252 STGLGPGNGFLDGCRMLKSQQKLNGIFVYSADDS 285
T + F+ GC L L G+F+++A+DS
Sbjct: 238 KT---TRDTFIRGCTKLLQTSSLPGVFIWNANDS 268
>UniRef100_Q41664 Nodulin homologous to narbonin [Vicia faba]
Length = 285
Score = 135 bits (339), Expect = 2e-30
Identities = 99/289 (34%), Positives = 150/289 (51%), Gaps = 27/289 (9%)
Query: 31 LFREYIGAQFKGVKFSDVPI---NPNVNFHFILSFAID-YDTSSPPSPTNGNFNIFWDTQ 86
+FREYIG + +D P + FHFIL FA + Y+ S S GNF W +
Sbjct: 5 VFREYIGVKPDSKSLADFPQITQTETIEFHFILGFATEKYNESRKGS---GNFEESWKDE 61
Query: 87 NLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN- 145
P +V +K ++P VKV +S+GG +G F P+ + WVSNAV SL II+ Y
Sbjct: 62 FFGPDKVRILKTKHPEVKVVISIGG---RGVETPFDPAEQNVWVSNAVKSLKLIIQKYKN 118
Query: 146 -----LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISF--ASIAPFDDAEVQNHYL 198
+DGID++YEH D F IG+LIT LK ++ SIAP ++ + YL
Sbjct: 119 ESGNLIDGIDINYEHIKWD-EAFPRLIGQLITELKKERDLNIHVVSIAPSENN--ASSYL 175
Query: 199 ALWKSYSNIIDYVNFQF-YAYDKSTTVAQFIDYYNKQSSNYNSEKVLVSFVSDGSTGLG- 256
L+ + + I+ V++QF + +T F+D Y + ++Y+ KVL F +D +
Sbjct: 176 NLYNANPDYINLVDYQFSNQLNPVSTDDAFVDIYERVVNDYHPRKVLPGFSTDPLDHINI 235
Query: 257 --PGNGFLDGCRMLKSQQKLNGIFVYSADDSK--ADGFRYEKEAQEVLA 301
+ F+ GC LK L G+F ++A+DS + F+ E +E+LA
Sbjct: 236 KITRDIFIGGCTKLKQTSSLPGVFFWNANDSNNGDEPFKVEHVLEELLA 284
>UniRef100_Q41660 Putative narbonin-like 2S protein [Vicia faba]
Length = 285
Score = 132 bits (333), Expect = 9e-30
Identities = 96/288 (33%), Positives = 147/288 (50%), Gaps = 25/288 (8%)
Query: 31 LFREYIGAQFKGVKFSDVPI---NPNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQN 87
+FREYIG + + D P + FHFIL FA + GNF W +
Sbjct: 5 IFREYIGVKRESETLEDFPQITQKETIEFHFILGFATEKYNEGRKG--TGNFEESWMDEF 62
Query: 88 LSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN-- 145
P +V +K ++P VKV +S+GG +G F P+ + WVSNAV SL II+ Y
Sbjct: 63 FGPDKVKNLKTKHPEVKVVISIGG---RGVETPFDPAEQNIWVSNAVKSLKLIIQKYKNE 119
Query: 146 ----LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISF--ASIAPFDDAEVQNHYLA 199
+DGID++YEH D F IG+LIT LK ++ SIAP ++ + YL
Sbjct: 120 SGNLIDGIDINYEHIKSD-EAFPRLIGQLITELKKERDLNIHVVSIAPSENN--ASSYLN 176
Query: 200 LWKSYSNIIDYVNFQFYAYDKS-TTVAQFIDYYNKQSSNYNSEKVLVSFVSDGSTGLG-- 256
L+ + + I+ V++QF + +T F+D Y + ++Y + KVL F++D +
Sbjct: 177 LYNANPDDINLVDYQFSNQLRHVSTEDAFVDIYKRVVNDYFTHKVLPVFITDPLDNMNTK 236
Query: 257 -PGNGFLDGCRMLKSQQKLNGIFVYSADDSK--ADGFRYEKEAQEVLA 301
+ F+ GC LK L G+F ++A+DS + F+ E +E+LA
Sbjct: 237 ITRDIFIGGCTKLKQTSSLPGVFFWNANDSNNGDEPFKVEHVLEELLA 284
>UniRef100_Q41704 Putative narbonin-like 2S protein [Vicia sativa]
Length = 285
Score = 131 bits (330), Expect = 2e-29
Identities = 96/288 (33%), Positives = 146/288 (50%), Gaps = 25/288 (8%)
Query: 31 LFREYIGAQFKGVKFSDVPI---NPNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQN 87
+FREYIG + + D P + FHFIL FA + GNF W +
Sbjct: 5 IFREYIGVKRESETLEDFPQITQKETIEFHFILGFATEKYNEGRKG--TGNFEESWMDEF 62
Query: 88 LSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN-- 145
P +V +K ++P VKV +S+GG +G F P+ + WVSNAV SL II+ Y
Sbjct: 63 FGPDKVKNLKTKHPEVKVVISIGG---RGVETPFDPAEQNIWVSNAVKSLKLIIQKYKNE 119
Query: 146 ----LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISF--ASIAPFDDAEVQNHYLA 199
+DGID++YEH D F IG+LIT LK ++ SIAP ++ + YL
Sbjct: 120 SGNLIDGIDINYEHIKSD-EAFPRLIGQLITELKKERDLNIHVVSIAPSENN--ASSYLN 176
Query: 200 LWKSYSNIIDYVNFQFYAYDKS-TTVAQFIDYYNKQSSNYNSEKVLVSFVSDGSTGLG-- 256
L+ + + I+ V++QF + +T F+D Y + ++Y + KVL F +D +
Sbjct: 177 LYNANPDDINLVDYQFSNQLRHVSTEDAFVDIYKRVVNDYFTHKVLPGFSTDPLDNMNTK 236
Query: 257 -PGNGFLDGCRMLKSQQKLNGIFVYSADDSK--ADGFRYEKEAQEVLA 301
+ F+ GC LK L G+F ++A+DS + F+ E +E+LA
Sbjct: 237 ITRDIFIGGCTKLKQTSSLPGVFFWNANDSNNGDEPFKVEHVLEELLA 284
>UniRef100_Q08884 Narbonin [Vicia narbonensis]
Length = 291
Score = 128 bits (322), Expect = 2e-28
Identities = 98/293 (33%), Positives = 147/293 (49%), Gaps = 30/293 (10%)
Query: 31 LFREYIGAQFKGVKFSDVPIN----PNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQ 86
+FREYIG + D P + FH+IL FAI+ S G F WD +
Sbjct: 5 IFREYIGVKPNSTTLHDFPTEIINTETLEFHYILGFAIESYYESGKG--TGTFEESWDVE 62
Query: 87 NLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN- 145
P +V +K ++P VKV +S+GG +G F P+ + WVSNA SL II+ Y+
Sbjct: 63 LFGPEKVKNLKRRHPEVKVVISIGG---RGVNTPFDPAEENVWVSNAKESLKLIIQKYSD 119
Query: 146 -----LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNN--VISFASIAPFDDAEVQNHYL 198
+DGID+ YEH D FA +G+LIT LK ++ I+ SIAP ++ +HY
Sbjct: 120 DSGNLIDGIDIHYEHIRSD-EPFATLMGQLITELKKDDDLNINVVSIAPSENN--SSHYQ 176
Query: 199 ALWKSYSNIIDYVNFQFYAYDKS-TTVAQFIDYYNKQSSNYNSEKVLVSFVS---DGSTG 254
L+ + + I++V++QF K +T F++ + +Y+ KVL F + D
Sbjct: 177 KLYNAKKDYINWVDYQFSNQQKPVSTDDAFVEIFKSLEKDYHPHKVLPGFSTDPLDTKHN 236
Query: 255 LGPGNGFLDGCRMLKSQQKLNGIFVYSADDS---KADG---FRYEKEAQEVLA 301
+ F+ GC L L G+F ++A+DS K DG F E Q++LA
Sbjct: 237 KITRDIFIGGCTRLVQTFSLPGVFFWNANDSVIPKRDGDKPFIVELTLQQLLA 289
>UniRef100_Q39888 Putative narbonin [Glycine max]
Length = 290
Score = 128 bits (321), Expect = 2e-28
Identities = 98/293 (33%), Positives = 143/293 (48%), Gaps = 30/293 (10%)
Query: 31 LFREYIGAQFKGVKFSDVPIN----PNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQ 86
+FREYIG + D P N+ FHFIL FA + S S GNF WD++
Sbjct: 5 IFREYIGVKPNSETLHDFPHEIIDTENLEFHFILGFATESYYESGKS--TGNFEESWDSE 62
Query: 87 NLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN- 145
+ V +K + P VKV +S+GG VQ F P+ + WV NA SL I + Y+
Sbjct: 63 SFGLENVKKLKERYPEVKVVISIGGRGVQ---TPFHPAEENVWVENAQESLKQIFQKYSN 119
Query: 146 -----LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNN--VISFASIAPFDDAEVQNHYL 198
+DGID++YEH D FA +G LIT LK ++ I+ SIAP + +HY
Sbjct: 120 ESGSMIDGIDINYEHISSD-EPFARLVGLLITELKKDDDLNINVVSIAPSETN--SSHYQ 176
Query: 199 ALWKSYSNIIDYVNFQFYAYDKST-TVAQFIDYYNKQSSNYNSEKVLVSFVSDGSTGLG- 256
L+ + + I++V++QF K F++ + +Y+ KVL F +D
Sbjct: 177 KLYNAKKDYINWVDYQFSNQHKPVHKDDHFVEIFKTVEKDYHLRKVLPGFSTDPDDAKHN 236
Query: 257 --PGNGFLDGCRMLKSQQKLNGIFVYSADDS---KADG---FRYEKEAQEVLA 301
+ F+ GC LK L G+F ++A DS + DG F E Q++LA
Sbjct: 237 KITRDVFIGGCTRLKQTSSLPGVFFWNAHDSVTPRLDGDKPFIVELTLQQLLA 289
>UniRef100_Q7DLM0 Narbonin [Vicia narbonensis]
Length = 291
Score = 128 bits (321), Expect = 2e-28
Identities = 98/293 (33%), Positives = 147/293 (49%), Gaps = 30/293 (10%)
Query: 31 LFREYIGAQFKGVKFSDVPIN----PNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQ 86
+FREYIG + D P + FH+IL FAI+ S G F WD +
Sbjct: 5 IFREYIGVKPNSTTLHDSPTEIVNTETLEFHYILGFAIESYYESGKG--TGTFEESWDVE 62
Query: 87 NLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN- 145
P +V +K ++P VKV +S+GG +G F P+ + WVSNA SL II+ Y+
Sbjct: 63 LFGPEKVKNLKRRHPEVKVVISIGG---RGVNTPFDPAEENVWVSNAKESLKLIIQKYSD 119
Query: 146 -----LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNN--VISFASIAPFDDAEVQNHYL 198
+DGID+ YEH D FA +G+LIT LK ++ I+ SIAP ++ +HY
Sbjct: 120 DSGNLIDGIDIHYEHIRSD-EPFATLMGQLITELKKDDDLNINVVSIAPSENN--SSHYQ 176
Query: 199 ALWKSYSNIIDYVNFQFYAYDKS-TTVAQFIDYYNKQSSNYNSEKVLVSFVS---DGSTG 254
L+ + + I++V++QF K +T F++ + +Y+ KVL F + D
Sbjct: 177 KLYNAKKDYINWVDYQFSNQQKPVSTDDAFVEIFKSLEKDYHPHKVLPGFSTDPLDTKHN 236
Query: 255 LGPGNGFLDGCRMLKSQQKLNGIFVYSADDS---KADG---FRYEKEAQEVLA 301
+ F+ GC L L G+F ++A+DS K DG F E Q++LA
Sbjct: 237 KITRDIFIGGCTRLVQTFSLPGVFFWNANDSVIPKRDGDKPFIVELTLQQLLA 289
>UniRef100_Q41675 Narbonin [Vicia narbonensis]
Length = 291
Score = 128 bits (321), Expect = 2e-28
Identities = 98/293 (33%), Positives = 147/293 (49%), Gaps = 30/293 (10%)
Query: 31 LFREYIGAQFKGVKFSDVPIN----PNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQ 86
+FREYIG + D P + FH+IL FAI+ S G F WD +
Sbjct: 5 IFREYIGVKPNSTTLHDFPTEIINTETLEFHYILGFAIESYYESGKG--TGTFEESWDVE 62
Query: 87 NLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN- 145
P +V +K ++P VKV +S+GG +G F P+ + WVSNA SL II+ Y+
Sbjct: 63 LFGPEKVKNLKRRHPEVKVVISIGG---RGVNTPFDPAEENVWVSNAKESLKLIIQKYSD 119
Query: 146 -----LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNN--VISFASIAPFDDAEVQNHYL 198
+DGID+ YEH D FA +G+LIT LK ++ I+ SIAP ++ +HY
Sbjct: 120 DSGNLIDGIDIHYEHIRSD-EPFATLMGQLITELKKDDDLNINVVSIAPSENN--SSHYQ 176
Query: 199 ALWKSYSNIIDYVNFQFYAYDKS-TTVAQFIDYYNKQSSNYNSEKVLVSFVS---DGSTG 254
L+ + + I++V++QF K +T F++ + +Y+ KVL F + D
Sbjct: 177 KLYNAKKDYINWVDYQFGNQQKPVSTDDAFVEIFKSLEKDYHPHKVLPGFSTDPLDTKHN 236
Query: 255 LGPGNGFLDGCRMLKSQQKLNGIFVYSADDS---KADG---FRYEKEAQEVLA 301
+ F+ GC L L G+F ++A+DS K DG F E Q++LA
Sbjct: 237 KITRDIFIGGCTRLVQTFSLPGVFFWNANDSVIPKRDGDKPFIVELTLQQLLA 289
>UniRef100_Q41680 Narbonin [Vicia pannonica]
Length = 291
Score = 125 bits (315), Expect = 1e-27
Identities = 90/278 (32%), Positives = 137/278 (48%), Gaps = 27/278 (9%)
Query: 31 LFREYIGAQFKGVKFSDVPIN----PNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQ 86
+FREYIG + D P N+ FHFIL FA + + G F WD++
Sbjct: 5 IFREYIGVKRDSPTLHDFPSEIIDTENLEFHFILGFATENYNQARKG--TGTFEESWDSE 62
Query: 87 NLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN- 145
+ V +K + P VKV +S+GG VQ F P+ + WV NA SL I + Y+
Sbjct: 63 SFGLENVKKLKERYPEVKVVISIGGRGVQ---TPFHPAEENVWVENAQESLKQIFQKYSN 119
Query: 146 -----LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNN--VISFASIAPFDDAEVQNHYL 198
+DGID++YEH D FA +G+LIT LK ++ I+ SIAP + +HY
Sbjct: 120 ESGSMIDGIDINYEHISSD-EPFARLVGQLITELKKDDDLNINVVSIAPSETN--SSHYQ 176
Query: 199 ALWKSYSNIIDYVNFQFYAYDKST-TVAQFIDYYNKQSSNYNSEKVLVSFVSDGSTGLG- 256
L+ + + I++V++QF K F++++ +Y+ KVL F +D
Sbjct: 177 KLYNAKKDYINWVDYQFSNQHKPVHKDDHFVEFFKTVEKDYHLRKVLPGFSTDPDDAKHN 236
Query: 257 --PGNGFLDGCRMLKSQQKLNGIFVYSADDS---KADG 289
+ F+ GC LK L G+F ++A DS ++DG
Sbjct: 237 KITRDVFIGGCTRLKQTSSLPGVFFWNAHDSVTPRSDG 274
>UniRef100_Q39486 Putative narbonin [Canavalia ensiformis]
Length = 287
Score = 124 bits (310), Expect = 4e-27
Identities = 94/289 (32%), Positives = 141/289 (48%), Gaps = 26/289 (8%)
Query: 31 LFREYIGAQFKGVKFSDVPIN----PNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQ 86
+FREYIG + D P + FHFIL FA + + GNF W +
Sbjct: 5 IFREYIGVKPDSTTLDDFPPEIIKTDTLEFHFILGFATE--KYNQKGNGTGNFEESWRDE 62
Query: 87 NLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN- 145
P +V +K ++P VKV +S+GG V+ F P+ WV AV SL II+ Y
Sbjct: 63 FFGPDKVKILKIKHPEVKVVISIGGRDVE---TPFDPAEQYVWVWKAVKSLKVIIQKYKN 119
Query: 146 -----LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNN--VISFASIAPFDDAEVQNHYL 198
+DGID++YE+ D F CI ++IT LKN++ I SIAP ++ + YL
Sbjct: 120 ESGNLIDGIDINYEYIKSD-ELFVNCISQVITELKNDDDLNIQVVSIAPSENN--ASSYL 176
Query: 199 ALWKSYSNIIDYVNFQF-YAYDKSTTVAQFIDYYNKQSSNYNSEKVLVSFVSDGSTGLG- 256
L+ + + I+ V++QF + +T F+D Y +Y + KVL F +D
Sbjct: 177 NLYNANPDYINLVDYQFSNQLNPVSTEDAFVDIYKSLVKDYFTHKVLPGFSTDPLDNTNT 236
Query: 257 --PGNGFLDGCRMLKSQQKLNGIFVYSADDSKADG--FRYEKEAQEVLA 301
+ F+ GC LK L G+F ++A+DS F+ E Q++LA
Sbjct: 237 KITRDIFIGGCTKLKQNSSLPGVFFWNANDSNNGDKPFKVELTLQQLLA 285
>UniRef100_Q41663 Nodulin homologous to narbonin [Vicia faba]
Length = 285
Score = 123 bits (308), Expect = 7e-27
Identities = 94/288 (32%), Positives = 143/288 (49%), Gaps = 25/288 (8%)
Query: 31 LFREYIGAQFKGVKFSDVPI---NPNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQN 87
+FREYIG + + +D P FHFIL FA + + S +GNF W +
Sbjct: 5 VFREYIGVKPESETLADFPQITQKETTEFHFILGFATENYSESRKG--SGNFVESWKDEF 62
Query: 88 LSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN-- 145
P +V +K ++P VKV +S+GG G F P+ WVSNAV SL II+ Y
Sbjct: 63 FGPDKVKILKRKHPEVKVVISIGG---HGLETPFDPAEQIVWVSNAVRSLKLIIQKYKNE 119
Query: 146 ----LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISF--ASIAPFDDAEVQNHYLA 199
+DGID++Y + D F IG+LI LK ++ SIAP ++ + Y
Sbjct: 120 SGNLIDGIDINYGNIKSD-EAFPRLIGQLIRELKKERDLNIHVVSIAPSENN--ASSYHK 176
Query: 200 LWKSYSNIIDYVNFQF-YAYDKSTTVAQFIDYYNKQSSNYNSEKVLVSFVSDGSTGLG-- 256
L+ + + I++V++QF + +T F+D Y +Y+ KVL F +D +
Sbjct: 177 LYNATRDDINWVDYQFSNQLNPVSTYDAFVDIYKSLVKDYHPHKVLPGFSTDPDDNMNTK 236
Query: 257 -PGNGFLDGCRMLKSQQKLNGIFVYSADDSK--ADGFRYEKEAQEVLA 301
+ FL GC LK L G+F ++A+DS + F+ E +E+LA
Sbjct: 237 ITRDIFLGGCTKLKQTSSLPGVFFWNANDSNNGDEPFQAEHVLEELLA 284
>UniRef100_Q41679 Narbonin [Vicia pannonica]
Length = 291
Score = 122 bits (305), Expect = 2e-26
Identities = 94/293 (32%), Positives = 140/293 (47%), Gaps = 30/293 (10%)
Query: 31 LFREYIGAQFKGVKFSDVPIN----PNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQ 86
+FREYIG + D P N+ FHFIL FA + + G F WD++
Sbjct: 5 IFREYIGVKRDSPTLHDFPSEIIGTENLEFHFILGFATENYNQARKG--TGTFEESWDSE 62
Query: 87 NLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN- 145
+ V K + P VKV +S+GG VQ F P+ + WV NA SL I + Y+
Sbjct: 63 SFGLENVKKSKERYPEVKVVISIGGRGVQ---TPFHPAEENVWVENAQESLKQIFQKYSN 119
Query: 146 -----LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNN--VISFASIAPFDDAEVQNHYL 198
++GID++YEH D FA +G+LIT LK ++ I+ SIAP + +HY
Sbjct: 120 ENGSMIEGIDINYEHISSD-EPFARLVGQLITELKKDDDLNINVVSIAPSETN--SSHYQ 176
Query: 199 ALWKSYSNIIDYVNFQFYAYDKST-TVAQFIDYYNKQSSNYNSEKVLVSFVSDGSTGLG- 256
L+ + + I++V++QF K F+ + +Y+ KVL F +D
Sbjct: 177 KLYNAKKDYINWVDYQFSNLHKPVHKDDDFVQIFKTVEKDYHLRKVLPGFSTDPDDAKHN 236
Query: 257 --PGNGFLDGCRMLKSQQKLNGIFVYSADDS---KADG---FRYEKEAQEVLA 301
+ F+ GC LK L G+F ++A DS + DG F E Q++LA
Sbjct: 237 KITRDVFIGGCTRLKQTSSLPGVFFWNAHDSVTPRLDGDKPFIVELSLQQLLA 289
>UniRef100_Q41662 Putative narbonin-like 2S protein [Vicia faba]
Length = 285
Score = 121 bits (303), Expect = 3e-26
Identities = 90/286 (31%), Positives = 137/286 (47%), Gaps = 21/286 (7%)
Query: 31 LFREYIGAQFKGVKFSDVPI---NPNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQN 87
+FREYIG + + D P + FHFIL FA + T + GNF W +
Sbjct: 5 IFREYIGVKPESETLQDFPQITQKETIEFHFILGFATEKYTEAKRG--TGNFEESWKDEF 62
Query: 88 LSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN-- 145
P V +K + P+VKV +S+GG G F P+ WVSNAV SL II+ Y
Sbjct: 63 FGPDNVKNLKTKYPDVKVVISIGG---SGLETPFDPAEQIVWVSNAVRSLKVIIQKYKND 119
Query: 146 ----LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISFASIAPFDDAEVQNHYLALW 201
+DGID++Y + D F IG+LIT LK ++ ++ + + Y L+
Sbjct: 120 SGNLIDGIDINYGNIKSD-QAFTRLIGQLITELKKERDLNIHVVSIAPSEKNASSYHKLY 178
Query: 202 KSYSNIIDYVNFQFYAYDKS-TTVAQFIDYYNKQSSNYNSEKVLVSFVSDGSTGLG---P 257
+ + I V++QF + +T F+D Y +Y+ KVL F +D +
Sbjct: 179 NATRDDITLVDYQFSNQLRHVSTYDAFVDIYKGLVKDYHPHKVLPGFSTDPLDNMNTKIT 238
Query: 258 GNGFLDGCRMLKSQQKLNGIFVYSADDSK--ADGFRYEKEAQEVLA 301
+ F+ GC LK L G+F ++A+DS + F+ E +E+LA
Sbjct: 239 RDIFIGGCTKLKQTSSLPGVFFWNANDSNNGDEPFKVEHVLEELLA 284
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 539,692,881
Number of Sequences: 2790947
Number of extensions: 23772398
Number of successful extensions: 69134
Number of sequences better than 10.0: 427
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 395
Number of HSP's that attempted gapping in prelim test: 68750
Number of HSP's gapped (non-prelim): 489
length of query: 304
length of database: 848,049,833
effective HSP length: 127
effective length of query: 177
effective length of database: 493,599,564
effective search space: 87367122828
effective search space used: 87367122828
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC149490.11


