

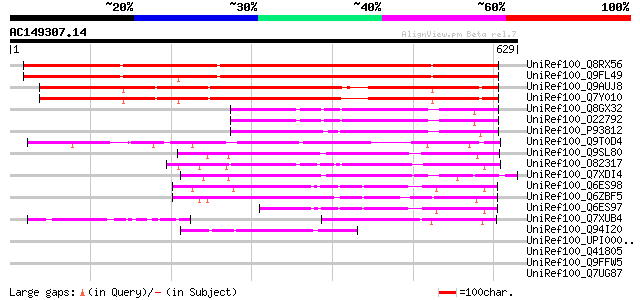
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149307.14 - phase: 0
(629 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8RX56 AT5g06970/MOJ9_14 [Arabidopsis thaliana] 830 0.0
UniRef100_Q9FL49 Similarity to unknown protein [Arabidopsis thal... 812 0.0
UniRef100_Q9AUJ8 Hypothetical protein OSJNBb0072E24.8 [Oryza sat... 672 0.0
UniRef100_Q7Y010 Hypothetical protein OSJNBb0070O09.17 [Oryza sa... 650 0.0
UniRef100_Q8GX32 Hypothetical protein At2g33420 [Arabidopsis tha... 205 4e-51
UniRef100_O22792 Hypothetical protein At2g33420 [Arabidopsis tha... 205 4e-51
UniRef100_P93812 F19P19.6 protein [Arabidopsis thaliana] 201 7e-50
UniRef100_Q9T0D4 Hypothetical protein AT4g11670 [Arabidopsis tha... 195 3e-48
UniRef100_Q9SL80 Hypothetical protein At2g20010 [Arabidopsis tha... 195 3e-48
UniRef100_O82317 Hypothetical protein At2g25800 [Arabidopsis tha... 190 9e-47
UniRef100_Q7XDI4 Hypothetical protein [Oryza sativa] 187 6e-46
UniRef100_Q6ES98 Hypothetical protein P0512H04.9-1 [Oryza sativa] 177 6e-43
UniRef100_Q6ZBF5 Hypothetical protein P0671F11.10 [Oryza sativa] 158 4e-37
UniRef100_Q6ES97 Hypothetical protein P0512H04.9-2 [Oryza sativa] 139 3e-31
UniRef100_Q7XUB4 OSJNBb0032E06.4 protein [Oryza sativa] 126 2e-27
UniRef100_Q94I20 Hypothetical protein OSJNBa0034E23.5 [Oryza sat... 71 1e-10
UniRef100_UPI00001D7AB7 UPI00001D7AB7 UniRef100 entry 45 0.008
UniRef100_Q41805 Extensin-like protein precursor [Zea mays] 44 0.011
UniRef100_Q9FFW5 Similarity to protein kinase [Arabidopsis thali... 44 0.019
UniRef100_Q7UG87 Hypothetical protein [Rhodopirellula baltica] 43 0.032
>UniRef100_Q8RX56 AT5g06970/MOJ9_14 [Arabidopsis thaliana]
Length = 1101
Score = 830 bits (2143), Expect = 0.0
Identities = 422/592 (71%), Positives = 497/592 (83%), Gaps = 10/592 (1%)
Query: 18 ENAIDLLQRYRRDRRVLLDFILSGSLIKKVVMPPGAVTLDDVDLDQVSIDYVLNCAKKSE 77
ENA+++LQRYRRDRR LLDF+L+GSLIKKV+MPPGAVTLDDVDLDQVS+DYV+NCAKK
Sbjct: 4 ENAVEILQRYRRDRRKLLDFMLAGSLIKKVIMPPGAVTLDDVDLDQVSVDYVINCAKKGG 63
Query: 78 MLELSEAIRDYHDHTGLPQMSDTGSVGEFYLVTDPESSGSPPKRAPPPVP--ISAVPPIA 135
MLEL+EAIRDYHDH GLP M+ G+ EF+L T PESSGSPPKRAPPP+P IS+ P+
Sbjct: 64 MLELAEAIRDYHDHIGLPYMNSVGTADEFFLATIPESSGSPPKRAPPPIPVLISSSSPMV 123
Query: 136 VSTPPPAYPTSPVASNISRSESLYSAQERELTVDDIEDFEDDDDTSMVEGLR-AKRTLND 194
+ P + SP A + RSES S + +ELTVDDI+DFEDDDD V R ++RT ND
Sbjct: 124 TN---PEWCESPSAPPLMRSESFDSPKAQELTVDDIDDFEDDDDLDEVGNFRISRRTAND 180
Query: 195 ASDLAVKLPPFSTGITDDDLRETAYEILLACAGATGGLIVPSKEKKKDRKSSSLIRKLGR 254
A+DL +LP F+TGITDDDLRETA+EILLACAGA+GGLIVPSKEKKK++ S LI+KLGR
Sbjct: 181 AADLVPRLPSFATGITDDDLRETAFEILLACAGASGGLIVPSKEKKKEKSRSRLIKKLGR 240
Query: 255 SKTGSIVSQSQNAPGLVGLLESMRVQLEISEAMDIRTKQGLLNALVGKAGKRMDTLLVPL 314
VSQSQ++ GLV LLE MR Q+EISEAMDIRT+QGLLNAL GK GKRMD+LLVPL
Sbjct: 241 KSES--VSQSQSSSGLVSLLEMMRGQMEISEAMDIRTRQGLLNALAGKVGKRMDSLLVPL 298
Query: 315 ELLCCVARTEFSDKKAFIRWQKRQLKVLEEGLVNHPVVGFGESGRKTNEMRILLAKIEES 374
ELLCCV+RTEFSDKKA++RWQKRQL +L EGL+N+PVVGFGESGRK +++ LL +IEES
Sbjct: 299 ELLCCVSRTEFSDKKAYLRWQKRQLNMLAEGLINNPVVGFGESGRKATDLKSLLLRIEES 358
Query: 375 EFLPSSSGELQRTECLRSLREIAIPLAERPARGDLTGEICHWADGYQFNVRLYEKLLLSV 434
E LPSS+GE+QR ECL+SLRE+AI LAERPARGDLTGE+CHWADGY NVRLYEKLLL V
Sbjct: 359 ESLPSSAGEVQRAECLKSLREVAISLAERPARGDLTGEVCHWADGYHLNVRLYEKLLLCV 418
Query: 435 FDMLDEGKLTEEVEEILELLKSTWRVLGITETIHHTCYAWVLFRQYVITREHRILLHALE 494
FD+L++GKLTEEVEEILELLKSTWRVLGITETIH+TCYAWVLFRQYVIT E +L HA++
Sbjct: 419 FDILNDGKLTEEVEEILELLKSTWRVLGITETIHYTCYAWVLFRQYVITSERGLLRHAIQ 478
Query: 495 QLNKIPLMEQRGQQERLHLKSLRSKVEGERDMSFLQAFLTPIQRWADKQLGDYHLHFSEG 554
QL KIPL EQRG QERLHLK+L+ +V+ E ++SFL++FL+PI+ WADKQLGDYHLHF+EG
Sbjct: 479 QLKKIPLKEQRGPQERLHLKTLKCRVDNE-EISFLESFLSPIRSWADKQLGDYHLHFAEG 537
Query: 555 SAIMEKIVAVAMITRRLLLEEPDTSTQSLPISDRDQIEVYITSSIKHAFTRV 606
S +ME V VAMIT RLLLEE D + S SDR+QIE Y+ SSIK+ FTR+
Sbjct: 538 SLVMEDTVTVAMITWRLLLEESDRAMHS-NSSDREQIESYVLSSIKNTFTRM 588
>UniRef100_Q9FL49 Similarity to unknown protein [Arabidopsis thaliana]
Length = 1105
Score = 812 bits (2097), Expect = 0.0
Identities = 422/627 (67%), Positives = 497/627 (78%), Gaps = 45/627 (7%)
Query: 18 ENAIDLLQRYRRDRRVLLDFILSGSLIKKVVMPPGAVTLDDVDLDQVSIDYVLNCAKKSE 77
ENA+++LQRYRRDRR LLDF+L+GSLIKKV+MPPGAVTLDDVDLDQVS+DYV+NCAKK
Sbjct: 4 ENAVEILQRYRRDRRKLLDFMLAGSLIKKVIMPPGAVTLDDVDLDQVSVDYVINCAKKGG 63
Query: 78 MLELSEAIRDYHDHTGLPQMSDTGSVGEFYLVTDPESSGSPPKRAPPPVP--ISAVPPIA 135
MLEL+EAIRDYHDH GLP M+ G+ EF+L T PESSGSPPKRAPPP+P IS+ P+
Sbjct: 64 MLELAEAIRDYHDHIGLPYMNSVGTADEFFLATIPESSGSPPKRAPPPIPVLISSSSPMV 123
Query: 136 VSTPPPAYPTSPVASNISRSESLYSAQERELTVDDIEDFEDDDDTSMVEGLR-AKRTLND 194
+ P + SP A + RSES S + +ELTVDDI+DFEDDDD V R ++RT ND
Sbjct: 124 TN---PEWCESPSAPPLMRSESFDSPKAQELTVDDIDDFEDDDDLDEVGNFRISRRTAND 180
Query: 195 ASDLAVKLPPFST-----------------------------------GITDDDLRETAY 219
A+DL +LP F+T GITDDDLRETA+
Sbjct: 181 AADLVPRLPSFATVARANYDWTISSTPTPMKSQGPADLLRKLQCISVSGITDDDLRETAF 240
Query: 220 EILLACAGATGGLIVPSKEKKKDRKSSSLIRKLGRSKTGSIVSQSQNAPGLVGLLESMRV 279
EILLACAGA+GGLIVPSKEKKK++ S LI+KLGR VSQSQ++ GLV LLE MR
Sbjct: 241 EILLACAGASGGLIVPSKEKKKEKSRSRLIKKLGRKSES--VSQSQSSSGLVSLLEMMRG 298
Query: 280 QLEISEAMDIRTKQGLLNALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRWQKRQL 339
Q+EISEAMDIRT+QGLLNAL GK GKRMD+LLVPLELLCCV+RTEFSDKKA++RWQKRQL
Sbjct: 299 QMEISEAMDIRTRQGLLNALAGKVGKRMDSLLVPLELLCCVSRTEFSDKKAYLRWQKRQL 358
Query: 340 KVLEEGLVNHPVVGFGESGRKTNEMRILLAKIEESEFLPSSSGELQRTECLRSLREIAIP 399
+L EGL+N+PVVGFGESGRK +++ LL +IEESE LPSS+GE+QR ECL+SLRE+AI
Sbjct: 359 NMLAEGLINNPVVGFGESGRKATDLKSLLLRIEESESLPSSAGEVQRAECLKSLREVAIS 418
Query: 400 LAERPARGDLTGEICHWADGYQFNVRLYEKLLLSVFDMLDEGKLTEEVEEILELLKSTWR 459
LAERPARGDLTGE+CHWADGY NVRLYEKLLL VFD+L++GKLTEEVEEILELLKSTWR
Sbjct: 419 LAERPARGDLTGEVCHWADGYHLNVRLYEKLLLCVFDILNDGKLTEEVEEILELLKSTWR 478
Query: 460 VLGITETIHHTCYAWVLFRQYVITREHRILLHALEQLNKIPLMEQRGQQERLHLKSLRSK 519
VLGITETIH+TCYAWVLFRQYVIT E +L HA++QL KIPL EQRG QERLHLK+L+ +
Sbjct: 479 VLGITETIHYTCYAWVLFRQYVITSERGLLRHAIQQLKKIPLKEQRGPQERLHLKTLKCR 538
Query: 520 VEGERDMSFLQAFLTPIQRWADKQLGDYHLHFSEGSAIMEKIVAVAMITRRLLLEEPDTS 579
V+ E ++SFL++FL+PI+ WADKQLGDYHLHF+EGS +ME V VAMIT RLLLEE D +
Sbjct: 539 VDNE-EISFLESFLSPIRSWADKQLGDYHLHFAEGSLVMEDTVTVAMITWRLLLEESDRA 597
Query: 580 TQSLPISDRDQIEVYITSSIKHAFTRV 606
S SDR+QIE Y+ SSIK+ FTR+
Sbjct: 598 MHS-NSSDREQIESYVLSSIKNTFTRM 623
>UniRef100_Q9AUJ8 Hypothetical protein OSJNBb0072E24.8 [Oryza sativa]
Length = 1049
Score = 672 bits (1733), Expect = 0.0
Identities = 355/578 (61%), Positives = 438/578 (75%), Gaps = 42/578 (7%)
Query: 38 ILSGSLIKKVVMPPGAVTLDDVDLDQVSIDYVLNCAKKSEMLELSEAIRDYHDHTGLPQM 97
+LSG+LIKKVVMPPGA++LDDVD+DQVS+DYVLNCAKK E L+L +AIR +HD P +
Sbjct: 1 MLSGNLIKKVVMPPGAISLDDVDIDQVSVDYVLNCAKKGEALDLGDAIRLFHDSLDYPYV 60
Query: 98 SDTGSVGEFYLVTDPESSGSPPKRAPPPVPISAVPPIAVSTP----PPAYPTSPVAS-NI 152
+++G+V EF+L+T PE SG P R PPPVP A P+ + P PP SPV++ N+
Sbjct: 61 NNSGTVEEFFLLTKPEYSGPAPAREPPPVPAIAPSPVVIPAPIVDPPPVAVHSPVSTTNL 120
Query: 153 SRSESLYSAQERELTVDDIEDFEDDDDTSMVEGLRA-KRTLNDASDLAVKLPPFSTGITD 211
S+S+S S E+ELT+DDIEDFED++D + RA +R +DA+DL+++LP F TGITD
Sbjct: 121 SKSQSFDSPTEKELTIDDIEDFEDEEDE--FDSRRASRRHQSDANDLSLRLPLFETGITD 178
Query: 212 DDLRETAYEILLACAGATGGLIVPSKEKKKDRKSSSLIRKLGRSKTGSIVSQSQNAPGLV 271
DDLRETAYEIL+A AGA+GGLIVP KEKKK++++ L+RKLGRSK+ S SQ+Q PGLV
Sbjct: 179 DDLRETAYEILVAAAGASGGLIVPQKEKKKEKRNK-LMRKLGRSKSESTQSQTQRQPGLV 237
Query: 272 GLLESMRVQLEISEAMDIRTKQGLLNALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAF 331
GLLE+MR QLEI+E+MDIRT+QGLLNA+VGK GKRMD LL+PLELLCC++R EFSD KA+
Sbjct: 238 GLLETMRAQLEITESMDIRTRQGLLNAMVGKVGKRMDNLLIPLELLCCISRAEFSDMKAY 297
Query: 332 IRWQKRQLKVLEEGLVNHPVVGFGESGRKTNEMRILLAKIEESEFLPSSSGELQRTECLR 391
+RWQKRQL +LEEGL+NHPVVGFGE GRK NE+R L KIEESE L S+ E+QRTECLR
Sbjct: 298 LRWQKRQLNMLEEGLINHPVVGFGELGRKVNELRNLFRKIEESESLQPSAVEVQRTECLR 357
Query: 392 SLREIAIPLAERPARGDLTGEICHWADGYQFNVRLYEKLLLSVFDMLDEGKLTEEVEEIL 451
SLRE+A L+ERPARGDLTGEI YQ +EVEEIL
Sbjct: 358 SLREVATSLSERPARGDLTGEI-----HYQL----------------------QEVEEIL 390
Query: 452 ELLKSTWRVLGITETIHHTCYAWVLFRQYVITREHRILLHALEQLNKIPLMEQRGQQERL 511
ELLKSTWR+LGITETIH TCYAWVLFRQ+V T E +L +E L KIPL EQRG QERL
Sbjct: 391 ELLKSTWRILGITETIHDTCYAWVLFRQFVFTGEQGLLKVVIEHLRKIPLKEQRGPQERL 450
Query: 512 HLKSLRSKVEGE---RDMSFLQAFLTPIQRWADKQLGDYHLHFSEGSAIMEKIVAVAMIT 568
HLKSLRS V+ E +D +F Q+FL+P+Q+W DK+L DYHLHFSEG ++M IV VAM+
Sbjct: 451 HLKSLRSSVDAEDSFQDFTFFQSFLSPVQKWVDKKLNDYHLHFSEGPSMMADIVTVAMLI 510
Query: 569 RRLLLEEPDTSTQSLPISDRDQIEVYITSSIKHAFTRV 606
RR+L EE + +S DRDQI+ YITSS+K AF ++
Sbjct: 511 RRILGEENNKGMES---PDRDQIDRYITSSVKSAFVKM 545
>UniRef100_Q7Y010 Hypothetical protein OSJNBb0070O09.17 [Oryza sativa]
Length = 1078
Score = 650 bits (1676), Expect = 0.0
Identities = 352/613 (57%), Positives = 434/613 (70%), Gaps = 83/613 (13%)
Query: 38 ILSGSLIKKVVMPPGAVTLDDVDLDQVSIDYVLNCAKKSEMLELSEAIRDYHDHTGLPQM 97
+LSG+LIKKVVMPPGA++LDDVD+DQVS+DYVLNCAKK E L+L +AIR +HD P +
Sbjct: 1 MLSGNLIKKVVMPPGAISLDDVDIDQVSVDYVLNCAKKGEALDLGDAIRLFHDSLDYPYV 60
Query: 98 SDTGSVGEFYLVTDPESSGSPPKRAPPPVPISAVPPIAVSTP----PPAYPTSPVAS-NI 152
+++G+V EF+L+T PE SG P R PPPVP A P+ + P PP SPV++ N+
Sbjct: 61 NNSGTVEEFFLLTKPEYSGPAPAREPPPVPAIAPSPVVIPAPIVDPPPVAVHSPVSTTNL 120
Query: 153 SRSESLYSAQERELTVDDIEDFEDDDDTSMVEGLRA-KRTLNDASDLAVKLPPFSTG--- 208
S+S+S S E+ELT+DDIEDFED++D + RA +R +DA+DL+++LP F TG
Sbjct: 121 SKSQSFDSPTEKELTIDDIEDFEDEEDE--FDSRRASRRHQSDANDLSLRLPLFETGNSD 178
Query: 209 --------------------------------ITDDDLRETAYEILLACAGATGGLIVPS 236
ITDDDLRETAYEIL+A AGA+GGLIVP
Sbjct: 179 DCYVIMKSSCWYGYIHWVKKVIDNGNCAVFPGITDDDLRETAYEILVAAAGASGGLIVPQ 238
Query: 237 KEKKKDRKSSSLIRKLGRSKTGSIVSQSQNAPGLVGLLESMRVQLEISEAMDIRTKQGLL 296
KEKKK++++ L+RKLGRSK+ S SQ+Q PGLVGLLE+MR QLEI+E+MDIRT+QGLL
Sbjct: 239 KEKKKEKRNK-LMRKLGRSKSESTQSQTQRQPGLVGLLETMRAQLEITESMDIRTRQGLL 297
Query: 297 NALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRWQKRQLKVLEEGLVNHPVVGFGE 356
NA+VGK GKRMD LL+PLELLCC++R EFSD KA++RWQKRQL +LEEGL+NHPVVGFGE
Sbjct: 298 NAMVGKVGKRMDNLLIPLELLCCISRAEFSDMKAYLRWQKRQLNMLEEGLINHPVVGFGE 357
Query: 357 SGRKTNEMRILLAKIEESEFLPSSSGELQRTECLRSLREIAIPLAERPARGDLTGEICHW 416
GRK NE+R L KIEESE L S+ E+QRTECLRSLRE+A L+ERPARGDLTG
Sbjct: 358 LGRKVNELRNLFRKIEESESLQPSAVEVQRTECLRSLREVATSLSERPARGDLTG----- 412
Query: 417 ADGYQFNVRLYEKLLLSVFDMLDEGKLTEEVEEILELLKSTWRVLGITETIHHTCYAWVL 476
EEVEEILELLKSTWR+LGITETIH TCYAWVL
Sbjct: 413 ----------------------------EEVEEILELLKSTWRILGITETIHDTCYAWVL 444
Query: 477 FRQYVITREHRILLHALEQLNKIPLMEQRGQQERLHLKSLRSKVEGE---RDMSFLQAFL 533
FRQ+V T E +L +E L KIPL EQRG QERLHLKSLRS V+ E +D +F Q+FL
Sbjct: 445 FRQFVFTGEQGLLKVVIEHLRKIPLKEQRGPQERLHLKSLRSSVDAEDSFQDFTFFQSFL 504
Query: 534 TPIQRWADKQLGDYHLHFSEGSAIMEKIVAVAMITRRLLLEEPDTSTQSLPISDRDQIEV 593
+P+Q+W DK+L DYHLHFSEG ++M IV VAM+ RR+L EE + +S DRDQI+
Sbjct: 505 SPVQKWVDKKLNDYHLHFSEGPSMMADIVTVAMLIRRILGEENNKGMES---PDRDQIDR 561
Query: 594 YITSSIKHAFTRV 606
YITSS+K AF ++
Sbjct: 562 YITSSVKSAFVKM 574
>UniRef100_Q8GX32 Hypothetical protein At2g33420 [Arabidopsis thaliana]
Length = 1039
Score = 205 bits (521), Expect = 4e-51
Identities = 123/344 (35%), Positives = 197/344 (56%), Gaps = 32/344 (9%)
Query: 275 ESMRVQLEISEAMDIRTKQGLLNALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRW 334
E MR Q++++E D R ++ LL LVG+ G+R +T+++PLELL + +EF D + W
Sbjct: 179 EIMRQQMKVTEQSDSRLRKTLLRTLVGQTGRRAETIILPLELLRHLKTSEFGDIHEYQLW 238
Query: 335 QKRQLKVLEEGLVNHPVVGFGESGRKTNEMRILLAKI-EESEFLPSSSGELQRTECLRSL 393
Q+RQLKVLE GL+ HP + KTN + L ++ +SE P + + T +R+L
Sbjct: 239 QRRQLKVLEAGLLLHPSIPLD----KTNNFAMRLREVVRQSETKPIDTSKTSDT--IRTL 292
Query: 394 REIAIPLAERPARGDLTGEICHWADGYQFNVRLYEKLLLSVFDMLDEGKLTEEVEEILEL 453
+ + L+ R G+ T ++CHWADGY N+ LY LL S+FD+ DE + +E++E+LEL
Sbjct: 293 TNVVVSLSWRGTNGNPT-DVCHWADGYPLNIHLYVALLQSIFDVRDETLVLDEIDELLEL 351
Query: 454 LKSTWRVLGITETIHHTCYAWVLFRQYVIT--REHRILLHALEQLNKIPLMEQRGQQERL 511
+K TW LGIT IH+ C+ WVLF QYV+T E +L + L ++ ++ +E L
Sbjct: 352 MKKTWSTLGITRPIHNLCFTWVLFHQYVVTSQMEPDLLGASHAMLAEVANDAKKLDREAL 411
Query: 512 HLKSLRSKVEGERDMSFLQAFLTPIQRWADKQLGDYHLHFSEGS-AIMEKIVAVAMITRR 570
++K L S L +Q W +K+L YH +F G+ ++E ++ +A+ + R
Sbjct: 412 YVKLLNST-------------LASMQGWTEKRLLSYHDYFQRGNVGLIENLLPLALSSSR 458
Query: 571 LLLEE--------PDTSTQSLPISDRDQIEVYITSSIKHAFTRV 606
+L E+ + L D+++ YI SSIK+AF++V
Sbjct: 459 ILGEDVTISQGKGQEKGDVKLVDHSGDRVDYYIRSSIKNAFSKV 502
>UniRef100_O22792 Hypothetical protein At2g33420 [Arabidopsis thaliana]
Length = 1039
Score = 205 bits (521), Expect = 4e-51
Identities = 123/344 (35%), Positives = 197/344 (56%), Gaps = 32/344 (9%)
Query: 275 ESMRVQLEISEAMDIRTKQGLLNALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRW 334
E MR Q++++E D R ++ LL LVG+ G+R +T+++PLELL + +EF D + W
Sbjct: 179 EIMRQQMKVTEQSDSRLRKTLLRTLVGQTGRRAETIILPLELLRHLKTSEFGDIHEYQLW 238
Query: 335 QKRQLKVLEEGLVNHPVVGFGESGRKTNEMRILLAKI-EESEFLPSSSGELQRTECLRSL 393
Q+RQLKVLE GL+ HP + KTN + L ++ +SE P + + T +R+L
Sbjct: 239 QRRQLKVLEAGLLLHPSIPLD----KTNNFAMRLREVVRQSETKPIDTSKTSDT--MRTL 292
Query: 394 REIAIPLAERPARGDLTGEICHWADGYQFNVRLYEKLLLSVFDMLDEGKLTEEVEEILEL 453
+ + L+ R G+ T ++CHWADGY N+ LY LL S+FD+ DE + +E++E+LEL
Sbjct: 293 TNVVVSLSWRGTNGNPT-DVCHWADGYPLNIHLYVALLQSIFDVRDETLVLDEIDELLEL 351
Query: 454 LKSTWRVLGITETIHHTCYAWVLFRQYVIT--REHRILLHALEQLNKIPLMEQRGQQERL 511
+K TW LGIT IH+ C+ WVLF QYV+T E +L + L ++ ++ +E L
Sbjct: 352 MKKTWSTLGITRPIHNLCFTWVLFHQYVVTSQMEPDLLGASHAMLAEVANDAKKLDREAL 411
Query: 512 HLKSLRSKVEGERDMSFLQAFLTPIQRWADKQLGDYHLHFSEGS-AIMEKIVAVAMITRR 570
++K L S L +Q W +K+L YH +F G+ ++E ++ +A+ + R
Sbjct: 412 YVKLLNST-------------LASMQGWTEKRLLSYHDYFQRGNVGLIENLLPLALSSSR 458
Query: 571 LLLEE--------PDTSTQSLPISDRDQIEVYITSSIKHAFTRV 606
+L E+ + L D+++ YI SSIK+AF++V
Sbjct: 459 ILGEDVTISQGKGQEKGDVKLVDHSGDRVDYYIRSSIKNAFSKV 502
>UniRef100_P93812 F19P19.6 protein [Arabidopsis thaliana]
Length = 1035
Score = 201 bits (510), Expect = 7e-50
Identities = 118/343 (34%), Positives = 194/343 (56%), Gaps = 31/343 (9%)
Query: 275 ESMRVQLEISEAMDIRTKQGLLNALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRW 334
E MR Q++++E D R ++ L+ LVG+ G+R +T+++PLELL V +EF D + W
Sbjct: 177 EIMRQQMKVTEQSDTRLRKTLMRTLVGQTGRRAETIILPLELLRHVKPSEFGDVHEYQIW 236
Query: 335 QKRQLKVLEEGLVNHPVVGFGESGRKTNEMRILLAKIEESEFLPSSSGELQRTECLRSLR 394
Q+RQLKVLE GL+ HP + ++ +R ++ + E S + ++ T C
Sbjct: 237 QRRQLKVLEAGLLIHPSIPLEKTNNFAMRLREIIRQSETKAIDTSKNSDIMPTLC----- 291
Query: 395 EIAIPLAERPARGDLTGEICHWADGYQFNVRLYEKLLLSVFDMLDEGKLTEEVEEILELL 454
+ L+ R A T +ICHWADGY N+ LY LL S+FD+ DE + +E++E+LEL+
Sbjct: 292 NLVASLSWRNATP--TTDICHWADGYPLNIHLYVALLQSIFDIRDETLVLDEIDELLELM 349
Query: 455 KSTWRVLGITETIHHTCYAWVLFRQYVIT--REHRILLHALEQLNKIPLMEQRGQQERLH 512
K TW +LGIT IH+ C+ WVLF QY++T E +L + L ++ ++ +E L+
Sbjct: 350 KKTWIMLGITRAIHNLCFTWVLFHQYIVTSQMEPDLLGASHAMLAEVANDAKKSDREALY 409
Query: 513 LKSLRSKVEGERDMSFLQAFLTPIQRWADKQLGDYHLHFSEGS-AIMEKIVAVAMITRRL 571
+K L S L +Q W +K+L YH +F G+ ++E ++ +A+ + ++
Sbjct: 410 VKLLTST-------------LASMQGWTEKRLLSYHDYFQRGNVGLIENLLPLALSSSKI 456
Query: 572 LLEEPDTSTQS--------LPISDRDQIEVYITSSIKHAFTRV 606
L E+ S + L S D+++ YI +SIK+AF++V
Sbjct: 457 LGEDVTISQMNGLEKGDVKLVDSSGDRVDYYIRASIKNAFSKV 499
>UniRef100_Q9T0D4 Hypothetical protein AT4g11670 [Arabidopsis thaliana]
Length = 998
Score = 195 bits (496), Expect = 3e-48
Identities = 166/619 (26%), Positives = 267/619 (42%), Gaps = 130/619 (21%)
Query: 23 LLQRYRRDRRVLLDFILSGSLIKKVVMPPGAVT-LDDVDLDQVSIDYVLNCAKKS----- 76
LLQRYR DRR L++F++S L+K++ P G+ T L DLD +S DYVL+C K
Sbjct: 4 LLQRYRNDRRKLMEFLMSSGLVKELRSPSGSPTSLSPADLDALSADYVLDCVKSEIAETL 63
Query: 77 --------EMLELSEAIRDYHDHTGLPQMSDTGSVGEFYLVTDPESSGSPPKRAPPPVPI 128
++++S+ Y+ + P + S ++LV+ P+ +GSPP R PPP
Sbjct: 64 HFVNTNAGGVVDVSKGREKYNFDSSYPVTIHSESGDSYFLVSSPDLAGSPPHRMPPP--- 120
Query: 129 SAVPPIAVSTPPPAYPTSPVASNISRSESLYSAQERELTVDDIEDFEDD----DDTSMVE 184
PV NI +S + + R + + D+ ++T ++
Sbjct: 121 ------------------PV--NIEKSSNNGADMSRHMDSSNTPSARDNYVFKEETPDIK 160
Query: 185 GLRAKRTLNDASDLAVKLPPFSTGITDDDLRETAYEILLAC----AGATGGLIVPSKEKK 240
++ + + + LPP TG++DDDLRE AYE+++A + T + ++
Sbjct: 161 PVKPIKII------PLGLPPLRTGLSDDDLREAAYELMIASMLLSSFLTNSVEAYPTHRR 214
Query: 241 KDRKSSSLIRKLGRSKTGSIVSQSQNAPGLVGLLESMRVQLEISEAMDIRTKQGLLNALV 300
K KSS L+ L R + Q N EIS MD ++ L+
Sbjct: 215 KIEKSSRLMLSLKRKDKPHLQPQISNTHS------------EISSKMDTCIRRNLVQLAT 262
Query: 301 GKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRWQKRQLKVLEEGLVNHPVVGFGESGRK 360
+ G+++D + L LL + +++F ++K +++W+ RQ +LEE L P + E
Sbjct: 263 LRTGEQIDLPQLALGLLVGIFKSDFPNEKLYMKWKTRQANLLEEVLCFSPSLEKNERAT- 321
Query: 361 TNEMRILLAKIEESEFLPSSSGELQRTECLRSLREIAIPLAERPARGDLTGEICHWADGY 420
MR LA I +S+ R E L S+R++A L+ P R + E +W Y
Sbjct: 322 ---MRKCLATIRDSKEWDVVVSASLRIEVLSSIRQVASKLSSLPGRCGIEEETYYWTAIY 378
Query: 421 QFNVRLYEKLLLSVFDMLDEGKLTEEVEEILELLKSTWRVLGITETIHHTCYAWVLFRQY 480
N+RLYEKLL VFD LDEG +E++
Sbjct: 379 HLNIRLYEKLLFGVFDTLDEGSTIQELQ-------------------------------- 406
Query: 481 VITREHRILLHALEQLNKIPLMEQRGQQERLHLKSL---RSKVEGERDMSFLQAFLTPIQ 537
K+ E +E L+L L R + + + ++A LT +
Sbjct: 407 -----------------KVTSAESGNPKEDLYLSHLVCSRQTIGTDIHLGLVKAILTSVS 449
Query: 538 RWADKQLGDYHLHFSEGSAIMEKIVAVAMIT-------RRLLLEEPDTSTQSLPISDRDQ 590
W D +L DYHLHF + +V +A R L + DT + + D+
Sbjct: 450 AWCDDKLQDYHLHFGKKPRDFGMLVRLASTVGLPPADCTRTELIKLDTLSDDV----SDK 505
Query: 591 IEVYITSSIKHAFTRVRVF 609
I+ Y+ +SIK A R F
Sbjct: 506 IQSYVQNSIKGACARAAHF 524
>UniRef100_Q9SL80 Hypothetical protein At2g20010 [Arabidopsis thaliana]
Length = 952
Score = 195 bits (496), Expect = 3e-48
Identities = 135/437 (30%), Positives = 224/437 (50%), Gaps = 57/437 (13%)
Query: 209 ITDDDLRETAYEILLACAGATGG--LIVPSKEKKKDRK------SSSLIRKLGRSKTGSI 260
+++ +LRETAYEIL+A +TG L + K DR S S L RS T +
Sbjct: 15 LSNSELRETAYEILVAACRSTGSRPLTYIPQSPKSDRSNGLTTASLSPSPSLHRSLTSTA 74
Query: 261 VSQSQNAPGL------------------------VGLLESMRVQLEISEAMDIRTKQGLL 296
S+ + A G+ V + E +RVQ+ ISE +D R ++ LL
Sbjct: 75 ASKVKKALGMKKRIGDGDGGAGESSSQPDRSKKSVTVGELVRVQMRISEQIDSRIRRALL 134
Query: 297 NALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRWQKRQLKVLEEGLVNHPVVGFGE 356
G+ G+R++ +++PLELL + ++F D++ + WQ+R LK+LE GL+ +P V +
Sbjct: 135 RIASGQLGRRVEMMVLPLELLQQLKASDFPDQEEYESWQRRNLKLLEAGLILYPCVPLSK 194
Query: 357 SGRKTNEMR-ILLAKIEESEFLPSSSGELQRTECLRSLREIAIPLAERPARGDLTGEICH 415
S + +++ I+ + +E +GE Q +LR + + LA R + E CH
Sbjct: 195 SDKSVQQLKQIIRSGLERPLDTGKITGETQ------NLRSLVMSLASRQNNNGIGSETCH 248
Query: 416 WADGYQFNVRLYEKLLLSVFDMLDEGKLTEEVEEILELLKSTWRVLGITETIHHTCYAWV 475
WADG+ N+R+Y+ LL S FD+ DE + EEV+E+LEL+K TW VLGI + IH+ C+ WV
Sbjct: 249 WADGFPLNLRIYQMLLESCFDVNDELLIVEEVDEVLELIKKTWPVLGINQMIHNVCFLWV 308
Query: 476 LFRQYVITRE-HRILLHALEQLNKIPLMEQRGQQERLHLKSLRSKVEGERDMSFLQAFLT 534
L +YV T + LL A L L +++ + L + L+
Sbjct: 309 LVNRYVSTGQVENDLLVAAHNL-------------ILEIENDAMETNDPEYSKILSSVLS 355
Query: 535 PIQRWADKQLGDYHLHFS-EGSAIMEKIVAVAMITRRLLLEEPDTS---TQSLPISDRDQ 590
+ W +K+L YH F+ + +E V++ ++ ++L E+ + + S RD+
Sbjct: 356 LVMDWGEKRLLAYHDTFNIDNVETLETTVSLGILVAKVLGEDISSEYRRKKKHVDSGRDR 415
Query: 591 IEVYITSSIKHAFTRVR 607
++ YI SS++ AF + +
Sbjct: 416 VDTYIRSSLRMAFQQTK 432
>UniRef100_O82317 Hypothetical protein At2g25800 [Arabidopsis thaliana]
Length = 993
Score = 190 bits (483), Expect = 9e-47
Identities = 147/460 (31%), Positives = 232/460 (49%), Gaps = 70/460 (15%)
Query: 195 ASDLAVKLPPFSTG-----ITDDDLRETAYEILLA-CAGATGGLI--------------- 233
+S ++ LPP G ++D DLR TAYEI +A C ATG +
Sbjct: 30 SSSMSSDLPPSPLGQLAVQLSDSDLRLTAYEIFVAACRSATGKPLSSAVSVAVLNQDSPN 89
Query: 234 ---------------VPSKEKKKDRKSSSLIRKLGRSKTGSIVSQSQNA---PGLVGLLE 275
SK KK SS G +K+ S S P VG E
Sbjct: 90 GSPASPAIQRSLTSTAASKMKKALGLRSSSSLSPGSNKSSGSASGSNGKSKRPTTVG--E 147
Query: 276 SMRVQLEISEAMDIRTKQGLLNALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRWQ 335
MR+Q+ +SEA+D R ++ L + G++++++++PLELL + ++F+D++ + W
Sbjct: 148 LMRIQMRVSEAVDSRVRRAFLRIAASQVGRKIESVVLPLELLQQLKSSDFTDQQEYDAWL 207
Query: 336 KRQLKVLEEGLVNHPVVGFGESGRKTNEMRILLAKIEESEFLPSSSGELQRTECLRSLRE 395
KR LKVLE GL+ HP V KTN + L I + P +G + E ++SLR
Sbjct: 208 KRSLKVLEAGLLLHPRVPLD----KTNSSQRLRQIIHGALDRPLETG--RNNEQMQSLRS 261
Query: 396 IAIPLAERPARGDLTGEICHWADGYQFNVRLYEKLLLSVFDMLDEGKLTEEVEEILELLK 455
+ LA R + G + + CHWADG FN+RLYE LL + FD D + EEV++++E +K
Sbjct: 262 AVMSLATR-SDGSFS-DSCHWADGSPFNLRLYELLLEACFDSNDATSMVEEVDDLMEHIK 319
Query: 456 STWRVLGITETIHHTCYAWVLFRQYVITREHRI-LLHALE-QLNKIPLMEQRGQQERLHL 513
TW +LGI + +H+ C+ W+LF +YV+T + + LLHA + QL ++
Sbjct: 320 KTWVILGINQMLHNLCFTWLLFSRYVVTGQVEMDLLHACDSQLAEV-------------A 366
Query: 514 KSLRSKVEGERDMSFLQAFLTPIQRWADKQLGDYHLHFSEGSA-IMEKIVAVAMITRRLL 572
K ++ + E L + L+ I WA+K+L YH F G+ ME IV++ + R+L
Sbjct: 367 KDAKTTKDPEYS-QVLSSTLSAILGWAEKRLLAYHDTFDRGNIHTMEGIVSLGVSAARIL 425
Query: 573 LEEPDTSTQSLPISD----RDQIEVYITSSIKHAFTRVRV 608
+E+ + + R +IE YI SS++ +F + +
Sbjct: 426 VEDISNEYRRRRKGEVDVARTRIETYIRSSLRTSFAQASI 465
>UniRef100_Q7XDI4 Hypothetical protein [Oryza sativa]
Length = 990
Score = 187 bits (476), Expect = 6e-46
Identities = 133/454 (29%), Positives = 227/454 (49%), Gaps = 63/454 (13%)
Query: 213 DLRETAYEIL-LACAGATGGLIVPSKEK---------------------KKDRKSSSLIR 250
+LRETAYEI ++C ++GG + E K R SSS
Sbjct: 47 ELRETAYEIFFMSCRSSSGGNTAGAAEVSSPVAGPRGGGGSRVKKALGLKARRLSSSSAA 106
Query: 251 KLGRSKTGSIVSQSQN--APGL----VGLLESMRVQLEISEAMDIRTKQGLLNALVGKAG 304
+ + +SQ+ +PG + E MR Q+ ++E D R ++ L+ A+VG+ G
Sbjct: 107 MVAQPMMVRTLSQTSGPASPGRGRRPMTSAEIMRQQMRVTEQSDARLRRTLMRAVVGQVG 166
Query: 305 KRMDTLLVPLELLCCVARTEFSDKKAFIRWQKRQLKVLEEGLVNHPVVGFGESGRKTNEM 364
+R DT+++PLELL + EF+D + + +WQ RQ+K+LE GL+ HP +
Sbjct: 167 RRPDTIVLPLELLRQLKPAEFADGEEYHQWQFRQVKLLEAGLILHPSLPLDRLNSAVLRF 226
Query: 365 RILLAKIEESEFLPSSSGELQRTECLRSLREIAIPLAERPARGDLTGEICHWADGYQFNV 424
R ++ E + S + RT L LA R G G+ CHWADGY NV
Sbjct: 227 REVMRATEIRAIDTAKSSDAMRT-----LTSAVHALAWRSGVGSGGGDACHWADGYPLNV 281
Query: 425 RLYEKLLLSVFDMLDEGKLTEEVEEILELLKSTWRVLGITETIHHTCYAWVLFRQYVITR 484
LY LL ++FD D + +EV+E+L+L++ TW LG+T +H+ C AW F+QYV+T
Sbjct: 282 LLYASLLHAIFDHRDCTVVLDEVDELLDLIRKTWPTLGVTRPVHNVCLAWAFFQQYVVTG 341
Query: 485 --EHRILLHALEQLNKIPLMEQRGQQERLHLKSLRSKVEGERDMSFLQAFLTPIQRWADK 542
E + AL L + + RG ++ ++ K+L L +Q W++K
Sbjct: 342 QVEPELAAAALAVLADV-AADARGTRDAVYGKALLGA-------------LGAMQEWSEK 387
Query: 543 QLGDYHLHFSEG-----SAIMEKIVAVAMITRRLLLEEPDTSTQSLPISD--RDQIEVYI 595
+L DYH + +G + +ME ++++++ +++ + D + + ++ D+++ YI
Sbjct: 388 RLLDYHDSYEKGIGGAPTEVMEILLSISLAAGKIIADR-DAAADADDAANFAGDRVDYYI 446
Query: 596 TSSIKHAFTRVRVFWALLFISDNVIFVMKMYEYL 629
S+K+AFT+V+ F+ FV+ + ++
Sbjct: 447 RCSMKNAFTKVK------FLGSESQFVLHQFMFV 474
>UniRef100_Q6ES98 Hypothetical protein P0512H04.9-1 [Oryza sativa]
Length = 985
Score = 177 bits (450), Expect = 6e-43
Identities = 130/455 (28%), Positives = 215/455 (46%), Gaps = 76/455 (16%)
Query: 203 PPFSTGITDDDLRETAYEILLAC------------------AGATGGLIVPSKEKKKDRK 244
P ++ DLRE AYE+L+A AG GG P+
Sbjct: 36 PDLGVPLSAADLREAAYEVLVASSRTTGGKPLTYIPQAAASAGGGGGPASPASASSLSSA 95
Query: 245 SSSLIRKLGRSKTGSIVSQSQNAPGLVGLLES-------------------------MRV 279
++S L RS T + S+ + A GL S MRV
Sbjct: 96 NASSSPSLQRSLTSAAASKMKKALGLRSSASSKGGSPGSGGGGKSVPPRRPATVGELMRV 155
Query: 280 QLEISEAMDIRTKQGLLNALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRWQKRQL 339
Q+ +SE D R ++GLL + G+R +++++PLE L ++ D + + WQ R L
Sbjct: 156 QMRVSEPADARIRRGLLRIAASQLGRRAESMVLPLEFLQQFKASDIPDPQEYEAWQSRNL 215
Query: 340 KVLEEGLVNHPVVGFGESGRKTNEMRILLAKIEESEFLPSSSGELQRTECLRSLREIAIP 399
K+LE GL+ HP+V +S +R ++ + P +G + +E ++ LR +
Sbjct: 216 KLLEAGLLVHPLVPLNKSDVSAQRLRQIIRGAYDR---PLETG--KNSESMQVLRSAVMS 270
Query: 400 LAERPARGDLTGEICHWADGYQFNVRLYEKLLLSVFDMLDEGKLTEEVEEILELLKSTWR 459
LA R G T + CHWADG+ N+ LY+ L+ + FD D+G + +E++E++ELLK TW
Sbjct: 271 LAGRSDDG--TSDGCHWADGFPLNLHLYQMLVEACFDN-DDGTVVDEIDEVMELLKKTWG 327
Query: 460 VLGITETIHHTCYAWVLFRQYVITREHRI-LLHALEQLNKIPLMEQRGQQERLHLKSLRS 518
+LGI + +H+ C+AW LF +V++ + I LL A E L +
Sbjct: 328 ILGINQMLHNLCFAWALFNHFVMSGQVDIELLSAAEN----------------QLAEVAK 371
Query: 519 KVEGERDMSF---LQAFLTPIQRWADKQLGDYHLHFSEGS-AIMEKIVAVAMITRRLLLE 574
+ +D ++ L + L+ I W +K+L YH F+ + M+ IV++ + R+L+E
Sbjct: 372 DAKTTKDPNYSKVLSSTLSSIMGWTEKRLLAYHETFNTSNIESMQGIVSIGVSAARVLVE 431
Query: 575 EPDTSTQSLPISD----RDQIEVYITSSIKHAFTR 605
+ + + R +IE YI SS++ AF +
Sbjct: 432 DISHEYRRRRKEETDVARSRIETYIRSSLRTAFAQ 466
>UniRef100_Q6ZBF5 Hypothetical protein P0671F11.10 [Oryza sativa]
Length = 975
Score = 158 bits (400), Expect = 4e-37
Identities = 121/435 (27%), Positives = 205/435 (46%), Gaps = 52/435 (11%)
Query: 203 PPFSTGITDDDLRETAYEILLACAGATGGLIV-----------------PSKEKKKDRK- 244
P ++ +LR TAYE+L+A + ATG + PS R
Sbjct: 45 PDLGVPLSAAELRATAYEVLVAASRATGAKPLTYITQSAASAASAAAPAPSLSSSIHRSL 104
Query: 245 ------SSSLIRK-LG-RSKTGSIVSQSQNAPGLVGLLESMRVQLEISEAMDIRTKQGLL 296
SSS ++K LG R + S S+ + A + E +RVQL ++E D R ++ LL
Sbjct: 105 ASTAAASSSKVKKALGLRRSSASSSSKRRGARRPATVAELVRVQLGVTEQADARIRRALL 164
Query: 297 NALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRWQKRQLKVLEEGLVNHPVVGFGE 356
G+ GK ++L++PLE L ++F D + WQ R LK+LE GL+ HP+V +
Sbjct: 165 RIAAGQLGKHAESLVLPLEFLQQFKASDFLDPHEYEAWQLRYLKLLEAGLLFHPLVPLKK 224
Query: 357 SGRKTNEMRILLAKIEESEFLPSSSGELQRTECLRSLREIAIPLAERPARGDLTGEICHW 416
S +R ++ + + +L C + L E T + CHW
Sbjct: 225 SDISALRLRQVIHGAYDKPVETEKNSKLLVELCSAARALAGRSLIE-------TFDECHW 277
Query: 417 ADGYQFNVRLYEKLLLSVFDMLDEGKLTEEVEEILELLKSTWRVLGITETIHHTCYAWVL 476
ADG+ N+ +Y+ L+ + FD ++G + +E++E++E+L TW +LGI + H+ C+AW L
Sbjct: 278 ADGFPLNLHIYQMLIEACFDS-EDGAVVDEIDEVVEMLTKTWPILGINQMFHNLCFAWAL 336
Query: 477 FRQYVITREHRILLHALEQLNKIPLMEQRGQQERLHLKSLRSKVEGERDMSFLQAFLTPI 536
F +V++ + I L+ G Q +K ++ + + L + + I
Sbjct: 337 FNHFVMSGQ-----------ADIELLSGAGIQLTEVVKDAKTTKDPDY-CDVLISTINSI 384
Query: 537 QRWADKQLGDYHLHFSEGSA-IMEKIVAVAMITRRLLLEEPD-----TSTQSLPISDRDQ 590
W +K+L YH FS + M+ IV++ + T ++L E+ Q + +
Sbjct: 385 MGWTEKRLLAYHETFSASNIDSMQGIVSIGVSTAKILAEDISHEYHRKRKQETDVVVHSK 444
Query: 591 IEVYITSSIKHAFTR 605
IE YI SS++ AF +
Sbjct: 445 IETYIRSSLRTAFAQ 459
>UniRef100_Q6ES97 Hypothetical protein P0512H04.9-2 [Oryza sativa]
Length = 800
Score = 139 bits (350), Expect = 3e-31
Identities = 92/305 (30%), Positives = 159/305 (51%), Gaps = 33/305 (10%)
Query: 310 LLVPLELLCCVARTEFSDKKAFIRWQKRQLKVLEEGLVNHPVVGFGESGRKTNEMRILLA 369
+++PLE L ++ D + + WQ R LK+LE GL+ HP+V +S +R ++
Sbjct: 1 MVLPLEFLQQFKASDIPDPQEYEAWQSRNLKLLEAGLLVHPLVPLNKSDVSAQRLRQIIR 60
Query: 370 KIEESEFLPSSSGELQRTECLRSLREIAIPLAERPARGDLTGEICHWADGYQFNVRLYEK 429
+ P +G+ +E ++ LR + LA R G T + CHWADG+ N+ LY+
Sbjct: 61 GAYDR---PLETGK--NSESMQVLRSAVMSLAGRSDDG--TSDGCHWADGFPLNLHLYQM 113
Query: 430 LLLSVFDMLDEGKLTEEVEEILELLKSTWRVLGITETIHHTCYAWVLFRQYVITREHRI- 488
L+ + FD D+G + +E++E++ELLK TW +LGI + +H+ C+AW LF +V++ + I
Sbjct: 114 LVEACFDN-DDGTVVDEIDEVMELLKKTWGILGINQMLHNLCFAWALFNHFVMSGQVDIE 172
Query: 489 LLHALEQLNKIPLMEQRGQQERLHLKSLRSKVEGERDMSF---LQAFLTPIQRWADKQLG 545
LL A E L + + +D ++ L + L+ I W +K+L
Sbjct: 173 LLSAAEN----------------QLAEVAKDAKTTKDPNYSKVLSSTLSSIMGWTEKRLL 216
Query: 546 DYHLHFSEGS-AIMEKIVAVAMITRRLLLEEPDTSTQSLPISD----RDQIEVYITSSIK 600
YH F+ + M+ IV++ + R+L+E+ + + R +IE YI SS++
Sbjct: 217 AYHETFNTSNIESMQGIVSIGVSAARVLVEDISHEYRRRRKEETDVARSRIETYIRSSLR 276
Query: 601 HAFTR 605
AF +
Sbjct: 277 TAFAQ 281
>UniRef100_Q7XUB4 OSJNBb0032E06.4 protein [Oryza sativa]
Length = 1005
Score = 126 bits (317), Expect = 2e-27
Identities = 74/227 (32%), Positives = 121/227 (52%), Gaps = 12/227 (5%)
Query: 388 ECLRSLREIAIPLAERPARGDLTGEICHWADGYQFNVRLYEKLLLSVFDMLDEGKLTEEV 447
E L + L E P + +L GE HW Y N RLYEKLL VFD+L++G+L EE
Sbjct: 211 EVLTIIERYNAKLCEAPKKFNLKGETYHWIQSYHLNFRLYEKLLCIVFDILEDGQLVEEA 270
Query: 448 EEILELLKSTWRVLGITETIHHTCYAWVLFRQYVITREHRILLHALEQLNKIPLMEQRGQ 507
+EILE +K TW +LGIT+ +H T +AWVLF+++ T E +L H Q K+ L +
Sbjct: 271 DEILETVKLTWTILGITQKLHDTLFAWVLFKKFAETGEILLLKHTCLQTQKLRLHNDAKE 330
Query: 508 QERLHLKSLRSKVE---GERDMSFLQAFLTPIQRWADKQLGDYHLHFSE-GSAIMEKIVA 563
E L+ S E G +S + + + I +W +QL +YH +F++ ++I E ++
Sbjct: 331 IE-LYTNSFVCSAEACGGNMALSLVDSAILKINKWCFRQLENYHSYFNKVDNSIFEGMLN 389
Query: 564 VAMITRRLLLEEPDTSTQSLPI-------SDRDQIEVYITSSIKHAF 603
+ +I+ ++ D +++ I + I + + SI+ A+
Sbjct: 390 LVVISETSRTDDDDDDEKAMLIGTPLDATQESKLIHILVVRSIQAAY 436
Score = 98.6 bits (244), Expect = 5e-19
Identities = 75/202 (37%), Positives = 98/202 (48%), Gaps = 30/202 (14%)
Query: 23 LLQRYRRDRRVLLDFILSGSLIKKVVMPPGAVTLDDVDLDQVSIDYVLNCAKKSEMLELS 82
LL+ YRRDRR LL FILS + + AV L VDLD VS DY L C + S
Sbjct: 6 LLEVYRRDRRALLGFILSSAGGR-------AVDLSRVDLDAVSADYALGCVASGVQFDAS 58
Query: 83 EAIRDYHDHTGLPQMSDTGSVGEFYLVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPA 142
EA R Y D P M + SV ++L++ PE SGSPP + A P I P
Sbjct: 59 EATRRYFDERRYPIMMGSPSVNSYFLLSRPERSGSPPNK--------AAPDIVPQAPAEE 110
Query: 143 YPTSPVASNISRSESLYSAQERELTVDDIEDFEDDDDTSMVEGLRAKRTLNDASDLAVKL 202
PT P+ ++ + A L D+ D S+ + + +N L++ L
Sbjct: 111 NPT-PIREHV----DFFRAAINILGTDN-----GTKDVSLAD--IYPKQVNKMDILSLGL 158
Query: 203 PPFSTGITDDDLRETAYEILLA 224
P S TDDD+RETAYE+LLA
Sbjct: 159 PKLS---TDDDIRETAYEVLLA 177
>UniRef100_Q94I20 Hypothetical protein OSJNBa0034E23.5 [Oryza sativa]
Length = 368
Score = 70.9 bits (172), Expect = 1e-10
Identities = 60/220 (27%), Positives = 98/220 (44%), Gaps = 9/220 (4%)
Query: 213 DLRETAYEILLACAGATGGLIVPSKEKKKDRKSSSLIRKLGRSKTGSIVSQSQNAPGLVG 272
+LR+TAYEI ++C ++GG ++ + SL + R + G S+ +NA GL
Sbjct: 131 ELRKTAYEIFMSCRSSSGGNTAGARGAAMEAAEVSL--PVARPRGGGGGSRIKNALGLKA 188
Query: 273 LLESMRVQLEISEAMDIRT-KQGLLNALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAF 331
S + ++ M +RT Q L AL G+ + M + + + + + ++
Sbjct: 189 RRLSSSA-VAATQPMMVRTLSQTLGPALPGRGRQLMTSAEIMRQQIRVTEQNNARLRRTL 247
Query: 332 IRWQKRQLKVLEEGLVNHPVVGFGESGRKTNEMRILLAKIEESEFLPSSSGELQRTECLR 391
+R Q+K+LE GL+ HP + R ++ E + + +R
Sbjct: 248 MRAIVGQVKLLEAGLILHPSLPLDRLNSAVLRFREVMRATEIRAI-----DTAKNSNAMR 302
Query: 392 SLREIAIPLAERPARGDLTGEICHWADGYQFNVRLYEKLL 431
+L LA R G G+ CHWADGY NV LY LL
Sbjct: 303 TLTSAVHALAWRSGVGSGGGDACHWADGYSLNVLLYISLL 342
>UniRef100_UPI00001D7AB7 UPI00001D7AB7 UniRef100 entry
Length = 334
Score = 44.7 bits (104), Expect = 0.008
Identities = 58/191 (30%), Positives = 78/191 (40%), Gaps = 46/191 (24%)
Query: 69 VLNCAKKSEMLELSEAIRDYHDHTG-LPQMSDTGSVGEFYLVTDP-------ESSGSP-- 118
+L C S LE+ EA H H L + S G V P S G+P
Sbjct: 77 LLTCLLPSSFLEMMEA--HSHGHASPLAAKGQSPSSGSQSPVVPPGAVSTGSSSPGTPQP 134
Query: 119 ----PKRAPPPVPISAVPPIAVSTPPPAYPTSPVA--SNISR-------SESL------Y 159
P A PP ++ VPP + + PPPA P++P S ISR SE+
Sbjct: 135 APQLPLNAAPPSSVTPVPP-SEALPPPACPSAPAPRRSIISRLFGTSPASEAAPPPPEPV 193
Query: 160 SAQERELTVDDIEDFEDDD--DTSMVE-----------GLRAKRTLNDASDLAVKLPPFS 206
A E TV +EDF DD D S +E G +A + +D+ A+ P
Sbjct: 194 PAAEAPATVQSVEDFVPDDRLDRSFLEDTTPARDEKKVGAKAAQQDSDSDGEALGGNPMV 253
Query: 207 TGITDD-DLRE 216
G DD DL++
Sbjct: 254 AGFQDDVDLKD 264
>UniRef100_Q41805 Extensin-like protein precursor [Zea mays]
Length = 1188
Score = 44.3 bits (103), Expect = 0.011
Identities = 19/34 (55%), Positives = 23/34 (66%)
Query: 114 SSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
S S PK +PPP P+S PPI S+PPPA +SP
Sbjct: 891 SPPSEPKSSPPPTPVSLPPPIVKSSPPPAMVSSP 924
Score = 40.8 bits (94), Expect = 0.12
Identities = 21/37 (56%), Positives = 23/37 (61%), Gaps = 1/37 (2%)
Query: 112 PESSGSPP-KRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P SS PP K PPP PIS+ PP S PPPA +SP
Sbjct: 1049 PVSSPPPPVKSPPPPAPISSPPPPVKSPPPPAPVSSP 1085
Score = 40.4 bits (93), Expect = 0.16
Identities = 20/37 (54%), Positives = 23/37 (62%), Gaps = 1/37 (2%)
Query: 112 PESSGSPP-KRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P SS PP K PPP P+S+ PP S PPPA +SP
Sbjct: 1017 PVSSPPPPVKSPPPPAPVSSPPPPVKSPPPPAPVSSP 1053
Score = 40.4 bits (93), Expect = 0.16
Identities = 19/36 (52%), Positives = 23/36 (63%), Gaps = 1/36 (2%)
Query: 112 PESSGSP-PKRAPPPVPISAVPPIAVSTPPPAYPTS 146
P SS P PK +PPP P+S+ PP V +PPP P S
Sbjct: 984 PVSSPPPAPKSSPPPAPMSSPPPPEVKSPPPPAPVS 1019
Score = 40.0 bits (92), Expect = 0.21
Identities = 17/32 (53%), Positives = 23/32 (71%)
Query: 119 PKRAPPPVPISAVPPIAVSTPPPAYPTSPVAS 150
PK +PPPV +S+ PP S+PPPA +SP A+
Sbjct: 928 PKSSPPPVVVSSPPPTVKSSPPPAPVSSPPAT 959
Score = 40.0 bits (92), Expect = 0.21
Identities = 20/37 (54%), Positives = 23/37 (62%), Gaps = 1/37 (2%)
Query: 112 PESSGSPP-KRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P SS PP K PPP P+S+ PP S PPPA +SP
Sbjct: 1033 PVSSPPPPVKSPPPPAPVSSPPPPVKSPPPPAPISSP 1069
Score = 40.0 bits (92), Expect = 0.21
Identities = 20/37 (54%), Positives = 23/37 (62%), Gaps = 1/37 (2%)
Query: 112 PESSGSPP-KRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P SS PP K PPP P+S+ PP S PPPA +SP
Sbjct: 1065 PISSPPPPVKSPPPPAPVSSPPPPVKSPPPPAPVSSP 1101
Score = 40.0 bits (92), Expect = 0.21
Identities = 20/38 (52%), Positives = 23/38 (59%), Gaps = 2/38 (5%)
Query: 112 PESSGSPP--KRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P SS PP K PPP P+S+ PP S PPPA +SP
Sbjct: 1000 PMSSPPPPEVKSPPPPAPVSSPPPPVKSPPPPAPVSSP 1037
Score = 40.0 bits (92), Expect = 0.21
Identities = 20/37 (54%), Positives = 23/37 (62%), Gaps = 1/37 (2%)
Query: 112 PESSGSPP-KRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P SS PP K PPP P+S+ PP S PPPA +SP
Sbjct: 1081 PVSSPPPPVKSPPPPAPVSSPPPPIKSPPPPAPVSSP 1117
Score = 38.5 bits (88), Expect = 0.60
Identities = 18/39 (46%), Positives = 21/39 (53%), Gaps = 3/39 (7%)
Query: 112 PESSGSPP---KRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P GSPP K PPP P+++ PP S PPP SP
Sbjct: 553 PAPVGSPPPPEKSPPPPAPVASPPPPVKSPPPPTLVASP 591
Score = 38.5 bits (88), Expect = 0.60
Identities = 15/28 (53%), Positives = 19/28 (67%)
Query: 119 PKRAPPPVPISAVPPIAVSTPPPAYPTS 146
PK +PPP P+S+ PP VS+PP P S
Sbjct: 732 PKSSPPPAPVSSPPPTPVSSPPALAPVS 759
Score = 38.5 bits (88), Expect = 0.60
Identities = 20/39 (51%), Positives = 24/39 (61%), Gaps = 7/39 (17%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPP-AYPTSPVA 149
P SS PP+++PPP PP A STPPP YPT P +
Sbjct: 644 PVSSPPPPEKSPPP------PPPAKSTPPPEEYPTPPTS 676
Score = 38.1 bits (87), Expect = 0.79
Identities = 17/32 (53%), Positives = 22/32 (68%), Gaps = 2/32 (6%)
Query: 118 PP--KRAPPPVPISAVPPIAVSTPPPAYPTSP 147
PP K +PPP P+S+ PP S+PPPA +SP
Sbjct: 973 PPEVKSSPPPTPVSSPPPAPKSSPPPAPMSSP 1004
Score = 37.4 bits (85), Expect = 1.3
Identities = 20/52 (38%), Positives = 28/52 (53%), Gaps = 10/52 (19%)
Query: 112 PESSGSPPKRAPPPVPISAVPP----------IAVSTPPPAYPTSPVASNIS 153
P SS K +PPP P+S+ PP + VS+PPPA +SP + +S
Sbjct: 757 PVSSPPSVKSSPPPAPLSSPPPAPQVKSSPPPVQVSSPPPAPKSSPPLAPVS 808
Score = 37.4 bits (85), Expect = 1.3
Identities = 18/39 (46%), Positives = 22/39 (56%), Gaps = 3/39 (7%)
Query: 112 PESSGSPPKR---APPPVPISAVPPIAVSTPPPAYPTSP 147
PE SPP +PPP P S+ PP +S+PPP SP
Sbjct: 974 PEVKSSPPPTPVSSPPPAPKSSPPPAPMSSPPPPEVKSP 1012
Score = 37.0 bits (84), Expect = 1.7
Identities = 20/43 (46%), Positives = 26/43 (59%), Gaps = 1/43 (2%)
Query: 112 PESSGSP-PKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNIS 153
P+SS P P PPP S+ PP VS+PPPA +SP + +S
Sbjct: 960 PKSSPPPAPVNLPPPEVKSSPPPTPVSSPPPAPKSSPPPAPMS 1002
Score = 37.0 bits (84), Expect = 1.7
Identities = 15/29 (51%), Positives = 21/29 (71%)
Query: 119 PKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
PK +PP V +S+ PP+ S+PPPA +SP
Sbjct: 830 PKSSPPHVVVSSPPPVVKSSPPPAPVSSP 858
Score = 37.0 bits (84), Expect = 1.7
Identities = 15/34 (44%), Positives = 21/34 (61%)
Query: 114 SSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
S + PK +PPP P++ PP S+PPP +SP
Sbjct: 955 SPPATPKSSPPPAPVNLPPPEVKSSPPPTPVSSP 988
Score = 37.0 bits (84), Expect = 1.7
Identities = 18/45 (40%), Positives = 25/45 (55%), Gaps = 3/45 (6%)
Query: 112 PESSGSPPKR---APPPVPISAVPPIAVSTPPPAYPTSPVASNIS 153
P SPP +PP P S+ PP+ VS+PPP +SP + +S
Sbjct: 910 PIVKSSPPPAMVSSPPMTPKSSPPPVVVSSPPPTVKSSPPPAPVS 954
Score = 37.0 bits (84), Expect = 1.7
Identities = 18/49 (36%), Positives = 27/49 (54%)
Query: 110 TDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNISRSESL 158
T + SP K +PP P+S+ P S+PPPA +SP + +S +L
Sbjct: 707 TPSKPPSSPEKPSPPKEPVSSPPQTPKSSPPPAPVSSPPPTPVSSPPAL 755
Score = 37.0 bits (84), Expect = 1.7
Identities = 19/43 (44%), Positives = 24/43 (55%), Gaps = 4/43 (9%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPP----AYPTSPVAS 150
P +S PP ++PPP + A PP V +PPP A P PV S
Sbjct: 571 PVASPPPPVKSPPPPTLVASPPPPVKSPPPPAPVASPPPPVKS 613
Score = 36.6 bits (83), Expect = 2.3
Identities = 20/40 (50%), Positives = 25/40 (62%), Gaps = 4/40 (10%)
Query: 112 PESSGSPPKRA-PPPVPISAVPPIAV---STPPPAYPTSP 147
P SS PP ++ PPP P+S+ PP V S PPPA +SP
Sbjct: 1097 PVSSPPPPIKSPPPPAPVSSPPPAPVKPPSLPPPAPVSSP 1136
Score = 36.6 bits (83), Expect = 2.3
Identities = 19/39 (48%), Positives = 21/39 (53%), Gaps = 3/39 (7%)
Query: 112 PESSGSPP---KRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P S SPP K PPP P+ + PP S PPPA SP
Sbjct: 537 PVSVVSPPPPVKSPPPPAPVGSPPPPEKSPPPPAPVASP 575
Score = 36.2 bits (82), Expect = 3.0
Identities = 17/43 (39%), Positives = 27/43 (62%), Gaps = 1/43 (2%)
Query: 112 PESSGSP-PKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNIS 153
P+SS P P +PPP P+S+ P +A + PP+ +SP + +S
Sbjct: 732 PKSSPPPAPVSSPPPTPVSSPPALAPVSSPPSVKSSPPPAPLS 774
Score = 35.8 bits (81), Expect = 3.9
Identities = 20/48 (41%), Positives = 25/48 (51%), Gaps = 8/48 (16%)
Query: 112 PESSGSPP---KRAPPPVPISAVPPIAVSTPPPA-----YPTSPVASN 151
P SPP K PPP P+++ PP S PPP P +PVAS+
Sbjct: 585 PTLVASPPPPVKSPPPPAPVASPPPPVKSPPPPTPVASPPPPAPVASS 632
>UniRef100_Q9FFW5 Similarity to protein kinase [Arabidopsis thaliana]
Length = 681
Score = 43.5 bits (101), Expect = 0.019
Identities = 20/57 (35%), Positives = 30/57 (52%)
Query: 95 PQMSDTGSVGEFYLVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASN 151
P S++ + L T P + +PP PPP P + PP+ S+PPP +SP S+
Sbjct: 12 PPSSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVSSPPPSS 68
Score = 35.4 bits (80), Expect = 5.1
Identities = 19/49 (38%), Positives = 27/49 (54%), Gaps = 7/49 (14%)
Query: 108 LVTDPESSGSPPKRAP------PPVPISAVPPIAVSTPPPAYP-TSPVA 149
+V+ P S SPP P P V S PP+ +++PPP+ P T+P A
Sbjct: 60 VVSSPPPSSSPPPSPPVITSPPPTVASSPPPPVVIASPPPSTPATTPPA 108
>UniRef100_Q7UG87 Hypothetical protein [Rhodopirellula baltica]
Length = 593
Score = 42.7 bits (99), Expect = 0.032
Identities = 61/260 (23%), Positives = 99/260 (37%), Gaps = 34/260 (13%)
Query: 36 DFILSGSLIKKVVMPPGAVTLDDVDLDQVSIDYVLNCAKKSEMLELS-----EAIRDYHD 90
D +L+ ++ + V P V ++ VD+ + V N A+ SE +E S +++ D D
Sbjct: 232 DIVLAPAVQEPVSPQPAEVEVEAVDVAPIETPVVAN-AEVSEPIEFSLNDATDSLSDADD 290
Query: 91 HTGLPQMSDTGSVGEFYLVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVAS 150
L SD+ GE + + S P K A VP PP+ T P P
Sbjct: 291 SISL-SFSDSDEPGENLVADELVSMPEPMKIA---VPTKIAPPVEQPTDESTLPRLPKPV 346
Query: 151 NISRSESLYSAQERELTV--DDIEDFEDDDDTSMVEGLRAKRTLNDASDLAVKLPPF--- 205
I + Q+R L V D E D M + + +AV+ PP
Sbjct: 347 QIKTPSEMMVQQQRHLPVKIDSPTQLERADSERM-----QNAPVRHRAAVAVEAPPMVAV 401
Query: 206 --STGITDDDLRETAYEILLACAGATGGLI---VPSKEKKKDRKSSSLIRKLGRSKTGSI 260
T + R +A I+ A + G VP+ E ++ + I+ T
Sbjct: 402 AKRTASVSSESRVSAARIVPAALASHRGEFHRSVPATEAERSLRIEDTIQCDANDVTSLT 461
Query: 261 VSQSQNAPGLVGLLESMRVQ 280
V G++E++RV+
Sbjct: 462 VD---------GVIEAVRVE 472
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.136 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,016,050,872
Number of Sequences: 2790947
Number of extensions: 43258384
Number of successful extensions: 253307
Number of sequences better than 10.0: 585
Number of HSP's better than 10.0 without gapping: 164
Number of HSP's successfully gapped in prelim test: 460
Number of HSP's that attempted gapping in prelim test: 242971
Number of HSP's gapped (non-prelim): 6176
length of query: 629
length of database: 848,049,833
effective HSP length: 134
effective length of query: 495
effective length of database: 474,062,935
effective search space: 234661152825
effective search space used: 234661152825
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 78 (34.7 bits)
Medicago: description of AC149307.14


