

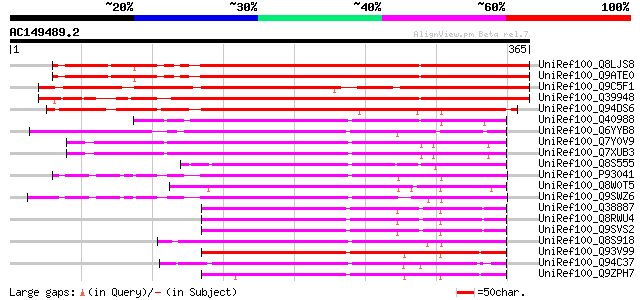
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149489.2 - phase: 0
(365 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LJS8 Homeodomain protein GhHOX1 [Gossypium hirsutum] 432 e-120
UniRef100_Q9ATE0 BNLGHi8377 [Gossypium hirsutum] 431 e-119
UniRef100_Q9C5F1 Homeobox protein GLABRA2 [Arabidopsis thaliana] 348 1e-94
UniRef100_Q39948 Homeodomain protein 1 [Helianthus annuus] 310 5e-83
UniRef100_Q94DS6 Putative homeobox protein GLABRA2 [Oryza sativa] 310 5e-83
UniRef100_Q40988 Homeobox protein [Phalaenopsis sp. SM9108] 203 5e-51
UniRef100_Q6YYB8 Putative OCL5 protein [Oryza sativa] 201 3e-50
UniRef100_Q7Y0V9 GL2-type homeodomain protein [Oryza sativa] 200 6e-50
UniRef100_Q7XUB3 OSJNBb0032E06.7 protein [Oryza sativa] 200 6e-50
UniRef100_Q8S555 Homeodomain protein HB2 [Picea abies] 197 5e-49
UniRef100_P93041 Homeodomain protein AHDP [Arabidopsis thaliana] 193 5e-48
UniRef100_Q8W0T5 OCL5 protein [Sorghum bicolor] 192 2e-47
UniRef100_Q9SWZ6 Anthocyaninless2 [Arabidopsis thaliana] 191 4e-47
UniRef100_Q38887 Meristem L1 layer homeobox protein [Arabidopsis... 189 1e-46
UniRef100_Q8RWU4 Putative L1-specific homeobox gene ATML1/ovule-... 189 1e-46
UniRef100_Q9SVS2 L1 specific homeobox gene ATML1/ovule-specific ... 189 1e-46
UniRef100_Q8S918 Roc1 [Oryza sativa] 188 2e-46
UniRef100_Q93V99 Protodermal factor2 [Arabidopsis thaliana] 187 4e-46
UniRef100_Q94C37 At1g05230/YUP8H12_16 [Arabidopsis thaliana] 182 1e-44
UniRef100_Q9ZPH7 T1J1.3 protein [Arabidopsis thaliana] 179 1e-43
>UniRef100_Q8LJS8 Homeodomain protein GhHOX1 [Gossypium hirsutum]
Length = 753
Score = 432 bits (1112), Expect = e-120
Identities = 234/342 (68%), Positives = 272/342 (79%), Gaps = 28/342 (8%)
Query: 31 NPPSTSKAKDFFPSPALSLSLAGIFRHGGGAAAEGEGGSISNMEVEEGEEGSTIGG---- 86
NPP+ KDFF SPALSLSLAGIFR G AA + +MEVEEG+EGS GG
Sbjct: 3 NPPT----KDFFASPALSLSLAGIFRDAGATAAAPTASA--SMEVEEGDEGSGGGGGSGS 56
Query: 87 --ERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYH 144
+ EISSE SGPA+S+S E D L+ D DDE D D K+KKKKRKKYH
Sbjct: 57 KKDDTVEISSENSGPARSRS----EDDLLDHD---DDENDAD-------KSKKKKRKKYH 102
Query: 145 RHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENS 204
RHT++QIR MEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENS
Sbjct: 103 RHTADQIREMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENS 162
Query: 205 LLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVERL 264
LLK E++KLR++NK +RETINKACC NCG+ TT +DG++ EEQQLRIENAKLKAEVE+L
Sbjct: 163 LLKQELDKLRDENKAMRETINKACCLNCGMATTAKDGSITAEEQQLRIENAKLKAEVEKL 222
Query: 265 RAALGKYASG-TMSPSCSTSHDQENIKSSLDFYTGIFCLDESRIMDVVNQAMEELIKMAT 323
R +GKY G + + SCS+ +DQEN +SSLDFYTGIF L++SRIM++VNQAMEEL KMAT
Sbjct: 223 RTVIGKYPPGASTTGSCSSGNDQEN-RSSLDFYTGIFGLEKSRIMEIVNQAMEELQKMAT 281
Query: 324 MGEPMWLRSLETGREILNYDEYMKEFADENSDHGRPKRSIEA 365
GEP+W+RS+ETGREILNYDEY+KE + E+S +GRPKRSIEA
Sbjct: 282 AGEPLWVRSVETGREILNYDEYVKELSVESSSNGRPKRSIEA 323
>UniRef100_Q9ATE0 BNLGHi8377 [Gossypium hirsutum]
Length = 758
Score = 431 bits (1107), Expect = e-119
Identities = 233/342 (68%), Positives = 271/342 (79%), Gaps = 28/342 (8%)
Query: 31 NPPSTSKAKDFFPSPALSLSLAGIFRHGGGAAAEGEGGSISNMEVEEGEEGSTIGG---- 86
NPP+ KDFF SPALSLSLAGIFR G AA + +MEVEEG+EGS GG
Sbjct: 8 NPPT----KDFFASPALSLSLAGIFRDAGATAAAPTASA--SMEVEEGDEGSGGGGGSGS 61
Query: 87 --ERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYH 144
+ EISSE SGPA+S+S E D L+ D DDE D D K+KKKKRKKYH
Sbjct: 62 KKDDTVEISSENSGPARSRS----EDDLLDHD---DDENDAD-------KSKKKKRKKYH 107
Query: 145 RHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENS 204
RHT++QIR MEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENS
Sbjct: 108 RHTADQIREMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENS 167
Query: 205 LLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVERL 264
LLK E++KLR++NK +RETINKACC NCG+ TT +DG + EEQQLRIENAKLKAEVE+L
Sbjct: 168 LLKQELDKLRDENKAMRETINKACCLNCGMATTAKDGFITAEEQQLRIENAKLKAEVEKL 227
Query: 265 RAALGKYASG-TMSPSCSTSHDQENIKSSLDFYTGIFCLDESRIMDVVNQAMEELIKMAT 323
R +GKY G + + SCS+ +DQEN +SSL+FYTGIF L++SRIM++VNQAMEEL KMAT
Sbjct: 228 RTVIGKYPPGASTTGSCSSGNDQEN-RSSLNFYTGIFALEKSRIMEIVNQAMEELQKMAT 286
Query: 324 MGEPMWLRSLETGREILNYDEYMKEFADENSDHGRPKRSIEA 365
GEP+W+RS+ETGREILNYDEY+KE + E+S +GRPKRSIEA
Sbjct: 287 AGEPLWVRSVETGREILNYDEYVKELSVESSSNGRPKRSIEA 328
>UniRef100_Q9C5F1 Homeobox protein GLABRA2 [Arabidopsis thaliana]
Length = 747
Score = 348 bits (893), Expect = 1e-94
Identities = 192/348 (55%), Positives = 241/348 (69%), Gaps = 36/348 (10%)
Query: 21 LSMCADMSNNNPPSTSKAKDFFPSPALSLSLAGIFRHGGGAAAEGEGGSISNMEVEEGEE 80
+SM DMS+ P KDFF SPALSLSLAGIFR+ + E + V++ +
Sbjct: 1 MSMAVDMSSKQP-----TKDFFSSPALSLSLAGIFRNASSGSTNPEEDFLGRRVVDDED- 54
Query: 81 GSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKR 140
R E+SSE SGP +S+S ++ EG++ +D+E E+++G G K+KR
Sbjct: 55 -------RTVEMSSENSGPTRSRSEEDLEGEDHDDEEEEEEDGAAGNKG-----TNKRKR 102
Query: 141 KKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQER 200
KKYHRHT++QIR MEALFKE+PHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQER
Sbjct: 103 KKYHRHTTDQIRHMEALFKETPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQER 162
Query: 201 HENSLLKSEIEKLREKNKTLRETINKA--CCPNCGVPTTNRDGTMATEEQQLRIENAKLK 258
HENSLLK+E+EKLRE+NK +RE+ +KA CPNCG L +EN+KLK
Sbjct: 163 HENSLLKAELEKLREENKAMRESFSKANSSCPNCG-----------GGPDDLHLENSKLK 211
Query: 259 AEVERLRAALGKYASGTMSP-SCSTSHDQENIKSSLDFYTGIFCLDESRIMDVVNQAMEE 317
AE+++LRAALG+ T P S S DQE+ SLDFYTG+F L++SRI ++ N+A E
Sbjct: 212 AELDKLRAALGR----TPYPLQASCSDDQEHRLGSLDFYTGVFALEKSRIAEISNRATLE 267
Query: 318 LIKMATMGEPMWLRSLETGREILNYDEYMKEFADENSDHGRPKRSIEA 365
L KMAT GEPMWLRS+ETGREILNYDEY+KEF + +++IEA
Sbjct: 268 LQKMATSGEPMWLRSVETGREILNYDEYLKEFPQAQASSFPGRKTIEA 315
>UniRef100_Q39948 Homeodomain protein 1 [Helianthus annuus]
Length = 682
Score = 310 bits (793), Expect = 5e-83
Identities = 182/350 (52%), Positives = 233/350 (66%), Gaps = 32/350 (9%)
Query: 21 LSMCADMSNN--NPPSTSKAKDFFPSPALSLSLAGIFRHGGGAAAEGEGGSISNMEVEEG 78
+S D +NN N P TS KD FPS +LSL+L G+FR G A V++
Sbjct: 1 MSSSNDNNNNFINNPLTS-TKDLFPSTSLSLTL-GLFRDGEDA-------------VDDR 45
Query: 79 EEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKK 138
+E + V EISSE SGP +S+S +DD+ + D DGD NK K
Sbjct: 46 KEEAA--NTTVTEISSETSGPVRSRS---------DDDDFDADPDVDDGDDASNKNNKSK 94
Query: 139 KRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQ 198
KRKKYHRHT++QIR MEALFKESPHPDEKQRQQLSK+LGL PRQVKFWFQNRRTQIK IQ
Sbjct: 95 KRKKYHRHTADQIREMEALFKESPHPDEKQRQQLSKRLGLHPRQVKFWFQNRRTQIKTIQ 154
Query: 199 ERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIENAKLK 258
ERHENSLLKSE++KL E+NK LRETI K C NCG ++++D +EQQLR+ENAKL+
Sbjct: 155 ERHENSLLKSELDKLGEENKLLRETIKKGTCTNCGFGSSSKDVHTYVDEQQLRVENAKLR 214
Query: 259 AEVERLR-AALGK-YASGTMSPSCSTSHDQENIKSSLDFYTGIFCLDESRIMDVVNQAM- 315
AE+E+LR + G + SCS +D EN KSSLD +G+F L++SR+ +
Sbjct: 215 AEIEKLRNISWGNIHREHHQRNSCSVGNDHEN-KSSLDLCSGLFGLEKSRVNGGCRSGLW 273
Query: 316 EELIKMATMGEPMWLRSLETGREILNYDEYMKEFADENSDHGRPKRSIEA 365
+ ++MA+ G P+W++SLETGREILNYDEY+ +F+ +S + + R IEA
Sbjct: 274 KSFVQMASAGAPLWVKSLETGREILNYDEYLTKFSTVDSSNVQRLRFIEA 323
>UniRef100_Q94DS6 Putative homeobox protein GLABRA2 [Oryza sativa]
Length = 779
Score = 310 bits (793), Expect = 5e-83
Identities = 180/344 (52%), Positives = 229/344 (66%), Gaps = 34/344 (9%)
Query: 27 MSNNNPPSTSKAKDFFPSPALSLSLAGIFRHGGGAAAEGEGGSISNMEVEEGEEGSTIGG 86
M N P + KDFF +PALSL+LAG+F G AA G G VEEG+E G
Sbjct: 1 MGTNRP--RPRTKDFFAAPALSLTLAGVFGRKNGPAASGGDG------VEEGDEEVQAAG 52
Query: 87 ERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRH 146
E EISSE +GP +S + G ED +DD+G+G NKK++RK YHRH
Sbjct: 53 EAAVEISSENAGPGCRQS--QSGGGSGEDGGHDDDDGEGS--------NKKRRRKNYHRH 102
Query: 147 TSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLL 206
T+EQIR+MEALFKESPHPDE+QRQQ+SKQLGL+ RQVKFWFQNRRTQIKA+QERHENSLL
Sbjct: 103 TAEQIRIMEALFKESPHPDERQRQQVSKQLGLSARQVKFWFQNRRTQIKAVQERHENSLL 162
Query: 207 KSEIEKLREKNKTLRETINK-ACCPNCGVPTTNRDGTMA-----TEEQQLRIENAKLKAE 260
KSE+EKL+++++ +RE K + C NCGV T+ D A T EQ+LR+E AKLKAE
Sbjct: 163 KSELEKLQDEHRAMRELAKKPSRCLNCGVVATSSDAAAAATAADTREQRLRLEKAKLKAE 222
Query: 261 VERLRAALGKYAS-GTMSPSCSTSHD--QENIKS-SLDFYTGIFCL---DESRIMDVVNQ 313
+ERLR GK A+ G SP CS S Q N +S L + G F D+ RI+++ +
Sbjct: 223 IERLRGTPGKSAADGIASPPCSASAGAMQTNSRSPPLHDHDGGFLRHDDDKPRILELATR 282
Query: 314 AMEELIKMATMGEPMWLRSLETGREILNYDEYMKEFADENSDHG 357
A++EL+ M + GEP+W+R +ETGR+ILNYDEY++ F DHG
Sbjct: 283 ALDELVGMCSSGEPVWVRGVETGRDILNYDEYVRLF---RRDHG 323
>UniRef100_Q40988 Homeobox protein [Phalaenopsis sp. SM9108]
Length = 768
Score = 203 bits (517), Expect = 5e-51
Identities = 119/290 (41%), Positives = 172/290 (59%), Gaps = 35/290 (12%)
Query: 88 RVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDD-EGDGDGDGDGVNKNKKKKRKKYHRH 146
++ E++++ + A+S + E D+ E G D+ EG G GD + N++ ++K+YHRH
Sbjct: 41 QLAELTAQATTTAESDMMRARE-DDFESKSGSDNIEG---GSGDEHDPNQRPRKKRYHRH 96
Query: 147 TSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLL 206
T QI+ MEA FKE PHPD+KQR+ LSK+LGL P QVKFWFQN+RTQ+K +R ENS L
Sbjct: 97 TQHQIQEMEAFFKECPHPDDKQRKALSKELGLEPLQVKFWFQNKRTQMKTQHDRQENSQL 156
Query: 207 KSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVERLRA 266
++E +KLR +N +E ++ A CPNCG P T G M+ +E LRIENA+L+ E++R+
Sbjct: 157 RAENDKLRNENLRYKEALSNASCPNCGGPATL--GEMSFDEHHLRIENARLREEIDRISG 214
Query: 267 ALGKYASGTMSPSCSTSHDQENIKSSLDFYTGIFCL------------------------ 302
KY M+ S + +SSLD G F L
Sbjct: 215 IAAKYVGKPMNSYPLLSPTLPS-RSSLDLGVGGFGLHSPTMGGDMFSPAELLRSVAGQPE 273
Query: 303 -DESRIMDVVNQAMEELIKMATMGEPMWLRS--LETGREILNYDEYMKEF 349
D+ ++++ AMEELI+MA +GEP+W S L+ G EILN +EY++ F
Sbjct: 274 VDKPMVIELAVAAMEELIRMAQLGEPLWTSSPGLDGGNEILNEEEYVQNF 323
>UniRef100_Q6YYB8 Putative OCL5 protein [Oryza sativa]
Length = 828
Score = 201 bits (511), Expect = 3e-50
Identities = 122/347 (35%), Positives = 184/347 (52%), Gaps = 32/347 (9%)
Query: 15 LLSSPTLSMCADMSNNNPPSTSKAKDFFPSPALSLSLAGIFRHGGGAAAEGEGGSISNME 74
++++ +C + N + + P +S ++ G GGG S S +
Sbjct: 56 MMANDAQELCDNDDGGNNGLRERERMMIPGRNMSPAMIGRPNGGGGGGGVAYASSSSALS 115
Query: 75 VEEGEEGSTIGGERV-EEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVN 133
+ + S G +E+ SG + D LE G G G G G +
Sbjct: 116 LGQATTTSESDGRAPRDELEMSKSGGS----------DNLESGGG----GGGGGSGGDQD 161
Query: 134 KNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQ 193
N++ ++K+YHRHT QI+ +EA FKE PHPD+KQR++LS++LGL P QVKFWFQN+RTQ
Sbjct: 162 PNQRPRKKRYHRHTQHQIQELEAFFKECPHPDDKQRKELSRELGLEPLQVKFWFQNKRTQ 221
Query: 194 IKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIE 253
+K ERHEN+ L++E EKLR +N +E + A CPNCG P G M+ +E LR+E
Sbjct: 222 MKTQHERHENNALRAENEKLRAENMRYKEALANASCPNCGGPAA--IGEMSFDEHHLRLE 279
Query: 254 NAKLKAEVERLRAALGKY----------ASGTMSPSCSTSHDQENIK-SSLDFYTGIFCL 302
NA+L+ E++R+ A KY A + PS + D I + D + F
Sbjct: 280 NARLRDEIDRISAIAAKYVGKPAAAVSAAYPPLPPSNRSPLDHMGIPGAGADVFGADF-- 337
Query: 303 DESRIMDVVNQAMEELIKMATMGEPMWLRSLETGREILNYDEYMKEF 349
D+ ++++ AMEEL++MA +GEP+W +L G E L +EY + F
Sbjct: 338 DKPLVIELAVAAMEELVRMAQLGEPLWAPAL--GGEALGEEEYARTF 382
>UniRef100_Q7Y0V9 GL2-type homeodomain protein [Oryza sativa]
Length = 813
Score = 200 bits (508), Expect = 6e-50
Identities = 128/356 (35%), Positives = 180/356 (49%), Gaps = 56/356 (15%)
Query: 41 FFPSPALSLSLAGIFRHGGGAAAEGEGGSISNMEVEEGEEGSTIGGERVEEISSEYSGPA 100
F SPALSL+LA A GG + G G GG R + +
Sbjct: 13 FTSSPALSLALAD------AVAGRNSGGGGKMVTAAHGGVGGGGGGGRAKARDALEVENE 66
Query: 101 KSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKE 160
S+S ++ D + + GD D D D + N K++K+YHRHT +QI+ +EA+FKE
Sbjct: 67 MSRSGSDHL-DVVSCGDAGGGGGDDDDDEDAEHGNPPKRKKRYHRHTPQQIQELEAMFKE 125
Query: 161 SPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTL 220
PHPDEKQR +LSK+LGL PRQVKFWFQNRRTQ+K ERHENSLLK E +KLR +N ++
Sbjct: 126 CPHPDEKQRAELSKRLGLEPRQVKFWFQNRRTQMKMQLERHENSLLKQENDKLRSENLSI 185
Query: 221 RETINKACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGTMSPSC 280
RE + A C CG P G ++ EE LR+ENA+LK E+ R+ A K+ ++S
Sbjct: 186 REATSNAVCVGCGGPAML--GEVSLEEHHLRVENARLKDELSRVCALAAKFLGKSISVMA 243
Query: 281 STSHDQEN-----------------------IKSSLDFY--------------------T 297
Q + I + DF +
Sbjct: 244 PPQMHQPHPVPGSSLELAVGGIGSMPSATMPISTITDFAGAMSSSMGTVITPMKSEAEPS 303
Query: 298 GIFCLDESRIMDVVNQAMEELIKMATMGEPMWLRSLET----GREILNYDEYMKEF 349
+ +D+S +++ AM+EL+KMA MG+P+W+ +E LN++EY+ F
Sbjct: 304 AMAGIDKSLFLELAMSAMDELVKMAQMGDPLWIPGASVPSSPAKESLNFEEYLNTF 359
>UniRef100_Q7XUB3 OSJNBb0032E06.7 protein [Oryza sativa]
Length = 806
Score = 200 bits (508), Expect = 6e-50
Identities = 128/356 (35%), Positives = 180/356 (49%), Gaps = 56/356 (15%)
Query: 41 FFPSPALSLSLAGIFRHGGGAAAEGEGGSISNMEVEEGEEGSTIGGERVEEISSEYSGPA 100
F SPALSL+LA A GG + G G GG R + +
Sbjct: 13 FTSSPALSLALAD------AVAGRNSGGGGKMVTAAHGGVGGGGGGGRAKARDALEVENE 66
Query: 101 KSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKE 160
S+S ++ D + + GD D D D + N K++K+YHRHT +QI+ +EA+FKE
Sbjct: 67 MSRSGSDHL-DVVSCGDAGGGGGDDDDDEDAEHGNPPKRKKRYHRHTPQQIQELEAMFKE 125
Query: 161 SPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTL 220
PHPDEKQR +LSK+LGL PRQVKFWFQNRRTQ+K ERHENSLLK E +KLR +N ++
Sbjct: 126 CPHPDEKQRAELSKRLGLEPRQVKFWFQNRRTQMKMQLERHENSLLKQENDKLRSENLSI 185
Query: 221 RETINKACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGTMSPSC 280
RE + A C CG P G ++ EE LR+ENA+LK E+ R+ A K+ ++S
Sbjct: 186 REATSNAVCVGCGGPAML--GEVSLEEHHLRVENARLKDELSRVCALAAKFLGKSISVMA 243
Query: 281 STSHDQEN-----------------------IKSSLDFY--------------------T 297
Q + I + DF +
Sbjct: 244 PPQMHQPHPVPGSSLELAVGGIGSMPSATMPISTITDFAGAMSSSMGTVITPMKSEAEPS 303
Query: 298 GIFCLDESRIMDVVNQAMEELIKMATMGEPMWLRSLET----GREILNYDEYMKEF 349
+ +D+S +++ AM+EL+KMA MG+P+W+ +E LN++EY+ F
Sbjct: 304 AMAGIDKSLFLELAMSAMDELVKMAQMGDPLWIPGASVPSSPAKESLNFEEYLNTF 359
>UniRef100_Q8S555 Homeodomain protein HB2 [Picea abies]
Length = 708
Score = 197 bits (500), Expect = 5e-49
Identities = 115/253 (45%), Positives = 151/253 (59%), Gaps = 32/253 (12%)
Query: 121 DEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAP 180
D G GD D D + +KKR YHRHT +QI+ +EALFKE PHPDEKQR +SK+L L
Sbjct: 11 DGGSGD-DQDAADNPPRKKR--YHRHTPQQIQELEALFKECPHPDEKQRLDISKRLNLET 67
Query: 181 RQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRD 240
RQVK WFQNRRTQ+K ERHENS+L+ E EKLR +N ++R+ + C NCG P
Sbjct: 68 RQVKLWFQNRRTQMKTQLERHENSILRQENEKLRSENLSIRDAMRNPICTNCGGPAVL-- 125
Query: 241 GTMATEEQQLRIENAKLKAEVERLRAALGKYASGTMSPSCSTSHDQENIKSSLDFYTG-- 298
G M+ EEQQLRIENA+LK E++RL A GK+ G PS + KSSLD G
Sbjct: 126 GEMSFEEQQLRIENARLKKELDRLCALAGKF-FGRPVPSMPSVPLMP--KSSLDLGVGGM 182
Query: 299 ----------------------IFCLDESRIMDVVNQAMEELIKMATMGEPMWLRSLETG 336
I ++ S + ++ +M+EL KMA E +W+ +L+ G
Sbjct: 183 PTSLPSGCADLMHGPAGGRTGNIIGIERSMLAELALASMDELFKMAQADETLWIPNLDAG 242
Query: 337 REILNYDEYMKEF 349
+E LNY+EYM++F
Sbjct: 243 KETLNYEEYMRQF 255
>UniRef100_P93041 Homeodomain protein AHDP [Arabidopsis thaliana]
Length = 745
Score = 193 bits (491), Expect = 5e-48
Identities = 128/342 (37%), Positives = 185/342 (53%), Gaps = 50/342 (14%)
Query: 31 NPPSTSKAKDFFPSPALSLSLAGIFRHGGGAAAEGEGGSISNMEVEEGEEGSTIGGERVE 90
+PP T K + S LSL+L R GE +N V G G T G V
Sbjct: 12 SPPLT---KSVYASSGLSLALEQPER----GTNRGEASMRNNNNVGGG--GDTFDGS-VN 61
Query: 91 EISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQ 150
S E ++S S D +E GED + +K ++K+YHRHT +Q
Sbjct: 62 RRSREEEHESRSGS------DNVEGISGEDQDA----------ADKPPRKKRYHRHTPQQ 105
Query: 151 IRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEI 210
I+ +E++FKE PHPDEKQR +LSK+L L RQVKFWFQNRRTQ+K ERHEN+LL+ E
Sbjct: 106 IQELESMFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRTQMKTQLERHENALLRQEN 165
Query: 211 EKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVERLRAALGK 270
+KLR +N ++RE + C NCG P G ++ EE LRIENA+LK E++R+ GK
Sbjct: 166 DKLRAENMSIREAMRNPICTNCGGPAML--GDVSLEEHHLRIENARLKDELDRVCNLTGK 223
Query: 271 YA--------SGTMSPSCSTSHDQENIKSSLDFYTGIFCL--------------DESRIM 308
+ + ++ + T+++ + DF G CL +S ++
Sbjct: 224 FLGHHHNHHYNSSLELAVGTNNNGGHFAFPPDFGGGGGCLPPQQQQSTVINGIDQKSVLL 283
Query: 309 DVVNQAMEELIKMATMGEPMWLRSLETGREILNYDEYMKEFA 350
++ AM+EL+K+A EP+W++SL+ R+ LN DEYM+ F+
Sbjct: 284 ELALTAMDELVKLAQSEEPLWVKSLDGERDELNQDEYMRTFS 325
>UniRef100_Q8W0T5 OCL5 protein [Sorghum bicolor]
Length = 803
Score = 192 bits (487), Expect = 2e-47
Identities = 107/271 (39%), Positives = 157/271 (57%), Gaps = 36/271 (13%)
Query: 113 LEDDEGEDDEGDGDGDGDGVNKNKKK------KRKKYHRHTSEQIRVMEALFKESPHPDE 166
+E G D+ G G G G + ++ ++K+YHRHT QI+ +EA FKE PHPD+
Sbjct: 67 MESKSGSDNMEGGAGSGSGGEELQEDLSLQPARKKRYHRHTQHQIQELEAFFKEYPHPDD 126
Query: 167 KQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINK 226
KQR++LS++LGL P QVKFWFQN+RTQ+K QERHEN L++E EKLR +N ++ +
Sbjct: 127 KQRKELSRELGLEPLQVKFWFQNKRTQMKTQQERHENMQLRAENEKLRAENARYKDALAN 186
Query: 227 ACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVERLRAALGKYA---SGTMSPSCS-- 281
A CPNCG P T G M+ +E LRIENA+L+ EV+R+ KY +G++ P+ S
Sbjct: 187 ASCPNCGGPATAVIGEMSFDEHHLRIENARLRDEVDRISTIAAKYVGKPAGSLLPNLSNI 246
Query: 282 --------------TSHDQENIKSSLDFYTGIFC------LDESRIMDVVNQAMEELIKM 321
+SH + D + G+ D+ ++++ AMEEL++M
Sbjct: 247 SSASMAPYPPPPPLSSH--HLLPGGTDMFGGLHLHGAAAGFDKGLVVELAVAAMEELVRM 304
Query: 322 ATMGEPMWLRSLETGR---EILNYDEYMKEF 349
A +GEP+W+ +L E LN +EY + F
Sbjct: 305 AQLGEPLWIPALVVDGATIETLNEEEYARGF 335
>UniRef100_Q9SWZ6 Anthocyaninless2 [Arabidopsis thaliana]
Length = 801
Score = 191 bits (484), Expect = 4e-47
Identities = 135/368 (36%), Positives = 185/368 (49%), Gaps = 67/368 (18%)
Query: 13 NTLLSSPTLSMCADMSNNNPPSTSKAKDFFPSPALSLSLAGIFRHGGGAAAEGEGGSISN 72
N L A S +PP T K + S LSL+L R GE +N
Sbjct: 34 NVLPGGAMAQAAAAASLFSPPLT---KSVYASSGLSLALEQPER----GTNRGEASMRNN 86
Query: 73 MEVEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGV 132
V G G T G V S E ++S S D +E GED +
Sbjct: 87 NNVGGG--GDTFDGS-VNRRSREEEHESRSGS------DNVEGISGEDQDA--------- 128
Query: 133 NKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRT 192
+K ++K+YHRHT +QI+ +E++FKE PHPDEKQR +LSK+L L RQVKFWFQNRRT
Sbjct: 129 -ADKPPRKKRYHRHTPQQIQELESMFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRT 187
Query: 193 QIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRI 252
Q+K ERHEN+LL+ E +KLR +N ++RE + C NCG P G ++ EE LRI
Sbjct: 188 QMKTQLERHENALLRQENDKLRAENMSIREAMRNPICTNCGGPAML--GDVSLEEHHLRI 245
Query: 253 ENAKLKAEVERLRAALGKYASGTMSPSCSTSHDQENIKSSL----------------DFY 296
ENA+LK E++R+ GK+ H + SSL DF
Sbjct: 246 ENARLKDELDRVCNLTGKFLG---------HHHNHHYNSSLELAVGTNNGGHFAFPPDFG 296
Query: 297 TGIFCL--------------DESRIMDVVNQAMEELIKMATMGEPMWLRSLETGREILNY 342
G CL +S ++++ AM+EL+K+A EP+W++SL+ R+ LN
Sbjct: 297 GGGGCLPPQQQQSTVINGIDQKSVLLELALTAMDELVKLAQSEEPLWVKSLDGERDELNQ 356
Query: 343 DEYMKEFA 350
DEYM+ F+
Sbjct: 357 DEYMRTFS 364
>UniRef100_Q38887 Meristem L1 layer homeobox protein [Arabidopsis thaliana]
Length = 718
Score = 189 bits (480), Expect = 1e-46
Identities = 104/248 (41%), Positives = 147/248 (58%), Gaps = 39/248 (15%)
Query: 136 KKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIK 195
+ K+K+YHRHT QI+ +E+ FKE PHPD+KQR++LS++L L P QVKFWFQN+RTQ+K
Sbjct: 15 RPNKKKRYHRHTQRQIQELESFFKECPHPDDKQRKELSRELSLEPLQVKFWFQNKRTQMK 74
Query: 196 AIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIENA 255
A ERHEN +LKSE +KLR +N ++ ++ A CPNCG P G M+ +EQ LRIENA
Sbjct: 75 AQHERHENQILKSENDKLRAENNRYKDALSNATCPNCGGPAA--IGEMSFDEQHLRIENA 132
Query: 256 KLKAEVERLRAALGKY------ASGTMSPSCSTSHDQENIKSSLDFYTGIFC-------- 301
+L+ E++R+ A KY A+ + P S+SH + SLD G F
Sbjct: 133 RLREEIDRISAIAAKYVGKPLMANSSSFPQLSSSHHIPS--RSLDLEVGNFGNNNNSHTG 190
Query: 302 --------------------LDESRIMDVVNQAMEELIKMATMGEPMWLRSLETGREILN 341
D+ I+++ AMEEL++MA G+P+W+ S + EILN
Sbjct: 191 FVGEMFGSSDILRSVSIPSEADKPMIVELAVAAMEELVRMAQTGDPLWVSS-DNSVEILN 249
Query: 342 YDEYMKEF 349
+EY + F
Sbjct: 250 EEEYFRTF 257
>UniRef100_Q8RWU4 Putative L1-specific homeobox gene ATML1/ovule-specific homeobox
protein A20 [Arabidopsis thaliana]
Length = 762
Score = 189 bits (480), Expect = 1e-46
Identities = 104/248 (41%), Positives = 147/248 (58%), Gaps = 39/248 (15%)
Query: 136 KKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIK 195
+ K+K+YHRHT QI+ +E+ FKE PHPD+KQR++LS++L L P QVKFWFQN+RTQ+K
Sbjct: 59 RPNKKKRYHRHTQRQIQELESFFKECPHPDDKQRKELSRELSLEPLQVKFWFQNKRTQMK 118
Query: 196 AIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIENA 255
A ERHEN +LKSE +KLR +N ++ ++ A CPNCG P G M+ +EQ LRIENA
Sbjct: 119 AQHERHENQILKSENDKLRAENNRYKDALSNATCPNCGGPAA--IGEMSFDEQHLRIENA 176
Query: 256 KLKAEVERLRAALGKY------ASGTMSPSCSTSHDQENIKSSLDFYTGIFC-------- 301
+L+ E++R+ A KY A+ + P S+SH + SLD G F
Sbjct: 177 RLREEIDRISAIAAKYVGKPLMANSSSFPQLSSSHHIPS--RSLDLEVGNFGNNNNSHTG 234
Query: 302 --------------------LDESRIMDVVNQAMEELIKMATMGEPMWLRSLETGREILN 341
D+ I+++ AMEEL++MA G+P+W+ S + EILN
Sbjct: 235 FVGEMFGSSDILRSVSIPSEADKPMIVELAVAAMEELVRMAQTGDPLWVSS-DNSVEILN 293
Query: 342 YDEYMKEF 349
+EY + F
Sbjct: 294 EEEYFRTF 301
>UniRef100_Q9SVS2 L1 specific homeobox gene ATML1/ovule-specific homeobox protein A20
[Arabidopsis thaliana]
Length = 718
Score = 189 bits (480), Expect = 1e-46
Identities = 104/248 (41%), Positives = 147/248 (58%), Gaps = 39/248 (15%)
Query: 136 KKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIK 195
+ K+K+YHRHT QI+ +E+ FKE PHPD+KQR++LS++L L P QVKFWFQN+RTQ+K
Sbjct: 15 RPNKKKRYHRHTQRQIQELESFFKECPHPDDKQRKELSRELSLEPLQVKFWFQNKRTQMK 74
Query: 196 AIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIENA 255
A ERHEN +LKSE +KLR +N ++ ++ A CPNCG P G M+ +EQ LRIENA
Sbjct: 75 AQHERHENQILKSENDKLRAENNRYKDALSNATCPNCGGPAA--IGEMSFDEQHLRIENA 132
Query: 256 KLKAEVERLRAALGKY------ASGTMSPSCSTSHDQENIKSSLDFYTGIFC-------- 301
+L+ E++R+ A KY A+ + P S+SH + SLD G F
Sbjct: 133 RLREEIDRISAIAAKYVGKPLMANSSSFPQLSSSHHIPS--RSLDLEVGNFGNNNNSHTG 190
Query: 302 --------------------LDESRIMDVVNQAMEELIKMATMGEPMWLRSLETGREILN 341
D+ I+++ AMEEL++MA G+P+W+ S + EILN
Sbjct: 191 FVGEMFGSSDILRSVSIPSEADKPMIVELAVAAMEELVRMAQTGDPLWVSS-DNSVEILN 249
Query: 342 YDEYMKEF 349
+EY + F
Sbjct: 250 EEEYFRTF 257
>UniRef100_Q8S918 Roc1 [Oryza sativa]
Length = 784
Score = 188 bits (477), Expect = 2e-46
Identities = 104/271 (38%), Positives = 156/271 (57%), Gaps = 32/271 (11%)
Query: 105 IDEYEGDELEDDEGEDDEGDGDG-DGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPH 163
+DE+E E+ +G GDG GD + N++ ++K+YHRHT QI+ MEA FKE PH
Sbjct: 75 VDEFESKSCS----ENVDGAGDGLSGDDQDPNQRPRKKRYHRHTQHQIQEMEAFFKECPH 130
Query: 164 PDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRET 223
PD+KQR++LS++LGL P QVKFWFQN+RTQ+K ERHEN+ L++E +KLR +N +E
Sbjct: 131 PDDKQRKELSRELGLEPLQVKFWFQNKRTQMKNQHERHENAQLRAENDKLRAENMRYKEA 190
Query: 224 INKACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGTMSPSCSTS 283
++ A CPNCG P G M+ +E LR+ENA+L+ E++R+ K+ S S
Sbjct: 191 LSSASCPNCGGPAAL--GEMSFDEHHLRVENARLRDEIDRISGIAAKHVGKPPIVSFSVL 248
Query: 284 HDQENIKSS----------------LDFYTGIFCL---------DESRIMDVVNQAMEEL 318
+ ++ LD + G L D+ I+++ AM+EL
Sbjct: 249 SSPLAVAAARSPLDLAGAYGVVTPGLDMFGGAGDLLRGVHPLDADKPMIVELAVAAMDEL 308
Query: 319 IKMATMGEPMWLRSLETGREILNYDEYMKEF 349
++MA + EP+W S E +L+ +EY + F
Sbjct: 309 VQMAQLDEPLWSSSSEPAAALLDEEEYARMF 339
>UniRef100_Q93V99 Protodermal factor2 [Arabidopsis thaliana]
Length = 743
Score = 187 bits (475), Expect = 4e-46
Identities = 100/237 (42%), Positives = 143/237 (60%), Gaps = 26/237 (10%)
Query: 136 KKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIK 195
+ K+K+YHRHT QI+ +E+ FKE PHPD+KQR++LS+ L L P QVKFWFQN+RTQ+K
Sbjct: 59 RPNKKKRYHRHTQRQIQELESFFKECPHPDDKQRKELSRDLNLEPLQVKFWFQNKRTQMK 118
Query: 196 AIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIENA 255
A ERHEN +LKS+ +KLR +N +E ++ A CPNCG P G M+ +EQ LRIENA
Sbjct: 119 AQSERHENQILKSDNDKLRAENNRYKEALSNATCPNCGGPAA--IGEMSFDEQHLRIENA 176
Query: 256 KLKAEVERLRAALGKYASGTM----------SPSCSTSHDQENIKSSLDFYTGIFC---- 301
+L+ E++R+ A KY + +PS S + N + F ++
Sbjct: 177 RLREEIDRISAIAAKYVGKPLGSSFAPLAIHAPSRSLDLEVGNFGNQTGFVGEMYGTGDI 236
Query: 302 ---------LDESRIMDVVNQAMEELIKMATMGEPMWLRSLETGREILNYDEYMKEF 349
D+ I+++ AMEEL++MA G+P+WL S + EILN +EY + F
Sbjct: 237 LRSVSIPSETDKPIIVELAVAAMEELVRMAQTGDPLWL-STDNSVEILNEEEYFRTF 292
>UniRef100_Q94C37 At1g05230/YUP8H12_16 [Arabidopsis thaliana]
Length = 721
Score = 182 bits (462), Expect = 1e-44
Identities = 112/263 (42%), Positives = 149/263 (56%), Gaps = 31/263 (11%)
Query: 106 DEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPD 165
DE++ + E+ EG D D ++ NKKK+ YHRHT QI+ MEA FKE PHPD
Sbjct: 35 DEFDSPNTKSGS-ENQEGGSGNDQDPLHPNKKKR---YHRHTQLQIQEMEAFFKECPHPD 90
Query: 166 EKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETIN 225
+KQR+QLS++L L P QVKFWFQN+RTQ+K ERHENS L++E EKLR N RE +
Sbjct: 91 DKQRKQLSRELNLEPLQVKFWFQNKRTQMKNHHERHENSHLRAENEKLRNDNLRYREALA 150
Query: 226 KACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGT------MSPS 279
A CPNCG PT G M+ +E QLR+ENA+L+ E++R+ A KY MSP
Sbjct: 151 NASCPNCGGPTAI--GEMSFDEHQLRLENARLREEIDRISAIAAKYVGKPVSNYPLMSPP 208
Query: 280 CSTSHDQE-------------NIKSSLDFYTGIFCLDESRIMDVVNQAMEELIKMATMGE 326
E N L T D+ I+D+ AMEEL++M + E
Sbjct: 209 PLPPRPLELAMGNIGGEAYGNNPNDLLKSITAPTESDKPVIIDLSVAAMEELMRMVQVDE 268
Query: 327 PMWLRSLETGREILNYDEYMKEF 349
P+W +SL +L+ +EY + F
Sbjct: 269 PLW-KSL-----VLDEEEYARTF 285
>UniRef100_Q9ZPH7 T1J1.3 protein [Arabidopsis thaliana]
Length = 772
Score = 179 bits (453), Expect = 1e-43
Identities = 101/252 (40%), Positives = 144/252 (57%), Gaps = 41/252 (16%)
Query: 136 KKKKRKKYHRHTSEQIRVMEAL---------------FKESPHPDEKQRQQLSKQLGLAP 180
+ K+K+YHRHT QI+ +E+L FKE PHPD+KQR++LS+ L L P
Sbjct: 54 RPNKKKRYHRHTQRQIQELESLKTFSYILSFLEFYRFFKECPHPDDKQRKELSRDLNLEP 113
Query: 181 RQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRD 240
QVKFWFQN+RTQ+KA ERHEN +LKS+ +KLR +N +E ++ A CPNCG P
Sbjct: 114 LQVKFWFQNKRTQMKAQSERHENQILKSDNDKLRAENNRYKEALSNATCPNCGGPAA--I 171
Query: 241 GTMATEEQQLRIENAKLKAEVERLRAALGKYASGTM----------SPSCSTSHDQENIK 290
G M+ +EQ LRIENA+L+ E++R+ A KY + +PS S + N
Sbjct: 172 GEMSFDEQHLRIENARLREEIDRISAIAAKYVGKPLGSSFAPLAIHAPSRSLDLEVGNFG 231
Query: 291 SSLDFYTGIFC-------------LDESRIMDVVNQAMEELIKMATMGEPMWLRSLETGR 337
+ F ++ D+ I+++ AMEEL++MA G+P+WL S +
Sbjct: 232 NQTGFVGEMYGTGDILRSVSIPSETDKPIIVELAVAAMEELVRMAQTGDPLWL-STDNSV 290
Query: 338 EILNYDEYMKEF 349
EILN +EY + F
Sbjct: 291 EILNEEEYFRTF 302
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.309 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 625,247,522
Number of Sequences: 2790947
Number of extensions: 28148690
Number of successful extensions: 209601
Number of sequences better than 10.0: 7804
Number of HSP's better than 10.0 without gapping: 4991
Number of HSP's successfully gapped in prelim test: 3013
Number of HSP's that attempted gapping in prelim test: 177797
Number of HSP's gapped (non-prelim): 18047
length of query: 365
length of database: 848,049,833
effective HSP length: 129
effective length of query: 236
effective length of database: 488,017,670
effective search space: 115172170120
effective search space used: 115172170120
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC149489.2


