

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149471.8 + phase: 0
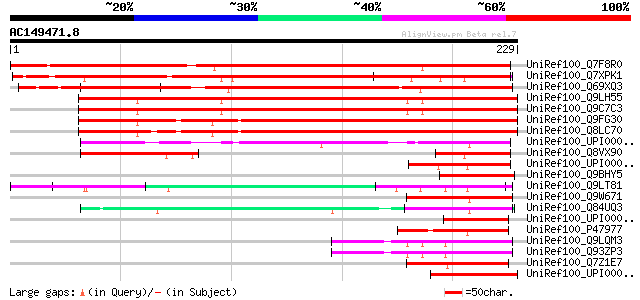
(229 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7F8R0 KH domain-containing protein-like [Oryza sativa] 256 2e-67
UniRef100_Q7XPK1 OSJNBa0087O24.9 protein [Oryza sativa] 243 4e-63
UniRef100_Q69XQ3 KH domain-containing protein / zinc finger prot... 234 2e-60
UniRef100_Q9LH55 Similarity to unknown protein [Arabidopsis thal... 181 1e-44
UniRef100_Q9C7C3 Hypothetical protein T21B14.5 [Arabidopsis thal... 181 1e-44
UniRef100_Q9FG30 Similarity to unknown protein [Arabidopsis thal... 176 6e-43
UniRef100_Q8LC70 Hypothetical protein [Arabidopsis thaliana] 176 6e-43
UniRef100_UPI0000343263 UPI0000343263 UniRef100 entry 91 2e-17
UniRef100_Q8VX90 Similarity to the EST C99174 [Pinus pinaster] 59 8e-08
UniRef100_UPI00002A92C8 UPI00002A92C8 UniRef100 entry 53 7e-06
UniRef100_Q9BHY5 Hypothetical zf-CCCH protein L5213.07 [Leishman... 51 2e-05
UniRef100_Q9LT81 Gb|AAC36178.1 [Arabidopsis thaliana] 51 3e-05
UniRef100_Q9W671 CCCH zinc finger protein C3H-1 [Xenopus laevis] 50 5e-05
UniRef100_Q84UQ3 Putative zinc finger protein [Oryza sativa] 50 6e-05
UniRef100_UPI000034C487 UPI000034C487 UniRef100 entry 49 8e-05
UniRef100_P47977 Zinc finger protein CTH2 [Saccharomyces cerevis... 48 2e-04
UniRef100_Q9LQM3 F5D14.12 protein [Arabidopsis thaliana] 47 3e-04
UniRef100_Q93ZP3 At1g32359/F27G20.10 [Arabidopsis thaliana] 47 3e-04
UniRef100_Q7Z1E7 FIP1-like protein [Trypanosoma cruzi] 47 3e-04
UniRef100_UPI000046C0E9 UPI000046C0E9 UniRef100 entry 47 4e-04
>UniRef100_Q7F8R0 KH domain-containing protein-like [Oryza sativa]
Length = 300
Score = 256 bits (655), Expect = 2e-67
Identities = 131/232 (56%), Positives = 161/232 (68%), Gaps = 11/232 (4%)
Query: 1 MINVGSSPAIPPIGRNPNVPQSFPDGSSPPVAKTRLCNKFNTAEGCKFGDKCHFAHGEWE 60
M N+G PP GR P + + PDG P KTRLCNK+NTAEGCK+GDKCHFAHGE E
Sbjct: 70 MTNLGGPAIAPPPGRMP-MGNAVPDGPPTPTVKTRLCNKYNTAEGCKWGDKCHFAHGERE 128
Query: 61 LGRPTVPAYEDTRAMGQMQSSSVGGRIEPPP---PAHGAAAGFGVSATATVSINATLAGA 117
LG+P + MG + G PPP PA A FG SATA +S++A+LAG
Sbjct: 129 LGKPMLMDSSMPPPMGPRPT----GHFAPPPMPSPAMSTPASFGASATAKISVDASLAGG 184
Query: 118 IIGKNDVNSKQICHITGAKLSIREHDSDPNLRNIELEGSFDQIKQASAMVHDLILNVSSV 177
IIG+ VN+KQI +TGAKL+IR+H+SD NL+NIELEG+FDQIK ASAMV +LI+++
Sbjct: 185 IIGRGGVNTKQISRVTGAKLAIRDHESDTNLKNIELEGTFDQIKNASAMVRELIVSIGGG 244
Query: 178 SGPPGKNI---TSQTSAPANNFKTKLCENFTKGSCTFGERCHFAHGTDELRK 226
+ P GK + + P +NFKTKLCENFTKGSCTFG+RCHFAHG +ELRK
Sbjct: 245 APPQGKKPVGGSHRGGGPGSNFKTKLCENFTKGSCTFGDRCHFAHGENELRK 296
Score = 43.9 bits (102), Expect = 0.003
Identities = 21/56 (37%), Positives = 30/56 (53%), Gaps = 4/56 (7%)
Query: 176 SVSGPPGKNITSQT---SAPANNFKTKLCENF-TKGSCTFGERCHFAHGTDELRKP 227
+++ PPG+ P KT+LC + T C +G++CHFAHG EL KP
Sbjct: 77 AIAPPPGRMPMGNAVPDGPPTPTVKTRLCNKYNTAEGCKWGDKCHFAHGERELGKP 132
>UniRef100_Q7XPK1 OSJNBa0087O24.9 protein [Oryza sativa]
Length = 309
Score = 243 bits (619), Expect = 4e-63
Identities = 132/237 (55%), Positives = 164/237 (68%), Gaps = 17/237 (7%)
Query: 2 INVGSSPAIPPIGRNPNVPQSFPDGSSPPVA--KTRLCNKFNTAEGCKFGDKCHFAHGEW 59
+N+G+ PA+P R P G+S P + KTR+C K+NTAEGCKFGDKCHFAHGE
Sbjct: 75 LNLGN-PAVPAPARAPM--DHAAGGNSHPASSGKTRMCTKYNTAEGCKFGDKCHFAHGER 131
Query: 60 ELGRPTVPAYEDTRAMGQMQSSSVGGRIEPPPPAH-GAAAG-FGVSATATVSINATLAGA 117
ELG+P ++E AM GGR EPPPPA G AG FG SATA +S++A+LAG
Sbjct: 132 ELGKPAYMSHES--AMAPPMGGRYGGRPEPPPPAAMGPPAGNFGASATAKISVDASLAGG 189
Query: 118 IIGKNDVNSKQICHITGAKLSIREHDSDPNLRNIELEGSFDQIKQASAMVHDLILNVSSV 177
IIGK VN+KQIC +TG KLSIR+H+SD NL+NIELEG+FDQIKQAS MV +LI +S
Sbjct: 190 IIGKGGVNTKQICRVTGVKLSIRDHESDSNLKNIELEGNFDQIKQASNMVGELIATISPS 249
Query: 178 SGPPGKNITSQTSAPA--------NNFKTKLCENFTKGSCTFGERCHFAHGTDELRK 226
+ ++ +APA +N+KTKLCENF KG+CTFG+RCHFAHG +E RK
Sbjct: 250 TPAKKPAGSAAGAAPAGRGGPGGRSNYKTKLCENFVKGTCTFGDRCHFAHGENEQRK 306
Score = 47.0 bits (110), Expect = 4e-04
Identities = 26/67 (38%), Positives = 36/67 (52%), Gaps = 4/67 (5%)
Query: 165 AMVHDLILNVSSVSGP---PGKNITSQTSAPANNFKTKLCENF-TKGSCTFGERCHFAHG 220
A+ L L +V P P + S PA++ KT++C + T C FG++CHFAHG
Sbjct: 70 AVAKTLNLGNPAVPAPARAPMDHAAGGNSHPASSGKTRMCTKYNTAEGCKFGDKCHFAHG 129
Query: 221 TDELRKP 227
EL KP
Sbjct: 130 ERELGKP 136
>UniRef100_Q69XQ3 KH domain-containing protein / zinc finger protein-like [Oryza
sativa]
Length = 295
Score = 234 bits (596), Expect = 2e-60
Identities = 125/227 (55%), Positives = 155/227 (68%), Gaps = 13/227 (5%)
Query: 5 GSSPAIPPIGRNPNVPQSFPDGSSPPVAKTRLCNKFNTAEGCKFGDKCHFAHGEWELGRP 64
G++ A PP GR P P + P+G KTR+CNK+NTAEGCK+G KCHFAHGE ELG+P
Sbjct: 74 GTAVAAPP-GRMPLGPGA-PNGPPTSSVKTRMCNKYNTAEGCKWGSKCHFAHGERELGKP 131
Query: 65 TVPAYEDTRAMGQMQSSSVGGRIEPPPPAHGAA--AGFGVSATATVSINATLAGAIIGKN 122
+ MG M + PP P + FG SATA +S++A+LAG IIGK
Sbjct: 132 MLLDNSMPHPMGSMPFEA------PPMPGPDIVPPSTFGASATAKISVDASLAGGIIGKG 185
Query: 123 DVNSKQICHITGAKLSIREHDSDPNLRNIELEGSFDQIKQASAMVHDLILNVSSVSGPPG 182
N+K I +TGAKL+IR+++S+PNL+NIELEG+FDQIK ASAMV +LI+ +S + PP
Sbjct: 186 GTNTKHISRMTGAKLAIRDNESNPNLKNIELEGTFDQIKHASAMVTELIVRISG-NAPPA 244
Query: 183 KN--ITSQTSAPANNFKTKLCENFTKGSCTFGERCHFAHGTDELRKP 227
KN S P +NFKTKLCENF KGSCTFG+RCHFAHG ELRKP
Sbjct: 245 KNPGRGSHAGGPGSNFKTKLCENFNKGSCTFGDRCHFAHGESELRKP 291
Score = 48.1 bits (113), Expect = 2e-04
Identities = 21/36 (58%), Positives = 24/36 (66%), Gaps = 1/36 (2%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTVPA 68
KT+LC FN C FGD+CHFAHGE EL +P A
Sbjct: 261 KTKLCENFNKGS-CTFGDRCHFAHGESELRKPPAAA 295
Score = 34.7 bits (78), Expect = 2.0
Identities = 12/25 (48%), Positives = 15/25 (60%)
Query: 32 AKTRLCNKFNTAEGCKFGDKCHFAH 56
+K + C KF + GC FG CHF H
Sbjct: 33 SKLKPCTKFFSTSGCPFGSSCHFLH 57
>UniRef100_Q9LH55 Similarity to unknown protein [Arabidopsis thaliana]
Length = 231
Score = 181 bits (459), Expect = 1e-44
Identities = 100/211 (47%), Positives = 132/211 (62%), Gaps = 13/211 (6%)
Query: 32 AKTRLCNKFNTAEGCKFGDKCHFAH----GEWELGRPTVPAYEDTRAMGQMQSSSVGGRI 87
+K++ C KF + GC FG+ CHF H G + + T + MQ S GGR
Sbjct: 20 SKSKPCTKFFSTSGCPFGENCHFLHYVPGGYNAVSQMTNMGPPIPQVSRNMQGSGNGGRF 79
Query: 88 EPPPPAH-GAAAGFGVSATATVSINATLAGAIIGKNDVNSKQICHITGAKLSIREHDSDP 146
+ G + FG SATA S++A+LAGAIIGK V+SKQIC TG KLSI++H+ DP
Sbjct: 80 SGRGESGPGHVSNFGDSATARFSVDASLAGAIIGKGGVSSKQICRQTGVKLSIQDHERDP 139
Query: 147 NLRNIELEGSFDQIKQASAMVHDLILNVSSVS-GPPGKNI-------TSQTSAPANNFKT 198
NL+NI LEG+ +QI +ASAMV DLI ++S + PPG + + P +NFKT
Sbjct: 140 NLKNIVLEGTLEQISEASAMVKDLIGRLNSAAKKPPGGGLGGGGGMGSEGKPHPGSNFKT 199
Query: 199 KLCENFTKGSCTFGERCHFAHGTDELRKPGM 229
K+CE F+KG+CTFG+RCHFAHG ELRK G+
Sbjct: 200 KICERFSKGNCTFGDRCHFAHGEAELRKSGI 230
>UniRef100_Q9C7C3 Hypothetical protein T21B14.5 [Arabidopsis thaliana]
Length = 248
Score = 181 bits (459), Expect = 1e-44
Identities = 100/211 (47%), Positives = 132/211 (62%), Gaps = 13/211 (6%)
Query: 32 AKTRLCNKFNTAEGCKFGDKCHFAH----GEWELGRPTVPAYEDTRAMGQMQSSSVGGRI 87
+K++ C KF + GC FG+ CHF H G + + T + MQ S GGR
Sbjct: 37 SKSKPCTKFFSTSGCPFGENCHFLHYVPGGYNAVSQMTNMGPPIPQVSRNMQGSGNGGRF 96
Query: 88 EPPPPAH-GAAAGFGVSATATVSINATLAGAIIGKNDVNSKQICHITGAKLSIREHDSDP 146
+ G + FG SATA S++A+LAGAIIGK V+SKQIC TG KLSI++H+ DP
Sbjct: 97 SGRGESGPGHVSNFGDSATARFSVDASLAGAIIGKGGVSSKQICRQTGVKLSIQDHERDP 156
Query: 147 NLRNIELEGSFDQIKQASAMVHDLILNVSSVS-GPPGKNI-------TSQTSAPANNFKT 198
NL+NI LEG+ +QI +ASAMV DLI ++S + PPG + + P +NFKT
Sbjct: 157 NLKNIVLEGTLEQISEASAMVKDLIGRLNSAAKKPPGGGLGGGGGMGSEGKPHPGSNFKT 216
Query: 199 KLCENFTKGSCTFGERCHFAHGTDELRKPGM 229
K+CE F+KG+CTFG+RCHFAHG ELRK G+
Sbjct: 217 KICERFSKGNCTFGDRCHFAHGEAELRKSGI 247
>UniRef100_Q9FG30 Similarity to unknown protein [Arabidopsis thaliana]
Length = 240
Score = 176 bits (445), Expect = 6e-43
Identities = 93/206 (45%), Positives = 128/206 (61%), Gaps = 11/206 (5%)
Query: 32 AKTRLCNKFNTAEGCKFGDKCHFAH-------GEWELGRPTVPAYEDTRAMGQMQSSSVG 84
+K++ C KF + GC FGD CHF H ++ P + +R M S G
Sbjct: 37 SKSKPCTKFFSTSGCPFGDNCHFLHYVPGGYNAAAQMTNLRPPVSQVSRNM--QGSGGPG 94
Query: 85 GRIEPP-PPAHGAAAGFGVSATATVSINATLAGAIIGKNDVNSKQICHITGAKLSIREHD 143
GR P G + FG S T+ +S++A+LAGAIIGK ++SKQIC TGAKLSI++H+
Sbjct: 95 GRFSGRGDPGSGPVSIFGAS-TSKISVDASLAGAIIGKGGIHSKQICRETGAKLSIKDHE 153
Query: 144 SDPNLRNIELEGSFDQIKQASAMVHDLILNVSSVSGPPGKNITSQTSAPANNFKTKLCEN 203
DPNL+ IELEG+F+QI AS MV +LI + SV P G P +N+KTK+C+
Sbjct: 154 RDPNLKIIELEGTFEQINVASGMVRELIGRLGSVKKPQGIGGPEGKPHPGSNYKTKICDR 213
Query: 204 FTKGSCTFGERCHFAHGTDELRKPGM 229
++KG+CT+G+RCHFAHG ELR+ G+
Sbjct: 214 YSKGNCTYGDRCHFAHGESELRRSGI 239
>UniRef100_Q8LC70 Hypothetical protein [Arabidopsis thaliana]
Length = 240
Score = 176 bits (445), Expect = 6e-43
Identities = 95/208 (45%), Positives = 130/208 (61%), Gaps = 15/208 (7%)
Query: 32 AKTRLCNKFNTAEGCKFGDKCHFAH---------GEWELGRPTVPAYEDTRAMGQMQSSS 82
+K++ C KF + GC FGD CHF H + + RP P + +R M S
Sbjct: 37 SKSKPCTKFFSTSGCPFGDNCHFLHYVPGGYNAAAQMKNLRP--PVSQVSRNM--QGSGG 92
Query: 83 VGGRIEPP-PPAHGAAAGFGVSATATVSINATLAGAIIGKNDVNSKQICHITGAKLSIRE 141
GGR P G + FG S T+ +S++A+LAGAIIGK ++SKQIC TGAKLSI++
Sbjct: 93 PGGRFSGRGDPGSGPVSIFGAS-TSKISVDASLAGAIIGKGGIHSKQICRETGAKLSIKD 151
Query: 142 HDSDPNLRNIELEGSFDQIKQASAMVHDLILNVSSVSGPPGKNITSQTSAPANNFKTKLC 201
H+ DPNL+ IELEG+F+QI AS MV +LI + SV P G P +N+KTK+C
Sbjct: 152 HERDPNLKIIELEGTFEQINVASGMVRELIGRLGSVKKPQGIGGPEGKPHPGSNYKTKIC 211
Query: 202 ENFTKGSCTFGERCHFAHGTDELRKPGM 229
+ ++KG+CT+G+RCHFAHG ELR+ G+
Sbjct: 212 DRYSKGNCTYGDRCHFAHGESELRRSGI 239
>UniRef100_UPI0000343263 UPI0000343263 UniRef100 entry
Length = 246
Score = 90.9 bits (224), Expect = 2e-17
Identities = 63/197 (31%), Positives = 96/197 (47%), Gaps = 33/197 (16%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTVPAYEDTRAMGQMQSSSVGGRIEPPPP 92
KTRLC F + EGC+FGD+C FAHG+ EL +D G M++
Sbjct: 76 KTRLCRNFQSPEGCRFGDRCSFAHGKGEL------RSDDLNGDGTMENLE---------- 119
Query: 93 AHGAAAGFGVSATATVSINATLAGAIIGKNDVNSKQICHITGAKLSI--REHDSDPNLRN 150
GFG V + + GAI+GK + Q+ +GAK+S+ E+ + R
Sbjct: 120 ---THQGFG--KAVLVPVPQSQVGAIVGKAGSSIAQVSAASGAKVSMLSAEYTNSDGDRL 174
Query: 151 IELEGSFDQIKQASAMVHDLILNVSSVSGPPGKNITSQTSAPANNFKTKLCENFTK-GSC 209
L GS +++A +++ + GK + + + P KTK+C + K GSC
Sbjct: 175 CRLVGSPFDVQRAKELIYHRL--------AVGK-VKKRDTKPPRPCKTKICALWQKDGSC 225
Query: 210 TFGERCHFAHGTDELRK 226
+G++CHFAHG EL+K
Sbjct: 226 MYGDKCHFAHGIAELQK 242
Score = 40.4 bits (93), Expect = 0.036
Identities = 17/37 (45%), Positives = 21/37 (55%)
Query: 25 DGSSPPVAKTRLCNKFNTAEGCKFGDKCHFAHGEWEL 61
D P KT++C + C +GDKCHFAHG EL
Sbjct: 204 DTKPPRPCKTKICALWQKDGSCMYGDKCHFAHGIAEL 240
>UniRef100_Q8VX90 Similarity to the EST C99174 [Pinus pinaster]
Length = 80
Score = 59.3 bits (142), Expect = 8e-08
Identities = 31/56 (55%), Positives = 36/56 (63%), Gaps = 3/56 (5%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTVPAY--EDTRAMGQMQSS-SVGG 85
KTRLC+ F T GC+FGDKCHFAHGE ELG+ A+ +D A G S VGG
Sbjct: 5 KTRLCSNFGTDTGCRFGDKCHFAHGEKELGKVNAVAHNLKDDLATGPFGSRFPVGG 60
Score = 49.3 bits (116), Expect = 8e-05
Identities = 21/35 (60%), Positives = 26/35 (74%), Gaps = 1/35 (2%)
Query: 193 ANNFKTKLCENF-TKGSCTFGERCHFAHGTDELRK 226
A+N+KT+LC NF T C FG++CHFAHG EL K
Sbjct: 1 ASNYKTRLCSNFGTDTGCRFGDKCHFAHGEKELGK 35
>UniRef100_UPI00002A92C8 UPI00002A92C8 UniRef100 entry
Length = 110
Score = 52.8 bits (125), Expect = 7e-06
Identities = 26/58 (44%), Positives = 36/58 (61%), Gaps = 12/58 (20%)
Query: 181 PGKNITSQTSAP-----------ANNFKTKLCENFT-KGSCTFGERCHFAHGTDELRK 226
P +SQTS+ A+NFKT +C+N+ KG C FG++C+FAHG DELR+
Sbjct: 44 PDSETSSQTSSDRRSECSVCIGGASNFKTSMCKNYREKGFCKFGDKCNFAHGKDELRR 101
Score = 41.6 bits (96), Expect = 0.016
Identities = 17/31 (54%), Positives = 21/31 (66%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWELGR 63
KT +C + CKFGDKC+FAHG+ EL R
Sbjct: 71 KTSMCKNYREKGFCKFGDKCNFAHGKDELRR 101
Score = 40.4 bits (93), Expect = 0.036
Identities = 17/28 (60%), Positives = 21/28 (74%), Gaps = 1/28 (3%)
Query: 200 LCENFT-KGSCTFGERCHFAHGTDELRK 226
+C+NF KG C FG +C+FAHG ELRK
Sbjct: 1 MCKNFRDKGFCKFGAKCNFAHGESELRK 28
Score = 37.4 bits (85), Expect = 0.31
Identities = 20/54 (37%), Positives = 24/54 (44%), Gaps = 9/54 (16%)
Query: 36 LCNKFNTAEGCKFGDKCHFAHGEWELGR---------PTVPAYEDTRAMGQMQS 80
+C F CKFG KC+FAHGE EL + P P D+ Q S
Sbjct: 1 MCKNFRDKGFCKFGAKCNFAHGESELRKLDEAMGLSLPARPQSPDSETSSQTSS 54
>UniRef100_Q9BHY5 Hypothetical zf-CCCH protein L5213.07 [Leishmania major]
Length = 364
Score = 51.2 bits (121), Expect = 2e-05
Identities = 19/34 (55%), Positives = 26/34 (75%)
Query: 195 NFKTKLCENFTKGSCTFGERCHFAHGTDELRKPG 228
N+KT+LC+ F +G C G +C+FAHG ELR+PG
Sbjct: 208 NYKTRLCKGFAEGHCNRGNQCNFAHGVGELRQPG 241
Score = 38.9 bits (89), Expect = 0.10
Identities = 28/69 (40%), Positives = 34/69 (48%), Gaps = 10/69 (14%)
Query: 33 KTRLCNKFNTAEG-CKFGDKCHFAHGEWEL-----GRPTVPAYEDTRAMGQM--QSSSVG 84
KTRLC F AEG C G++C+FAHG EL G P P + + M Q +
Sbjct: 210 KTRLCKGF--AEGHCNRGNQCNFAHGVGELRQPGGGEPIPPPPQQQQHMPQQLPPAGLAA 267
Query: 85 GRIEPPPPA 93
G PPP A
Sbjct: 268 GGSAPPPMA 276
>UniRef100_Q9LT81 Gb|AAC36178.1 [Arabidopsis thaliana]
Length = 386
Score = 50.8 bits (120), Expect = 3e-05
Identities = 30/74 (40%), Positives = 38/74 (50%), Gaps = 13/74 (17%)
Query: 1 MINVGSSPAIPPIGRNPNVPQSFPDGSSPPVA-------------KTRLCNKFNTAEGCK 47
+I+VG++ A P N+ + GS P A KTRLC KF+ C
Sbjct: 226 VISVGATAADQPSDTASNLIEVNRQGSIPVPAPMNNGGVVKTVYWKTRLCMKFDITGQCP 285
Query: 48 FGDKCHFAHGEWEL 61
FGDKCHFAHG+ EL
Sbjct: 286 FGDKCHFAHGQAEL 299
Score = 50.8 bits (120), Expect = 3e-05
Identities = 59/224 (26%), Positives = 80/224 (35%), Gaps = 28/224 (12%)
Query: 20 PQSFPDGSSPPVAK--------TRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTVPAYE- 70
P P + PPV K TR+C KF A C+ G+ C+FAHG +L +P E
Sbjct: 85 PWMVPSLNPPPVNKGTANIFYKTRMCAKFR-AGTCRNGELCNFAHGIEDLRQPPSNWQEI 143
Query: 71 -DTRAMGQMQSSSVGGRIEPPPPAHGAAAGFGVSATATVSINATLAGAIIGKNDVNSKQI 129
GQ + E P+ + + L +
Sbjct: 144 VGPPPAGQDRERERERERERERPSLAPVVNNNWEDDQKIILRMKLCRKFCFGEECPYGDR 203
Query: 130 CHITGAKLSIREHDSDPNLRNIELEGSFDQIKQASAMVHDLILNVSSVSGPPGKNITSQT 189
C+ LS DS + + Q S +LI V+ P
Sbjct: 204 CNFIHEDLSKFREDSGKLRESSVISVGATAADQPSDTASNLI-EVNRQGSIP-------V 255
Query: 190 SAPANN--------FKTKLCENFT-KGSCTFGERCHFAHGTDEL 224
AP NN +KT+LC F G C FG++CHFAHG EL
Sbjct: 256 PAPMNNGGVVKTVYWKTRLCMKFDITGQCPFGDKCHFAHGQAEL 299
Score = 46.2 bits (108), Expect = 7e-04
Identities = 26/72 (36%), Positives = 37/72 (51%), Gaps = 10/72 (13%)
Query: 166 MVHDLILN-VSSVSGPPGKN---ITSQTSAPANN------FKTKLCENFTKGSCTFGERC 215
+V D + N SS P N + S P N +KT++C F G+C GE C
Sbjct: 65 LVDDNLFNPASSFPQPSSSNPWMVPSLNPPPVNKGTANIFYKTRMCAKFRAGTCRNGELC 124
Query: 216 HFAHGTDELRKP 227
+FAHG ++LR+P
Sbjct: 125 NFAHGIEDLRQP 136
>UniRef100_Q9W671 CCCH zinc finger protein C3H-1 [Xenopus laevis]
Length = 313
Score = 50.1 bits (118), Expect = 5e-05
Identities = 22/49 (44%), Positives = 31/49 (62%), Gaps = 1/49 (2%)
Query: 180 PPGKNITSQTSAPANNFKTKLCENFTK-GSCTFGERCHFAHGTDELRKP 227
PP K AP+ +KT+LC F++ G+C +G +C FAHG ELR+P
Sbjct: 88 PPLKTALPALPAPSPRYKTELCRTFSETGTCKYGAKCQFAHGKIELREP 136
Score = 43.9 bits (102), Expect = 0.003
Identities = 25/58 (43%), Positives = 31/58 (53%), Gaps = 3/58 (5%)
Query: 8 PAIPPIGRNPNVPQSFPDGSSP-PVAKTRLCNKFNTAEGCKFGDKCHFAHGEWELGRP 64
PA PP P + + P +P P KT LC F+ CK+G KC FAHG+ EL P
Sbjct: 81 PAPPP--GFPPLKTALPALPAPSPRYKTELCRTFSETGTCKYGAKCQFAHGKIELREP 136
Score = 35.4 bits (80), Expect = 1.2
Identities = 25/75 (33%), Positives = 30/75 (39%), Gaps = 13/75 (17%)
Query: 30 PVAKTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTVPAYEDTRAMGQMQSSSVGG---- 85
P KT LC+KF C +G +C+F H P + T QS S G
Sbjct: 140 PKYKTELCHKFYLYGECPYGSRCNFIHH---------PREQGTSQHILRQSLSYSGVPTR 190
Query: 86 RIEPPPPAHGAAAGF 100
R PPPP A F
Sbjct: 191 RGSPPPPGLPDPAAF 205
>UniRef100_Q84UQ3 Putative zinc finger protein [Oryza sativa]
Length = 367
Score = 49.7 bits (117), Expect = 6e-05
Identities = 50/206 (24%), Positives = 78/206 (37%), Gaps = 25/206 (12%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPT------VPAYEDTRAMGQMQSSSVGGR 86
KT+LC KF A C + C+FAHG EL +P V A+E+ + +
Sbjct: 89 KTKLCCKFR-AGTCPYVTNCNFAHGMEELRKPPPNWQEIVAAHEEATEAREEHQIPIMTS 147
Query: 87 IEPPPPAHGAAAGFGVSATATVSINATLAGAIIGKNDVNSKQICHITGAKLSIREHDS-- 144
P G G + + + + RE +
Sbjct: 148 SGPTAGGDAGCGGGGGGGSGRAYKGRHCKKFYTDEGCPYGDACTFLHDEQSKARESVAIS 207
Query: 145 -DPNLRNIELEGSFDQIKQASAMVHDLILNVSSVSGPPGKNITSQTSAPANNFKTKLCEN 203
P++ GS++ A+A + ++ +GP K +N+KT++C
Sbjct: 208 LSPSVGGGGGGGSYNSAAAAAASA-----SAAAGNGPMQK---------PSNWKTRICNK 253
Query: 204 FTK-GSCTFGERCHFAHGTDELRKPG 228
+ G C FG +CHFAHG EL K G
Sbjct: 254 WEMTGYCPFGSKCHFAHGAAELHKYG 279
Score = 48.1 bits (113), Expect = 2e-04
Identities = 22/49 (44%), Positives = 28/49 (56%)
Query: 179 GPPGKNITSQTSAPANNFKTKLCENFTKGSCTFGERCHFAHGTDELRKP 227
G G N + + FKTKLC F G+C + C+FAHG +ELRKP
Sbjct: 71 GEGGGNTSKSRAIGKMFFKTKLCCKFRAGTCPYVTNCNFAHGMEELRKP 119
Score = 44.3 bits (103), Expect = 0.002
Identities = 18/29 (62%), Positives = 20/29 (68%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWEL 61
KTR+CNK+ C FG KCHFAHG EL
Sbjct: 247 KTRICNKWEMTGYCPFGSKCHFAHGAAEL 275
Score = 42.4 bits (98), Expect = 0.009
Identities = 32/106 (30%), Positives = 40/106 (37%), Gaps = 17/106 (16%)
Query: 26 GSSPPVAKTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTVPAYEDTRAMGQMQSSSVGG 85
G S K R C KF T EGC +GD C F H E R +V S SVGG
Sbjct: 164 GGSGRAYKGRHCKKFYTDEGCPYGDACTFLHDEQSKARESVAI---------SLSPSVGG 214
Query: 86 RIEPPPPAHGAAAGFGVSATATVSINATLAGAIIGKNDVNSKQICH 131
G + +A A S +A + K +IC+
Sbjct: 215 --------GGGGGSYNSAAAAAASASAAAGNGPMQKPSNWKTRICN 252
>UniRef100_UPI000034C487 UPI000034C487 UniRef100 entry
Length = 70
Score = 49.3 bits (116), Expect = 8e-05
Identities = 20/29 (68%), Positives = 23/29 (78%)
Query: 197 KTKLCENFTKGSCTFGERCHFAHGTDELR 225
KT+LC+NF GSC FGERC +AHG ELR
Sbjct: 38 KTELCKNFEAGSCRFGERCSYAHGLHELR 66
Score = 35.0 bits (79), Expect = 1.5
Identities = 15/29 (51%), Positives = 19/29 (64%), Gaps = 1/29 (3%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWEL 61
KT LC F A C+FG++C +AHG EL
Sbjct: 38 KTELCKNFE-AGSCRFGERCSYAHGLHEL 65
>UniRef100_P47977 Zinc finger protein CTH2 [Saccharomyces cerevisiae]
Length = 285
Score = 47.8 bits (112), Expect = 2e-04
Identities = 24/51 (47%), Positives = 33/51 (64%), Gaps = 3/51 (5%)
Query: 176 SVSGPPGKNITSQTSAPANNFKTKLCENFT-KGSCTFGERCHFAHGTDELR 225
S + P K+ +T P +KT+LCE+FT KGSC +G +C FAHG EL+
Sbjct: 152 SSAQPKVKSQVQET--PKQLYKTELCESFTLKGSCPYGSKCQFAHGLGELK 200
Score = 37.0 bits (84), Expect = 0.40
Identities = 15/29 (51%), Positives = 16/29 (54%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWEL 61
KT LC F C +G KC FAHG EL
Sbjct: 171 KTELCESFTLKGSCPYGSKCQFAHGLGEL 199
Score = 36.6 bits (83), Expect = 0.52
Identities = 16/29 (55%), Positives = 19/29 (65%), Gaps = 1/29 (3%)
Query: 195 NFKTKLCENFTK-GSCTFGERCHFAHGTD 222
NF+TK C N+ K G C +G RC F HG D
Sbjct: 207 NFRTKPCVNWEKLGYCPYGRRCCFKHGDD 235
>UniRef100_Q9LQM3 F5D14.12 protein [Arabidopsis thaliana]
Length = 384
Score = 47.4 bits (111), Expect = 3e-04
Identities = 35/99 (35%), Positives = 47/99 (47%), Gaps = 20/99 (20%)
Query: 146 PNLRNIELEGSFDQIKQASAMVHDLILNVSSVS-GPPGKNI--------TSQTSAPANN- 195
PNL + + + D +A AM D N S + GPP K + TSA +N
Sbjct: 28 PNLGDSAVWATEDDYNRAWAMNPD---NTSGDNNGPPNKKTRGSPSSSSATTTSAASNRT 84
Query: 196 -------FKTKLCENFTKGSCTFGERCHFAHGTDELRKP 227
FKTKLC F G+C + C+FAH +ELR+P
Sbjct: 85 KAIGKMFFKTKLCCKFRAGTCPYITNCNFAHTVEELRRP 123
Score = 44.7 bits (104), Expect = 0.002
Identities = 19/31 (61%), Positives = 21/31 (67%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWELGR 63
KTR+CNK+ C FG KCHFAHG EL R
Sbjct: 262 KTRICNKWEITGYCPFGAKCHFAHGAAELHR 292
Score = 42.7 bits (99), Expect = 0.007
Identities = 54/215 (25%), Positives = 86/215 (39%), Gaps = 32/215 (14%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTVPAYEDTRAMGQMQSSSVGGRIEPPPP 92
KT+LC KF A C + C+FAH EL RP P +++ A + + S G + P
Sbjct: 93 KTKLCCKFR-AGTCPYITNCNFAHTVEELRRPP-PNWQEIVAAHEEERS---GGMGTPTV 147
Query: 93 AHGAAAGFGVSATATVSINATLAGAIIGKN--DVNSKQICHITGAKLSIREHDSDPNLRN 150
+ + VS A + G++ +++ C G + ++ N +
Sbjct: 148 SVVEIPREEFQIPSLVSSTAESGRSFKGRHCKKFYTEEGCPY-GESCTFLHDEASRNRES 206
Query: 151 IELE----------------GSFDQIKQASAMVHDLILNVSSVSGPPGKNITSQTSAPAN 194
+ + GS +S +V V G G Q P+N
Sbjct: 207 VAISLGPGGYGSGGGGGSGGGSVGGGGSSSNVV------VLGGGGGSGSGSGIQILKPSN 260
Query: 195 NFKTKLCENFT-KGSCTFGERCHFAHGTDELRKPG 228
+KT++C + G C FG +CHFAHG EL + G
Sbjct: 261 -WKTRICNKWEITGYCPFGAKCHFAHGAAELHRFG 294
>UniRef100_Q93ZP3 At1g32359/F27G20.10 [Arabidopsis thaliana]
Length = 384
Score = 47.4 bits (111), Expect = 3e-04
Identities = 35/99 (35%), Positives = 47/99 (47%), Gaps = 20/99 (20%)
Query: 146 PNLRNIELEGSFDQIKQASAMVHDLILNVSSVS-GPPGKNI--------TSQTSAPANN- 195
PNL + + + D +A AM D N S + GPP K + TSA +N
Sbjct: 28 PNLGDSAVWATEDDYNRAWAMNPD---NTSGDNNGPPNKKTRGSPSSSSATTTSAASNRT 84
Query: 196 -------FKTKLCENFTKGSCTFGERCHFAHGTDELRKP 227
FKTKLC F G+C + C+FAH +ELR+P
Sbjct: 85 KAIGKMFFKTKLCCKFRAGTCPYITNCNFAHTVEELRRP 123
Score = 44.7 bits (104), Expect = 0.002
Identities = 19/31 (61%), Positives = 21/31 (67%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWELGR 63
KTR+CNK+ C FG KCHFAHG EL R
Sbjct: 262 KTRICNKWEITGYCPFGAKCHFAHGAAELHR 292
Score = 42.7 bits (99), Expect = 0.007
Identities = 54/215 (25%), Positives = 86/215 (39%), Gaps = 32/215 (14%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTVPAYEDTRAMGQMQSSSVGGRIEPPPP 92
KT+LC KF A C + C+FAH EL RP P +++ A + + S G + P
Sbjct: 93 KTKLCCKFR-AGTCPYITNCNFAHTVEELRRPP-PNWQEIVAAHEEERS---GGMGTPTV 147
Query: 93 AHGAAAGFGVSATATVSINATLAGAIIGKN--DVNSKQICHITGAKLSIREHDSDPNLRN 150
+ + VS A + G++ +++ C G + ++ N +
Sbjct: 148 SVVEIPREEFQIPSLVSSTAESGRSFKGRHCKKFYTEEGCPY-GESCTFLHDEASRNRES 206
Query: 151 IELE----------------GSFDQIKQASAMVHDLILNVSSVSGPPGKNITSQTSAPAN 194
+ + GS +S +V V G G Q P+N
Sbjct: 207 VAISLGPGGYGSGGGGGSGGGSVGGGGSSSNVV------VLGGGGGSGSGSGIQILKPSN 260
Query: 195 NFKTKLCENFT-KGSCTFGERCHFAHGTDELRKPG 228
+KT++C + G C FG +CHFAHG EL + G
Sbjct: 261 -WKTRICNKWEITGYCPFGAKCHFAHGAAELHRFG 294
>UniRef100_Q7Z1E7 FIP1-like protein [Trypanosoma cruzi]
Length = 280
Score = 47.4 bits (111), Expect = 3e-04
Identities = 22/47 (46%), Positives = 30/47 (63%), Gaps = 1/47 (2%)
Query: 180 PPGKNITSQTSAPANNF-KTKLCENFTKGSCTFGERCHFAHGTDELR 225
PP + + +AP +NF KTKLC+ F G C G++C +AHG ELR
Sbjct: 163 PPQGSSHAYYAAPRDNFLKTKLCQRFALGRCVKGDQCSYAHGHGELR 209
Score = 34.3 bits (77), Expect = 2.6
Identities = 32/115 (27%), Positives = 47/115 (40%), Gaps = 18/115 (15%)
Query: 5 GSSPAIPPIGRNPNVPQSFPDGSSPPVAKTRLCNKFNTAEGCKFGDKCHFAHGEWELGRP 64
G+S +PP G + ++ KT+LC +F C GD+C +AHG EL R
Sbjct: 157 GASYYVPPQGSS----HAYYAAPRDNFLKTKLCQRFALGR-CVKGDQCSYAHGHGEL-RS 210
Query: 65 TVPAYEDTRAMGQMQSSSVGGRIEPPPPAH---GAAAGFGVSATATVSINATLAG 116
P + + Q S + PP H G G+ T + T+AG
Sbjct: 211 APPVQQQQQQQQQPPSCIL-------PPFHVEQGYMPSMGMPPTGV--LEPTVAG 256
>UniRef100_UPI000046C0E9 UPI000046C0E9 UniRef100 entry
Length = 341
Score = 47.0 bits (110), Expect = 4e-04
Identities = 20/39 (51%), Positives = 27/39 (68%)
Query: 191 APANNFKTKLCENFTKGSCTFGERCHFAHGTDELRKPGM 229
A N +KT LCE+F+KG C G+ C +AHG +ELR+ M
Sbjct: 98 ATENLYKTALCESFSKGKCFSGQFCRYAHGQNELRENPM 136
Score = 33.9 bits (76), Expect = 3.4
Identities = 13/31 (41%), Positives = 20/31 (63%)
Query: 196 FKTKLCENFTKGSCTFGERCHFAHGTDELRK 226
+KTK+C + KG C + C +AH +ELR+
Sbjct: 33 YKTKICPWYIKGKCERRKTCLYAHAQNELRE 63
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.132 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 419,308,829
Number of Sequences: 2790947
Number of extensions: 17268688
Number of successful extensions: 42465
Number of sequences better than 10.0: 575
Number of HSP's better than 10.0 without gapping: 315
Number of HSP's successfully gapped in prelim test: 264
Number of HSP's that attempted gapping in prelim test: 41000
Number of HSP's gapped (non-prelim): 1716
length of query: 229
length of database: 848,049,833
effective HSP length: 123
effective length of query: 106
effective length of database: 504,763,352
effective search space: 53504915312
effective search space used: 53504915312
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 72 (32.3 bits)
Medicago: description of AC149471.8


