

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149305.8 - phase: 0 /pseudo
(439 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
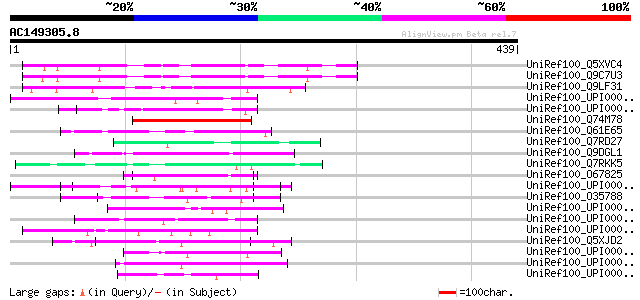
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q5XVC4 Hypothetical protein [Arabidopsis thaliana] 125 3e-27
UniRef100_Q9C7U3 Hypothetical protein TEL3N.1 [Arabidopsis thali... 125 3e-27
UniRef100_Q9LF31 Hypothetical protein T20K14_120 [Arabidopsis th... 118 4e-25
UniRef100_UPI0000436183 UPI0000436183 UniRef100 entry 57 8e-07
UniRef100_UPI00001A1808 UPI00001A1808 UniRef100 entry 56 2e-06
UniRef100_Q74M78 NEQ435 [Nanoarchaeum equitans] 54 7e-06
UniRef100_Q61E65 Hypothetical protein CBG12202 [Caenorhabditis b... 54 1e-05
UniRef100_Q7RD27 Drosophila melanogaster CG2972 gene product [Pl... 54 1e-05
UniRef100_Q9DGL1 Retinitis pigmentosa GTPase regulator-like prot... 53 2e-05
UniRef100_Q7RKK5 Hypothetical protein [Plasmodium yoelii yoelii] 53 2e-05
UniRef100_O67825 Translation initiation factor IF-2 [Aquifex aeo... 52 3e-05
UniRef100_UPI0000436182 UPI0000436182 UniRef100 entry 52 4e-05
UniRef100_O35788 Cyclic nucleotide-gated channel beta subunit [R... 52 4e-05
UniRef100_UPI0000364C4A UPI0000364C4A UniRef100 entry 51 6e-05
UniRef100_UPI0000498854 UPI0000498854 UniRef100 entry 51 6e-05
UniRef100_UPI00004659AA UPI00004659AA UniRef100 entry 51 6e-05
UniRef100_Q5XJD2 Hypothetical protein [Brachydanio rerio] 51 6e-05
UniRef100_UPI0000439AE7 UPI0000439AE7 UniRef100 entry 51 7e-05
UniRef100_UPI0000428FBC UPI0000428FBC UniRef100 entry 51 7e-05
UniRef100_UPI000046C3AD UPI000046C3AD UniRef100 entry 51 7e-05
>UniRef100_Q5XVC4 Hypothetical protein [Arabidopsis thaliana]
Length = 488
Score = 125 bits (313), Expect = 3e-27
Identities = 106/302 (35%), Positives = 146/302 (48%), Gaps = 61/302 (20%)
Query: 12 ENLNPNEFSNRTPLQKS----LTKGDNNCVNVV--VVSPQKRIRQRKFVVAKKK-KNDQS 64
EN NPN S+ TPL+KS K N V V SP+ RIR+R+FVV KK + +++
Sbjct: 33 ENSNPNFVSHSTPLEKSSKSSAQKNPKWKPNPVPAVFSPRNRIRERRFVVVKKNSRKEKN 92
Query: 65 PRKTVLCKCGEN--DDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEV 122
+V CKCG + KCVC AY LR SQE FF + E+
Sbjct: 93 DSASVDCKCGAKTISNMKKCVCIAYETLRASQEEFFNNRR--------------ESVSEI 138
Query: 123 IEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMH 182
E++ + D GNE+ + + + S++KRRREKV+EEAR S+PE GKVMH
Sbjct: 139 GESSQNLED-GNEQVEFGDSDETRV---------SLMKRRREKVLEEARMSIPEFGKVMH 188
Query: 183 LVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQPVKDGDEQSEVVSSCCDD 242
LVKAFE+L K ++EEE D+ + +KW LPG+ S+ S +
Sbjct: 189 LVKAFEKLTCFPLSKVTSKEEE--DQIKQPLKWELPGM------------SQPKCSESET 234
Query: 243 DDFDF*TTWIGSKG--FSYIFLGLWKWKKCV*QEFFWW*KKQKKSW-NHLVLLEEGGGRR 299
D F + +++ S G + LGL + V SW N + L GGRR
Sbjct: 235 DQFTWSSSFYPSSGLILTATNLGLEQPHASV-----------SSSWDNSVSSLNSNGGRR 283
Query: 300 SR 301
R
Sbjct: 284 GR 285
>UniRef100_Q9C7U3 Hypothetical protein TEL3N.1 [Arabidopsis thaliana]
Length = 488
Score = 125 bits (313), Expect = 3e-27
Identities = 106/302 (35%), Positives = 146/302 (48%), Gaps = 61/302 (20%)
Query: 12 ENLNPNEFSNRTPLQK----SLTKGDNNCVNVV--VVSPQKRIRQRKFVVAKKK-KNDQS 64
EN NPN S+ TPL+K S K N V V SP+ RIR+R+FVV KK + +++
Sbjct: 33 ENSNPNFVSHSTPLEKPSKSSAQKNPKWKPNPVPAVFSPRNRIRERRFVVVKKNSRKEKN 92
Query: 65 PRKTVLCKCGEN--DDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEV 122
+V CKCG + KCVC AY LR SQE FF + E+
Sbjct: 93 DSASVDCKCGAKTISNMKKCVCIAYETLRASQEEFFNNRR--------------ESVSEI 138
Query: 123 IEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMH 182
E++ + D GNE+ + + + S++KRRREKV+EEAR S+PE GKVMH
Sbjct: 139 GESSQNLED-GNEQVEFGDSDETRV---------SLMKRRREKVLEEARMSIPEFGKVMH 188
Query: 183 LVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQPVKDGDEQSEVVSSCCDD 242
LVKAFE+L K ++EEE D+ + +KW LPG+ S+ S +
Sbjct: 189 LVKAFEKLTCFPLSKVTSKEEE--DQIKQPLKWELPGM------------SQPKCSESET 234
Query: 243 DDFDF*TTWIGSKG--FSYIFLGLWKWKKCV*QEFFWW*KKQKKSW-NHLVLLEEGGGRR 299
D F + +++ S G + LGL + V SW N + L GGRR
Sbjct: 235 DQFTWSSSFYPSSGLILTATNLGLEQPHASV-----------SSSWDNSVSSLNSNGGRR 283
Query: 300 SR 301
R
Sbjct: 284 GR 285
>UniRef100_Q9LF31 Hypothetical protein T20K14_120 [Arabidopsis thaliana]
Length = 494
Score = 118 bits (295), Expect = 4e-25
Identities = 96/270 (35%), Positives = 132/270 (48%), Gaps = 45/270 (16%)
Query: 12 ENLNPN---EFSNRTPLQKSLTKGDNNCVNV------VVVSPQKRIRQRKFVVAKKKKND 62
EN NPN +++PL KS K + V SP+ RIR+RKFVV KK +
Sbjct: 32 ENANPNISQASPSKSPLMKSAKKSAQKNPSPSPKPTQAVFSPRNRIRERKFVVVAKKNSR 91
Query: 63 QSPRKTVL---------CKCGENDDGS-KCVCEAYRNLRESQEGFFEKENFFDGEEKNDE 112
+ ++ + CKCGE G+ KCVC AY LR SQE FF+ + +
Sbjct: 92 KGKKEPAITESKAAEIDCKCGERKKGNMKCVCVAYETLRASQEEFFKN--------RIES 143
Query: 113 EKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARN 172
E+ + LEE ++GN DE EG E EK S+M KR R KV+EEAR
Sbjct: 144 EEDKEALEECC-------NLGNG----DELEGKESGEPEKTGVSTM-KRSRAKVVEEARQ 191
Query: 173 SVPENGKVMHLVKAFERLLSIKKEKEKNEEEE---ENDKKNKVMKWALPGLQFQQPVKDG 229
SVP +GKVM+LV+AFE+L K N+ EE E D K V L G + + P
Sbjct: 192 SVPVSGKVMNLVEAFEKLTCFSNSKTANKIEENQTEEDTKKPVKLEFLEGEKEKNPWSSS 251
Query: 230 DEQSEVVSSCCD---DDDFDF*TTWIGSKG 256
SE+V + + D + ++W ++G
Sbjct: 252 FCPSEMVLTAKNLGLDPNASISSSWDSTRG 281
>UniRef100_UPI0000436183 UPI0000436183 UniRef100 entry
Length = 236
Score = 57.4 bits (137), Expect = 8e-07
Identities = 55/232 (23%), Positives = 102/232 (43%), Gaps = 34/232 (14%)
Query: 1 MTPEKKKPLEIENLNPNEFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIRQRKFVVAKKKK 60
M +++ +E E+ +++ + + + N+ +K ++RK ++KK
Sbjct: 21 MDKKEEGKIEEEDGRKGRIIKEEKMEEMMMEKEEEEGNIKKEGKKKERKRRKIRKKEEKK 80
Query: 61 NDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLE 120
+S RK + K E D + + +E E+E + EEK ++EK E +
Sbjct: 81 ERRSRRKEEMKKNEEED----------MEMMDMKEEIKEEEEEEEEEEKKEQEKTEETMM 130
Query: 121 EVIEANLIIHDIGNEERKIDEE--EGVEEENEEKGNCSSMVKR----------------R 162
+ E I + G + R I EE E + E EE+GN K+ R
Sbjct: 131 DKKEEGKIEEEGGRKGRIIKEEKMEEMMMEKEEEGNIKKEGKKKERKRRKIRKKEEKKER 190
Query: 163 REKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
R + EE + + E+ ++M + + IK+E+E+ EEEE+ K+N+ K
Sbjct: 191 RSRRKEEMKKNEEEDMEMMDMKE------EIKEEEEEEEEEEKVKKRNRKQK 236
Score = 42.7 bits (99), Expect = 0.020
Identities = 34/113 (30%), Positives = 57/113 (50%), Gaps = 10/113 (8%)
Query: 109 KNDEEKGENVLEEVIEANLIIHDIGNEERKIDE---EEGVEEENEEKGNCSSMVK---RR 162
K ++EK E + + E I + G + R I E EE + E+ EE+GN K R+
Sbjct: 10 KKEQEKTEETMMDKKEEGKIEEEDGRKGRIIKEEKMEEMMMEKEEEEGNIKKEGKKKERK 69
Query: 163 REKVMEEARNSVPENGKVMHLVKAFE---RLLSIKKE-KEKNEEEEENDKKNK 211
R K+ ++ + + + K E ++ +K+E KE+ EEEEE +KK +
Sbjct: 70 RRKIRKKEEKKERRSRRKEEMKKNEEEDMEMMDMKEEIKEEEEEEEEEEKKEQ 122
Score = 40.4 bits (93), Expect = 0.10
Identities = 36/157 (22%), Positives = 68/157 (42%), Gaps = 14/157 (8%)
Query: 85 EAYRNLRESQEGFFEKENFFDGEEKN-----DEEKGENVLEEVIEANLIIHDIGNEERKI 139
E R R +E +E D +E+ D KG + EE +E ++ + EE I
Sbjct: 2 EKKRRRRRKKEQEKTEETMMDKKEEGKIEEEDGRKGRIIKEEKMEEMMMEKE--EEEGNI 59
Query: 140 DEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEK 199
+E +E K K RR + EE + + E+ ++M + + KE+E+
Sbjct: 60 KKEGKKKERKRRKIRKKEEKKERRSRRKEEMKKNEEEDMEMMDMKEEI-------KEEEE 112
Query: 200 NEEEEENDKKNKVMKWALPGLQFQQPVKDGDEQSEVV 236
EEEEE ++ K + + + + ++G + ++
Sbjct: 113 EEEEEEKKEQEKTEETMMDKKEEGKIEEEGGRKGRII 149
>UniRef100_UPI00001A1808 UPI00001A1808 UniRef100 entry
Length = 290
Score = 56.2 bits (134), Expect = 2e-06
Identities = 46/156 (29%), Positives = 77/156 (48%), Gaps = 15/156 (9%)
Query: 59 KKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENV 118
+K ++S +K + K E D E + E ++ EKE + EEK +EEK E+
Sbjct: 6 EKGEKSKKKRIKKKKKEKDKKE----EKKKKKNEEED---EKEEKNEEEEKKEEEK-EDE 57
Query: 119 LEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENG 178
+E + N+ + E++K +EE+ EEE EE+ M + E+ EE + E
Sbjct: 58 EKEGKKKNVKEDNKEEEKKKEEEEKEKEEEKEEEEKEKKMEEEEEEETKEEEKEEKKEEE 117
Query: 179 KVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
K+ + K + KE EK+E++E+ KK + MK
Sbjct: 118 KMKEVKK-------VDKEVEKDEKKEKKKKKKEKMK 146
Score = 51.6 bits (122), Expect = 4e-05
Identities = 49/180 (27%), Positives = 84/180 (46%), Gaps = 18/180 (10%)
Query: 43 SPQKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKEN 102
S +KRI+++K KK++ + + K +N++ K E +E ++ +++N
Sbjct: 11 SKKKRIKKKKKEKDKKEEKKKKKNEEEDEKEEKNEEEEKKEEEKEDEEKEGKKKNVKEDN 70
Query: 103 FFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRR 162
+ ++K +EEK + +E E +E+K++EEE E + EEK K +
Sbjct: 71 KEEEKKKEEEEKEKEEEKEEEE----------KEKKMEEEEEEETKEEEKEEKKEEEKMK 120
Query: 163 R-EKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEE-------EEENDKKNKVMK 214
+KV +E + K K + KKEK+K EE EEE DKK +V K
Sbjct: 121 EVKKVDKEVEKDEKKEKKKKKKEKMKKEKREEKKEKKKEEEEKKKKKREEEEDKKEEVKK 180
Score = 47.0 bits (110), Expect = 0.001
Identities = 42/158 (26%), Positives = 75/158 (46%), Gaps = 11/158 (6%)
Query: 58 KKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGEN 117
KKKN + K K E + E + E ++ E+E EE+ +E+K E
Sbjct: 62 KKKNVKEDNKEEEKKKEEEEKEK----EEEKEEEEKEKKMEEEEEEETKEEEKEEKKEEE 117
Query: 118 VLEEVIEANLIIH-DIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPE 176
++EV + + + D E++K +E+ +E+ EEK + +++K EE E
Sbjct: 118 KMKEVKKVDKEVEKDEKKEKKKKKKEKMKKEKREEKKEKKKEEEEKKKKKREE------E 171
Query: 177 NGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
K + K E+ KKE+E+ +EE++ D+K + K
Sbjct: 172 EDKKEEVKKEEEKKGEKKKEEEEKKEEKKKDEKKEEKK 209
Score = 45.4 bits (106), Expect = 0.003
Identities = 44/169 (26%), Positives = 80/169 (47%), Gaps = 26/169 (15%)
Query: 46 KRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFD 105
K +++ V K +K ++ +K K + ++ + E ++ +E E+E+ +
Sbjct: 120 KEVKKVDKEVEKDEKKEKKKKKKEKMKKEKREEKKEKKKEEEEKKKKKRE---EEEDKKE 176
Query: 106 GEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREK 165
+K +E+KGE EE EE+K EE+ +E+ EEK KRRR++
Sbjct: 177 EVKKEEEKKGEKKKEE-------------EEKK--EEKKKDEKKEEK-------KRRRKR 214
Query: 166 VMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
E + P+ G+ K E K+E++K E+EEE +K+ + K
Sbjct: 215 RRRE-EEAPPQGGEKEEEKKKKEEEKENKEEEKKEEKEEEENKEEEKKK 262
Score = 38.9 bits (89), Expect = 0.29
Identities = 42/161 (26%), Positives = 64/161 (39%), Gaps = 24/161 (14%)
Query: 45 QKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFF 104
+K+ ++++ KKKK ++ K K E G K +E +E EK+
Sbjct: 152 EKKEKKKEEEEKKKKKREEEEDKKEEVKKEEEKKGEK--------KKEEEEKKEEKKKDE 203
Query: 105 DGEEKNDEEKGENVLEEVIEANLIIHDIGN--EERKIDEEEGVEEENEEKGNCSSMVKRR 162
EEK K EE + EE K ++EE +EE EE+ N K+
Sbjct: 204 KKEEKKRRRKRRRREEEAPPQGGEKEEEKKKKEEEKENKEEEKKEEKEEEENKEEEKKKE 263
Query: 163 REKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEE 203
K+ EE + EN + KK+KEK + E
Sbjct: 264 ENKIEEEEEEKMEENEE--------------KKKKEKKKVE 290
Score = 37.7 bits (86), Expect = 0.66
Identities = 45/211 (21%), Positives = 86/211 (40%), Gaps = 15/211 (7%)
Query: 4 EKKKPLEIENLNPNEFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIRQRKFVVAKKKKNDQ 63
EK+K +E E + + ++ + V+ V +K+ +++K KK+K +
Sbjct: 92 EKEKKMEEEEEEETKEEEKEEKKEEEKMKEVKKVDKEVEKDEKKEKKKK----KKEKMKK 147
Query: 64 SPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEK--ENFFDGEEKNDEEKGENVLEE 121
R+ K E ++ K E + +E + EK E + EEK +E+K + EE
Sbjct: 148 EKREEKKEKKKEEEEKKKKKREEEEDKKEEVKKEEEKKGEKKKEEEEKKEEKKKDEKKEE 207
Query: 122 VIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVM 181
+R+ EEE + E++ + + K E+ E K
Sbjct: 208 KKR---------RRKRRRREEEAPPQGGEKEEEKKKKEEEKENKEEEKKEEKEEEENKEE 258
Query: 182 HLVKAFERLLSIKKEKEKNEEEEENDKKNKV 212
K ++ ++EK + EE++ +K KV
Sbjct: 259 EKKKEENKIEEEEEEKMEENEEKKKKEKKKV 289
>UniRef100_Q74M78 NEQ435 [Nanoarchaeum equitans]
Length = 240
Score = 54.3 bits (129), Expect = 7e-06
Identities = 28/103 (27%), Positives = 63/103 (60%)
Query: 107 EEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKV 166
E+K +E +++ E++I+ + I D+ + E KI+E++ +++++ NCSS+++ ++K+
Sbjct: 122 EKKLEELTVDSIFEKIIKNGISITDVLDSEFKIEEKKEIKKKSLPTPNCSSIIEEYKQKI 181
Query: 167 MEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKK 209
++ K+ L++ + ++IK E E+ EE+EE D K
Sbjct: 182 DFLIEENMALRKKISQLLEMQKLTITIKIETERKEEKEEIDIK 224
>UniRef100_Q61E65 Hypothetical protein CBG12202 [Caenorhabditis briggsae]
Length = 284
Score = 53.5 bits (127), Expect = 1e-05
Identities = 48/184 (26%), Positives = 89/184 (48%), Gaps = 28/184 (15%)
Query: 45 QKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFF 104
Q+R+++R+ KKKK + R+ V K + ++ E + ++ E EK++
Sbjct: 25 QRRVKRRRR--RKKKKRRKKNRRRVKKKEEKKEEKK----EEKKEEKKEDEKKDEKKSDM 78
Query: 105 DGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRRE 164
E+K DEEK ++ ++ EE+K DE+E +E+ +EK K +E
Sbjct: 79 KDEDKKDEEKKDDKEKK-------------EEKKEDEKEKEDEKKDEK-------KDEKE 118
Query: 165 KVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALPGL--QF 222
+ +E + ++ K K + KKE+EK EE++E +KK + + G+ +F
Sbjct: 119 EDKKEEKKEEKKDDKEKKEEKKDQEKKEEKKEEEKKEEKKEEEKKEEKKEEPKAGIYYKF 178
Query: 223 QQPV 226
Q P+
Sbjct: 179 QSPL 182
Score = 39.3 bits (90), Expect = 0.23
Identities = 32/147 (21%), Positives = 74/147 (49%), Gaps = 26/147 (17%)
Query: 88 RNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEE 147
R +++ +E EK+ E+K DE+K E ++++ D +EE+K D+E+ E+
Sbjct: 45 RRVKKKEEKKEEKKEEKKEEKKEDEKKDEK------KSDMKDEDKKDEEKKDDKEKKEEK 98
Query: 148 ENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEEND 207
+ +EK + ++ +E ++ E+ K KKE++K+++E++ +
Sbjct: 99 KEDEK--------EKEDEKKDEKKDEKEEDKK------------EEKKEEKKDDKEKKEE 138
Query: 208 KKNKVMKWALPGLQFQQPVKDGDEQSE 234
KK++ K + ++ K+ +++ E
Sbjct: 139 KKDQEKKEEKKEEEKKEEKKEEEKKEE 165
>UniRef100_Q7RD27 Drosophila melanogaster CG2972 gene product [Plasmodium yoelii
yoelii]
Length = 491
Score = 53.5 bits (127), Expect = 1e-05
Identities = 48/181 (26%), Positives = 71/181 (38%), Gaps = 24/181 (13%)
Query: 91 RESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNE--ERKIDEEEGVEEE 148
R Q FEK + EE + E EN +E + + D+G E E +++ EE EEE
Sbjct: 140 RNGQPDTFEKRQVGENEENEENEVEENETDEDVREDETDEDVGEEVKENEVEVEEAEEEE 199
Query: 149 NEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDK 208
NE + + E+ EE + V E+ +E + EEE+E D+
Sbjct: 200 NEVEDGEADETDEGVEEEAEEVEDEVGED--------------ETDEEVNEAEEEDETDE 245
Query: 209 KNKVMKWALPGLQFQQPVKDGDEQSEVVSSCCDDDDFDF*TTWIGSKGFSYIFLGLWKWK 268
K L ++ V + DE DDD D W+ F L + K K
Sbjct: 246 GEDKSKRILGVETIKEEVVEVDENK--------DDDDDGGGKWVNVNNFDVFNLNVDKNK 297
Query: 269 K 269
K
Sbjct: 298 K 298
>UniRef100_Q9DGL1 Retinitis pigmentosa GTPase regulator-like protein [Fugu rubripes]
Length = 791
Score = 53.1 bits (126), Expect = 2e-05
Identities = 46/190 (24%), Positives = 88/190 (46%), Gaps = 11/190 (5%)
Query: 57 KKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGE 116
K++ N++ T K GE+++ + E + E Q EKE EE N+EE+
Sbjct: 517 KEESNEEEQSSTE--KEGESNEEEQSSTEKEESNEEEQSST-EKE-----EESNEEEQSS 568
Query: 117 NVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPE 176
EE E + + E + +EE+ +EE EE G + E+ +E E
Sbjct: 569 TEKEESNEEEEEEEESESNEEESEEEDQKDEEEEETGEEDEEEEEESEEQNQEEEAEEEE 628
Query: 177 NGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQPVKDGDEQSEVV 236
+G+ + E + EKE+ +EEEE +K+ + + ++ ++ ++ +E+ E
Sbjct: 629 DGETDVQEEEEEEE---ETEKEEEDEEEETEKEEEDEEEETDRVEEEEDAEENEEEEEEE 685
Query: 237 SSCCDDDDFD 246
S+ +D++ D
Sbjct: 686 SNSEEDEESD 695
Score = 42.4 bits (98), Expect = 0.027
Identities = 41/192 (21%), Positives = 79/192 (40%), Gaps = 28/192 (14%)
Query: 57 KKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGE 116
K++ N++ T + ++ S E E +E E + E++ DEE+ E
Sbjct: 544 KEESNEEEQSSTEKEEESNEEEQSSTEKEESNEEEEEEEESESNEEESEEEDQKDEEEEE 603
Query: 117 NVLEEVIEANLIIHDIGNEERKIDEEE----GVEEENEEKGNCSSMVKRRREKVMEEARN 172
E+ E + N+E + +EEE V+EE EE+ + E+ +E +
Sbjct: 604 TGEEDEEEEEE--SEEQNQEEEAEEEEDGETDVQEEEEEEEETEKEEEDEEEETEKEEED 661
Query: 173 SVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQPVKDGDEQ 232
E +V +++ E+NEEEEE + ++ + DG+E+
Sbjct: 662 EEEETDRVEE-----------EEDAEENEEEEEEESNSE-----------EDEESDGEEE 699
Query: 233 SEVVSSCCDDDD 244
+ S +D++
Sbjct: 700 EDEESDSEEDEE 711
Score = 41.2 bits (95), Expect = 0.059
Identities = 40/167 (23%), Positives = 68/167 (39%), Gaps = 25/167 (14%)
Query: 58 KKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKG-- 115
+++ ++S + + E +DG V E E+++ ++E + EE+++EE+
Sbjct: 609 EEEEEESEEQNQEEEAEEEEDGETDVQEEEEEEEETEKEEEDEEEETEKEEEDEEEETDR 668
Query: 116 --------ENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVM 167
EN EE E+N + + E + DEE EE+ EE+ E
Sbjct: 669 VEEEEDAEENEEEEEEESNSEEDEESDGEEEEDEESDSEEDEEEESESEEEEGEESESEE 728
Query: 168 EEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
EE S E + E+ EEEEE KK + +K
Sbjct: 729 EEGEESESEEE---------------EGEESDREEEEEIKKKKRPVK 760
Score = 38.5 bits (88), Expect = 0.38
Identities = 35/151 (23%), Positives = 63/151 (41%), Gaps = 17/151 (11%)
Query: 75 ENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGN 134
E+++GS E + +EG E+ + EE+ D + + E +
Sbjct: 426 ESEEGSDHESERKKASESEEEGEEEEADSLSEEEEGDSKSDAEESDAESETE---GEGAE 482
Query: 135 EERKIDEEEGVEEENE------EKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAF- 187
EE+ E+EG E E E+ N +E+ EE ++S + G+ ++
Sbjct: 483 EEQSSTEKEGESNEEEQSSTEKEESNEEEQSSTEKEESNEEEQSSTEKEGESNEEEQSST 542
Query: 188 -------ERLLSIKKEKEKNEEEEENDKKNK 211
E S +KE+E NEEE+ + +K +
Sbjct: 543 EKEESNEEEQSSTEKEEESNEEEQSSTEKEE 573
Score = 38.1 bits (87), Expect = 0.50
Identities = 31/117 (26%), Positives = 53/117 (44%), Gaps = 15/117 (12%)
Query: 92 ESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEE 151
E ++ EKE + EE++ EK E+ EE E+ + +EEE E E
Sbjct: 482 EEEQSSTEKEGESNEEEQSSTEKEESNEEE---------QSSTEKEESNEEEQSSTEKEG 532
Query: 152 KGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDK 208
+ N +E+ EE ++S + + + E S +++E NEEEEE ++
Sbjct: 533 ESNEEEQSSTEKEESNEEEQSSTEKEEE------SNEEEQSSTEKEESNEEEEEEEE 583
Score = 35.8 bits (81), Expect = 2.5
Identities = 35/147 (23%), Positives = 61/147 (40%), Gaps = 13/147 (8%)
Query: 74 GENDDGSKCVCEAYRNLRESQEGFFEKE-------NFFDGEEKNDEEKG----ENVLEEV 122
GE + + E E ++ EKE + + EE N+EE+ E E
Sbjct: 478 GEGAEEEQSSTEKEGESNEEEQSSTEKEESNEEEQSSTEKEESNEEEQSSTEKEGESNEE 537
Query: 123 IEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMH 182
+++ + EE+ E+E EE NEE+ + + + E+ EE S E +
Sbjct: 538 EQSSTEKEESNEEEQSSTEKE--EESNEEEQSSTEKEESNEEEEEEEESESNEEESEEED 595
Query: 183 LVKAFERLLSIKKEKEKNEEEEENDKK 209
E + E+E+ E EE+N ++
Sbjct: 596 QKDEEEEETGEEDEEEEEESEEQNQEE 622
Score = 35.0 bits (79), Expect = 4.2
Identities = 27/93 (29%), Positives = 41/93 (44%), Gaps = 14/93 (15%)
Query: 92 ESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEE 151
E +E E++ DGEE+ DEE EE EE + +EEEG E E+EE
Sbjct: 682 EEEESNSEEDEESDGEEEEDEESDSEEDEE-------------EESESEEEEGEESESEE 728
Query: 152 -KGNCSSMVKRRREKVMEEARNSVPENGKVMHL 183
+G S + E+ E + + + + L
Sbjct: 729 EEGEESESEEEEGEESDREEEEEIKKKKRPVKL 761
Score = 35.0 bits (79), Expect = 4.2
Identities = 28/125 (22%), Positives = 56/125 (44%), Gaps = 8/125 (6%)
Query: 85 EAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEG 144
E N+ +S + ++E D + + E++ ++ +E+ E G + EEE
Sbjct: 334 EEEENVTQSSDKASDEEKEGDSDTLHPEDESQSDVEDEEE--------GESRSEGQEEEE 385
Query: 145 VEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEE 204
EEE EE+ + ++ + EK E V E + ++ E + K+ +E EE
Sbjct: 386 EEEEEEEERSSTAESESEDEKKGERKERDVEEEEGESEVEESEEGSDHESERKKASESEE 445
Query: 205 ENDKK 209
E +++
Sbjct: 446 EGEEE 450
Score = 34.7 bits (78), Expect = 5.5
Identities = 37/147 (25%), Positives = 61/147 (41%), Gaps = 23/147 (15%)
Query: 85 EAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIG-NEERKIDEEE 143
E+ ++ + +EG E EE+ +EE+ E E+ G +ER ++EEE
Sbjct: 363 ESQSDVEDEEEGESRSEG---QEEEEEEEEEEEERSSTAESESEDEKKGERKERDVEEEE 419
Query: 144 GVEE-ENEEKGNCSSMVKRRREKVMEEARN------SVPENGKVMHLVK----------- 185
G E E E+G+ +++ + EE S E G +
Sbjct: 420 GESEVEESEEGSDHESERKKASESEEEGEEEEADSLSEEEEGDSKSDAEESDAESETEGE 479
Query: 186 -AFERLLSIKKEKEKNEEEEENDKKNK 211
A E S +KE E NEEE+ + +K +
Sbjct: 480 GAEEEQSSTEKEGESNEEEQSSTEKEE 506
Score = 33.9 bits (76), Expect = 9.5
Identities = 34/145 (23%), Positives = 63/145 (43%), Gaps = 20/145 (13%)
Query: 76 NDDGSKCVCEAYRNLRESQEGFFEKENFFDGEE---KNDEEKGENVLEEVIEANLIIHDI 132
+ D S+ V + +ES +G E+E + EE K +++ +V EE E D
Sbjct: 286 SQDKSESVTANEEDEKESIQGTEEEEEEEEEEEEDQKEEQKTSADVSEEEEENVTQSSDK 345
Query: 133 GNEERK----------IDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMH 182
++E K + + VE+E E + + E+ EE R+S E+
Sbjct: 346 ASDEEKEGDSDTLHPEDESQSDVEDEEEGESRSEGQEEEEEEEEEEEERSSTAES----- 400
Query: 183 LVKAFERLLSIKKEKEKNEEEEEND 207
++ + +KE++ EEE E++
Sbjct: 401 --ESEDEKKGERKERDVEEEEGESE 423
>UniRef100_Q7RKK5 Hypothetical protein [Plasmodium yoelii yoelii]
Length = 1549
Score = 53.1 bits (126), Expect = 2e-05
Identities = 62/291 (21%), Positives = 113/291 (38%), Gaps = 59/291 (20%)
Query: 6 KKPLEIENLNPNEFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIRQRKFVVAKKKKNDQSP 65
KK + + + ++ K+L++ N +K + K V K +ND S
Sbjct: 265 KKKNRLSSYDSETLKKQSKKNKNLSENSNK-----KEGNKKSVLNNKKNVKGKNENDYST 319
Query: 66 RKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEA 125
E+D K + N++ + E EN EE+ DEE+G EE
Sbjct: 320 --------SEDDTNKKNKNKKSGNVKNKGKTIIENEN----EEEEDEEEGNEEAEE---- 363
Query: 126 NLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVK 185
+E + +EEE E+E EE + RR ++ ++ + N + +VK
Sbjct: 364 ---------DEEEANEEEDEEDEEEESDEGYKRNRLRRSNSVKNKKSGINRNNRNSKIVK 414
Query: 186 AFERLLSIKK----------------EKEKNEEEEENDK---------KNKVMKWALPGL 220
+ + S K + E+NEEEEE ++ N+ K L
Sbjct: 415 KNKNIKSNKDKCVSNNKSGKSKIKNVKNEENEEEEEKEESSYFDEYIYSNEERKAILEEK 474
Query: 221 QFQQPVKDGDEQSEVVSSCCDDDDFDF*TTWIGSKGFSYIFLGLWKWKKCV 271
+ ++ +K+G + ++ DD+ +F I + + + L L +K V
Sbjct: 475 ERKKQIKEGYIKKMKLNKYADDETIEF----INNPNYIMVLLFLTIYKNIV 521
>UniRef100_O67825 Translation initiation factor IF-2 [Aquifex aeolicus]
Length = 805
Score = 52.0 bits (123), Expect = 3e-05
Identities = 39/112 (34%), Positives = 60/112 (52%), Gaps = 8/112 (7%)
Query: 107 EEKNDEEKGENVL-EEVIE---ANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRR 162
EEK +EEK E V+ EEV+E +I+ +I EE+K EEE +EE + K + ++K
Sbjct: 80 EEKKEEEKKEEVIVEEVVEEKKPEVIVEEI--EEKK--EEEEKKEEEKPKKSVEELIKEI 135
Query: 163 REKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
EK +E E + V+ E +KE++K E++EE K K+ K
Sbjct: 136 LEKKEKEKEKKKVEKERKEEKVRVVEVKKEERKEEKKEEKKEEEKPKIKMSK 187
Score = 47.4 bits (111), Expect = 8e-04
Identities = 29/113 (25%), Positives = 60/113 (52%), Gaps = 2/113 (1%)
Query: 99 EKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSM 158
E E + EEK +EEK + +EE+I+ L + E++K+++E E+ +
Sbjct: 108 EIEEKKEEEEKKEEEKPKKSVEELIKEILEKKEKEKEKKKVEKERKEEKVRVVEVKKEER 167
Query: 159 VKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNK 211
+ ++E+ EE + + + K +++ E +++KEK+K E+ E+ KK +
Sbjct: 168 KEEKKEEKKEEEKPKIKMSKKEREIMRKLEH--AVEKEKKKQEKREKEKKKKE 218
Score = 40.4 bits (93), Expect = 0.10
Identities = 32/114 (28%), Positives = 56/114 (49%), Gaps = 6/114 (5%)
Query: 101 ENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVK 160
E +D +EE+ E V+ E +A + + EE+K EE + EE E+ +V+
Sbjct: 51 EMIYDAFGIKEEEEKEEVVTEQAQAPAEVEEKKEEEKK---EEVIVEEVVEEKKPEVIVE 107
Query: 161 RRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
EK EE + E K V+ + + KKEKEK +++ E ++K + ++
Sbjct: 108 EIEEKKEEEEKK---EEEKPKKSVEELIKEILEKKEKEKEKKKVEKERKEEKVR 158
>UniRef100_UPI0000436182 UPI0000436182 UniRef100 entry
Length = 401
Score = 51.6 bits (122), Expect = 4e-05
Identities = 47/196 (23%), Positives = 92/196 (45%), Gaps = 18/196 (9%)
Query: 45 QKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFF 104
+K+ R+R+ + K++K ++ R+ K E +D + + +E E+E
Sbjct: 98 KKKERKRRKIRKKEEKKERRSRRKEEMKKNEEED---------MEMMDMKEEIKEEE--- 145
Query: 105 DGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVE---EENEEKGNCSSMVK- 160
+ EE+ ++EK E + + E I + G + R I EE+ E E+ EE+GN K
Sbjct: 146 EEEEEEEKEKTEETMMDKKEEGKIEEEDGRKGRIIKEEKMEEMMMEKEEEEGNIKKEGKK 205
Query: 161 --RRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALP 218
R+R K+ ++ + + + K E + + KE+ +EEEE +++ + +
Sbjct: 206 KERKRRKIRKKEEKKERRSRRKEEMKKNEEEDMEMMDMKEEIKEEEEEEEEEEKKTTSSC 265
Query: 219 GLQFQQPVKDGDEQSE 234
G Q ++ K +E E
Sbjct: 266 GEQEEEEGKIKEEMKE 281
Score = 50.8 bits (120), Expect = 7e-05
Identities = 48/195 (24%), Positives = 87/195 (44%), Gaps = 22/195 (11%)
Query: 55 VAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEK 114
+ K+ K + R+ + K + + S+ E +N E E KE + EE+ +EE+
Sbjct: 93 IKKEGKKKERKRRKIRKKEEKKERRSRRKEEMKKNEEEDMEMMDMKEEIKEEEEEEEEEE 152
Query: 115 GENVLEEVIEANLIIHDIGNEERKIDEEEGVEEE--NEEKGNCSSMVKRRRE-KVMEEAR 171
E E +++ EE KI+EE+G + EEK M K E + +E +
Sbjct: 153 KEKTEETMMDKK--------EEGKIEEEDGRKGRIIKEEKMEEMMMEKEEEEGNIKKEGK 204
Query: 172 NSVPENGKVMHLVKAFERLLSIKKEKEKNEEE--EENDKKNKVMKWALPGLQFQQPVKDG 229
+ K+ + ER K+E +KNEEE E D K ++ ++ ++
Sbjct: 205 KKERKRRKIRKKEEKKERRSRRKEEMKKNEEEDMEMMDMKEEIK---------EEEEEEE 255
Query: 230 DEQSEVVSSCCDDDD 244
+E+ + SSC + ++
Sbjct: 256 EEEKKTTSSCGEQEE 270
Score = 48.9 bits (115), Expect = 3e-04
Identities = 55/241 (22%), Positives = 101/241 (41%), Gaps = 30/241 (12%)
Query: 1 MTPEKKKPLEIENLNPNEFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIRQRKFVVAKKKK 60
M +++ +E E+ +++ + + + N+ +K ++RK ++KK
Sbjct: 161 MDKKEEGKIEEEDGRKGRIIKEEKMEEMMMEKEEEEGNIKKEGKKKERKRRKIRKKEEKK 220
Query: 61 NDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEE------------ 108
+S RK + K E D + E + E +E +K GE+
Sbjct: 221 ERRSRRKEEMKKNEEEDMEMMDMKEEIKEEEEEEEEEEKKTTSSCGEQEEEEGKIKEEMK 280
Query: 109 --------KNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVE---EENEEKGNCSS 157
K ++EK E + + E I + G + R I EE+ E E+ EE+GN
Sbjct: 281 EKKRRRRRKKEQEKTEETMMDKKEEGKIEEEDGRKGRIIKEEKMEEMMMEKEEEEGNIKK 340
Query: 158 MVK---RRREKVMEEARNSVPENGKVMHLVKAFER---LLSIKKE-KEKNEEEEENDKKN 210
K R+R K+ ++ + + + K E ++ +K+E KE+ EEEEE +K
Sbjct: 341 EGKKKERKRRKIRKKEEKKERRSRRKEEMKKNEEEDMEMMDMKEEIKEEEEEEEEEEKVK 400
Query: 211 K 211
K
Sbjct: 401 K 401
Score = 41.2 bits (95), Expect = 0.059
Identities = 36/133 (27%), Positives = 62/133 (46%), Gaps = 12/133 (9%)
Query: 91 RESQEGFFE---KENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDE---EEG 144
+E +EG + KE K ++EK E + + E I + G + R I E EE
Sbjct: 23 QEEEEGKIKEEMKEKKRRRRRKKEQEKTEETMMDKKEEGKIEEEDGRKGRMIKEEKMEEM 82
Query: 145 VEEENEEKGNCSSMVK---RRREKVMEEARNSVPENGKVMHLVKAFE---RLLSIKKEKE 198
+ E+ EE G K R+R K+ ++ + + + K E ++ +K+E +
Sbjct: 83 MMEKEEEGGTIKKEGKKKERKRRKIRKKEEKKERRSRRKEEMKKNEEEDMEMMDMKEEIK 142
Query: 199 KNEEEEENDKKNK 211
+ EEEEE ++K K
Sbjct: 143 EEEEEEEEEEKEK 155
Score = 39.3 bits (90), Expect = 0.23
Identities = 32/122 (26%), Positives = 57/122 (46%), Gaps = 18/122 (14%)
Query: 92 ESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEE 151
+ +EG E+E+ G +E+ E ++E+ E I + +ERK + EE+
Sbjct: 57 KKEEGKIEEEDGRKGRMIKEEKMEEMMMEKEEEGGTIKKEGKKKERKRRKIRKKEEK--- 113
Query: 152 KGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNK 211
K RR + EE + + E+ ++M + + KE+E+ EEEEE +K +
Sbjct: 114 --------KERRSRRKEEMKKNEEEDMEMMDMKEEI-------KEEEEEEEEEEKEKTEE 158
Query: 212 VM 213
M
Sbjct: 159 TM 160
>UniRef100_O35788 Cyclic nucleotide-gated channel beta subunit [Rattus norvegicus]
Length = 1339
Score = 51.6 bits (122), Expect = 4e-05
Identities = 50/201 (24%), Positives = 89/201 (43%), Gaps = 32/201 (15%)
Query: 45 QKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFF 104
+++ + K KKK+ ++ ++ + GE ++ + + +E +EG EKE
Sbjct: 394 EEKEEEEKREEEKKKEKEEEKKEKEEEENGEEEEKEE------KEEKEEEEGKEEKEEKE 447
Query: 105 DGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEK----GNCSSMVK 160
+ EEK +EEK E EE EE++ +EEE EEE EE +C +
Sbjct: 448 EKEEKEEEEKEEKEEEE------------KEEKEEEEEEEEEEEEEEPIVLLDSCLVVQA 495
Query: 161 RRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEK-------NEEEEENDKKNKVM 213
+ +E A+ PE + L + E KKE+E+ EEEEE +++ +
Sbjct: 496 DVDQCQLERAQ---PETASIQELPEEEEEKEEEKKEEEEEKEEEEEKEEEEEKEEEGEAT 552
Query: 214 KWALPGLQFQQPVKDGDEQSE 234
+P + ++ D +E
Sbjct: 553 NSTVPATKEHPELQVEDTDAE 573
Score = 48.9 bits (115), Expect = 3e-04
Identities = 43/135 (31%), Positives = 59/135 (42%), Gaps = 24/135 (17%)
Query: 77 DDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEE 136
+D S+ + E R L + QE E+ + EE+ EEK E EE E E+
Sbjct: 350 EDESEAMVEMPRELPQIQEQQEEENEEKEEEEEEKEEKEEKEEEEEKEEE---EKREEEK 406
Query: 137 RKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKE 196
+K EEE E+E EE G E+ EE E GK K+E
Sbjct: 407 KKEKEEEKKEKEEEENGE---------EEEKEEKEEKEEEEGK------------EEKEE 445
Query: 197 KEKNEEEEENDKKNK 211
KE+ EE+EE +K+ K
Sbjct: 446 KEEKEEKEEEEKEEK 460
Score = 42.0 bits (97), Expect = 0.035
Identities = 41/180 (22%), Positives = 78/180 (42%), Gaps = 12/180 (6%)
Query: 44 PQKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENF 103
PQ + +Q + K+++ ++ K + E ++ K E + E ++ E+EN
Sbjct: 364 PQIQEQQEEENEEKEEEEEEKEEKEEKEEEEEKEEEEKREEEKKKEKEEEKKEKEEEENG 423
Query: 104 FDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGN--------- 154
+ E++ EEK E +E E + EE++ EEE EE+ EE+
Sbjct: 424 EEEEKEEKEEKEEEEGKEEKEEKEEKEEKEEEEKEEKEEEEKEEKEEEEEEEEEEEEEEP 483
Query: 155 ---CSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNK 211
S + + + + + PE + L + E KKE+E+ +EEEE ++ +
Sbjct: 484 IVLLDSCLVVQADVDQCQLERAQPETASIQELPEEEEEKEEEKKEEEEEKEEEEEKEEEE 543
Score = 39.3 bits (90), Expect = 0.23
Identities = 28/93 (30%), Positives = 44/93 (47%), Gaps = 5/93 (5%)
Query: 142 EEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNE 201
+E EEENEEK + + EK EE + + + K E KKEKE+ E
Sbjct: 367 QEQQEEENEEKEEEEEEKEEKEEKEEEEEKEEEEKREEEKKKEKEEE-----KKEKEEEE 421
Query: 202 EEEENDKKNKVMKWALPGLQFQQPVKDGDEQSE 234
EE +K+ K K G + ++ ++ +E+ E
Sbjct: 422 NGEEEEKEEKEEKEEEEGKEEKEEKEEKEEKEE 454
Score = 37.4 bits (85), Expect = 0.86
Identities = 44/185 (23%), Positives = 75/185 (39%), Gaps = 22/185 (11%)
Query: 75 ENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGN 134
E ++ K E + +E +E EKE EE+ +EK E E+ E N +
Sbjct: 371 EEENEEKEEEEEEKEEKEEKEEEEEKEEEEKREEEKKKEKEEEKKEKEEEENGEEEEKEE 430
Query: 135 EERKIDEEEGVE-----EENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFER 189
+E K +EEEG E EE EEK K EK +E E + + +
Sbjct: 431 KEEK-EEEEGKEEKEEKEEKEEKEEEEKEEKEEEEKEEKEEEEEEEEEEEEEEPIVLLDS 489
Query: 190 LLSIKKE----------------KEKNEEEEENDKKNKVMKWALPGLQFQQPVKDGDEQS 233
L ++ + +E EEEEE +++ K + + ++ ++ +E+
Sbjct: 490 CLVVQADVDQCQLERAQPETASIQELPEEEEEKEEEKKEEEEEKEEEEEKEEEEEKEEEG 549
Query: 234 EVVSS 238
E +S
Sbjct: 550 EATNS 554
>UniRef100_UPI0000364C4A UPI0000364C4A UniRef100 entry
Length = 655
Score = 51.2 bits (121), Expect = 6e-05
Identities = 47/166 (28%), Positives = 76/166 (45%), Gaps = 19/166 (11%)
Query: 85 EAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEG 144
EA R L + ++ F E + EE+ +EE G+ EE L + E ++++EE
Sbjct: 249 EAKRRLLKEKKAFEEAKQGMGEEEEEEEEPGQEEKEEYRPGKLRLSFEELERQRVEEERK 308
Query: 145 VEEENEEKGNCSSMVKRRREKVMEEARNSV----------PENGKVMHLVKA---FERLL 191
E+ +K MV+ RR EAR S+ ++ + +H K FE LL
Sbjct: 309 KAEDEAKK----RMVEERR--AFAEARKSMAADEEEGMEKSDSQEALHPKKLEINFEELL 362
Query: 192 SIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQPVKDGDEQSEVVS 237
K+E E+ + EE KK + K L+ + + +E S+VVS
Sbjct: 363 KQKEEAERRRKGEERKKKLEQEKQEFEQLRQEMGEDEVNESSDVVS 408
>UniRef100_UPI0000498854 UPI0000498854 UniRef100 entry
Length = 512
Score = 51.2 bits (121), Expect = 6e-05
Identities = 38/160 (23%), Positives = 79/160 (48%), Gaps = 12/160 (7%)
Query: 57 KKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGE 116
+KK+ ++ RK + E ++ + E R +E +E E+ + EE+ +E+ E
Sbjct: 163 RKKREEEERRKEEEERRKEEEERRQQQEEEERRQQEEEE---ERRRQEEEEERKRQEEEE 219
Query: 117 NVLEEVIEANLIIHD--IGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSV 174
++ E + H+ I E+KI E+E EE ++K ++ + K+ E+ R +
Sbjct: 220 ERKKQEQERKIQEHERKIQEHEKKIQEQE--EERKKQKEEQDRKIQEQERKIQEQERKTT 277
Query: 175 PENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
+ K+ + ++ +E+E+N+++EE DKK + K
Sbjct: 278 EQEKKIQEQERKIKQ-----QEEERNKQKEEQDKKIQEQK 312
Score = 36.2 bits (82), Expect = 1.9
Identities = 27/114 (23%), Positives = 53/114 (45%), Gaps = 13/114 (11%)
Query: 97 FFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCS 156
+ E++ + E + +EE+ EE +++ +EE +EE EE+
Sbjct: 160 YLERKKREEEERRKEEEERRKEEEE------------RRQQQEEEERRQQEEEEERRRQE 207
Query: 157 SMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSI-KKEKEKNEEEEENDKK 209
+R+R++ EE + E H K E I ++E+E+ +++EE D+K
Sbjct: 208 EEEERKRQEEEEERKKQEQERKIQEHERKIQEHEKKIQEQEEERKKQKEEQDRK 261
>UniRef100_UPI00004659AA UPI00004659AA UniRef100 entry
Length = 285
Score = 51.2 bits (121), Expect = 6e-05
Identities = 56/225 (24%), Positives = 92/225 (40%), Gaps = 24/225 (10%)
Query: 12 ENLNPNEFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIRQRKFVVAKKKKNDQSPR---KT 68
E L+ N N Q ++ N +N + K K K++ND++ KT
Sbjct: 48 EKLDTNPEKNEEDSQCAVENSTNKNINSIEEKETKLPDYTKTDEIIKRENDETNENVDKT 107
Query: 69 VLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKN--DEEKGENVLEEVIEAN 126
C E D K ++ NL + + + +N FD +EK+ DE E E I
Sbjct: 108 HNTSC-ETKDADKST-DSISNLEQDSKDLKKSDNKFDAKEKDEKDEIPEEAATSEPIGKG 165
Query: 127 LIIHDIGNEERK----IDEEEGVEEENEEKGNC---------SSMVKRRREKVMEEAR-- 171
+I NEE K + +EE E N+ GN + K +EK +++ +
Sbjct: 166 IIETSGNNEEEKDKKIVSKEEESENRNKNTGNTDTENVENHDQNESKEPKEKTLKKKKRV 225
Query: 172 --NSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
S +N K + F R I K+ ++N+ +E + +NK K
Sbjct: 226 FFGSYKKNMKKKNYASNFLRDEKIGKKNKRNKNKENDKNRNKKNK 270
>UniRef100_Q5XJD2 Hypothetical protein [Brachydanio rerio]
Length = 535
Score = 51.2 bits (121), Expect = 6e-05
Identities = 54/179 (30%), Positives = 79/179 (43%), Gaps = 37/179 (20%)
Query: 38 NVVVVSPQKRIRQRKFVVAKKKKNDQSPRKTV--------LCKCGENDDGSKCVCEAYRN 89
N+V ++ +R FV KKK PRK V L + GE + + E +
Sbjct: 121 NLVAPEEEEDEGERHFV--KKKGEFLPPRKKVTEIWAKEMLEEEGEYLEDEEEEEEEEMD 178
Query: 90 LRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEEN 149
E +E + E+E +D EE +EE+ E LEE E L EE++++EEE EE+
Sbjct: 179 TDEEEEDYEEEEEEYD-EELEEEEEEEEELEEEEEEELE----EEEEKELEEEEEDEEDE 233
Query: 150 EEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDK 208
EEK V +E+ EE K+EKEK EEEE+ ++
Sbjct: 234 EEKEEPGVKVVEEKEEEDEE----------------------EAKEEKEKKEEEEDEEE 270
Score = 47.4 bits (111), Expect = 8e-04
Identities = 44/175 (25%), Positives = 78/175 (44%), Gaps = 7/175 (4%)
Query: 75 ENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGN 134
E ++ + E L E +E E+E + +E+ EE G V+EE E + +
Sbjct: 200 EEEEEEELEEEEEEELEEEEEKELEEEEEDEEDEEEKEEPGVKVVEEKEEEDE--EEAKE 257
Query: 135 EERKIDEEEGVEE---ENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLL 191
E+ K +EEE EE E EE ++ + + +K E E+ + K+ E+
Sbjct: 258 EKEKKEEEEDEEEAEAEEEEDEEAATEIAAKEKKKEEAGAKEEDEDEEEGKEEKSEEKEE 317
Query: 192 SIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQPVK--DGDEQSEVVSSCCDDDD 244
+ E++EE+EE +KK K A + ++ K +GDE +D++
Sbjct: 318 EEPEAMEEDEEKEEAEKKEKQEAGAEEEDEDEEEAKEMEGDEAETSKEEAPEDEE 372
Score = 41.6 bits (96), Expect = 0.045
Identities = 52/202 (25%), Positives = 81/202 (39%), Gaps = 17/202 (8%)
Query: 54 VVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQ-EGFFEKENFFDGEEKNDE 112
+ AK+KK +++ K E++D + E E + E E E + E+K +
Sbjct: 285 IAAKEKKKEEAGAKE------EDEDEEEGKEEKSEEKEEEEPEAMEEDEEKEEAEKKEKQ 338
Query: 113 EKG-ENVLEEVIEANLIIHD---IGNEERKIDEEEGVEEENE---EKGNCSSMVKRRREK 165
E G E E+ EA + D EE DEEE EEE E E+ K E
Sbjct: 339 EAGAEEEDEDEEEAKEMEGDEAETSKEEAPEDEEEKEEEEPETDKEEAPEDEEGKEEEED 398
Query: 166 VMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALPGLQFQ-- 223
EEA ++ PE E + +++ +K+ E +E KV +
Sbjct: 399 EKEEAASAAPEKKAAPIPAAPKEAEEADEEDDDKDAEAQEEPPAGKVTPAETDETETPIA 458
Query: 224 -QPVKDGDEQSEVVSSCCDDDD 244
P D D++ + + DDDD
Sbjct: 459 VTPTTDDDQKDDQIEKKEDDDD 480
Score = 41.2 bits (95), Expect = 0.059
Identities = 47/158 (29%), Positives = 63/158 (39%), Gaps = 18/158 (11%)
Query: 92 ESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENE- 150
E +E +KE G E+ DE++ E E EA + EE DEEE EEE E
Sbjct: 327 EEKEEAEKKEKQEAGAEEEDEDEEEAKEMEGDEA-----ETSKEEAPEDEEEKEEEEPET 381
Query: 151 --EKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDK 208
E+ K E EEA ++ PE K + A KE E +EE+D
Sbjct: 382 DKEEAPEDEEGKEEEEDEKEEAASAAPEK-KAAPIPAA---------PKEAEEADEEDDD 431
Query: 209 KNKVMKWALPGLQFQQPVKDGDEQSEVVSSCCDDDDFD 246
K+ + P + D E V+ DDD D
Sbjct: 432 KDAEAQEEPPAGKVTPAETDETETPIAVTPTTDDDQKD 469
Score = 40.8 bits (94), Expect = 0.077
Identities = 37/131 (28%), Positives = 58/131 (44%), Gaps = 13/131 (9%)
Query: 92 ESQEGFFEKENFFDGEEKN-DEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENE 150
E++E +KE D EE +EE+ E E+ + G +E DEEEG EE++E
Sbjct: 254 EAKEEKEKKEEEEDEEEAEAEEEEDEEAATEIAAKEKKKEEAGAKEEDEDEEEGKEEKSE 313
Query: 151 EKGN-----------CSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEK 199
EK K+ +++ E + E K M +A +++E+
Sbjct: 314 EKEEEEPEAMEEDEEKEEAEKKEKQEAGAEEEDEDEEEAKEMEGDEAETSKEEAPEDEEE 373
Query: 200 NEEEE-ENDKK 209
EEEE E DK+
Sbjct: 374 KEEEEPETDKE 384
>UniRef100_UPI0000439AE7 UPI0000439AE7 UniRef100 entry
Length = 284
Score = 50.8 bits (120), Expect = 7e-05
Identities = 42/143 (29%), Positives = 73/143 (50%), Gaps = 17/143 (11%)
Query: 99 EKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEK---GNC 155
E++ D E++ +EE+ E V+E+ H+ EE +EEE V E++EEK
Sbjct: 107 EEQKVKDKEKEVEEEEEEEVVEQ--------HE---EEEVEEEEEEVAEQHEEKEVEKEE 155
Query: 156 SSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEE---ENDKKNKV 212
+V+ + EK +EE V E + + K E ++ ++EKE EEEE E ++ +V
Sbjct: 156 EEVVEHQEEKEVEEEEEEVAEQHEEKEVEKEEEEVVEHQEEKEVEEEEEEVAEQHEEKEV 215
Query: 213 MKWALPGLQFQQPVKDGDEQSEV 235
K ++ Q+ + +E+ EV
Sbjct: 216 EKEEEEVVEHQEEKEVEEEEEEV 238
Score = 45.4 bits (106), Expect = 0.003
Identities = 43/168 (25%), Positives = 81/168 (47%), Gaps = 19/168 (11%)
Query: 46 KRIRQRKFVVAKKKKNDQSPRKTVLCKCGEND--DGSKCVCEAYRNLRESQEGFFEKENF 103
K+ + + V K+K+ ++ + V+ + E + + + V E + +E E+E
Sbjct: 103 KKEEEEQKVKDKEKEVEEEEEEEVVEQHEEEEVEEEEEEVAEQHEEKEVEKE---EEEVV 159
Query: 104 FDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKI---DEEEGVEEENEEKGNCSSMVK 160
EEK EE+ E V E+ E ++ EE ++ EE+ VEEE EE + +
Sbjct: 160 EHQEEKEVEEEEEEVAEQHEEK-----EVEKEEEEVVEHQEEKEVEEEEEE------VAE 208
Query: 161 RRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDK 208
+ EK +E+ V E+ + + + E + +EKEK +E+E+ +K
Sbjct: 209 QHEEKEVEKEEEEVVEHQEEKEVEEEEEEVAEQHEEKEKEDEDEKEEK 256
Score = 45.1 bits (105), Expect = 0.004
Identities = 47/201 (23%), Positives = 92/201 (45%), Gaps = 34/201 (16%)
Query: 43 SPQKRIRQRKFVVAKKKKNDQSPRKTVLC------KCGENDDGSKCVCEAYRNLRESQEG 96
S +K R+R+ + +K + + RK+ K G+ ++ + V + + + E +E
Sbjct: 65 SRRKMRRKRRSSIRRKSRRKRRRRKSRSSIRRKNRKRGKKEEEEQKVKDKEKEVEEEEEE 124
Query: 97 FFEKENFFDGEEKNDEEKGENVLEEVIEAN---LIIHDIGNEERKIDEEEG-VEEENEEK 152
+++ + E+ +EE E E+ +E ++ H EE++++EEE V E++EEK
Sbjct: 125 EVVEQHEEEEVEEEEEEVAEQHEEKEVEKEEEEVVEHQ---EEKEVEEEEEEVAEQHEEK 181
Query: 153 G---NCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIK--------------- 194
+V+ + EK +EE V E + + K E ++ +
Sbjct: 182 EVEKEEEEVVEHQEEKEVEEEEEEVAEQHEEKEVEKEEEEVVEHQEEKEVEEEEEEVAEQ 241
Query: 195 ---KEKEKNEEEEENDKKNKV 212
KEKE +E+EE D+ +V
Sbjct: 242 HEEKEKEDEDEKEEKDEVQEV 262
>UniRef100_UPI0000428FBC UPI0000428FBC UniRef100 entry
Length = 1552
Score = 50.8 bits (120), Expect = 7e-05
Identities = 48/163 (29%), Positives = 71/163 (43%), Gaps = 16/163 (9%)
Query: 92 ESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEE---- 147
E +EG E+E D EE+ +EE+ E EE E + G EE DEEEG EE
Sbjct: 705 EEEEG--EEEEGEDEEEEGEEEEEEEEEEEEEEEEEEEKEEGEEEEGEDEEEGEEEEEEE 762
Query: 148 ---------ENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKE 198
E+EE+G E+V EE E G+ + E ++EKE
Sbjct: 763 EEEGEEKEGEDEEEGEEEEEEVEDEEEVGEEEEEEEEEEGEEEEEGEDKEVEEEEEEEKE 822
Query: 199 KNE-EEEENDKKNKVMKWALPGLQFQQPVKDGDEQSEVVSSCC 240
+ E EEEE +++ + W L + Q + S + ++ C
Sbjct: 823 EEEGEEEEEEEEEEDNSWLLEETEIQDVKERCSSTSLMQAATC 865
Score = 45.4 bits (106), Expect = 0.003
Identities = 41/153 (26%), Positives = 73/153 (46%), Gaps = 9/153 (5%)
Query: 88 RNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEE 147
+N R ++G +KE +GEE+ +E++ + EE +E + + +EE + +EEE EE
Sbjct: 643 KNRRSEEKGKGKKE---EGEEEEEEKEVKQQQEEQVEDEKLEEEEEDEEEEGEEEEEEEE 699
Query: 148 ----ENEEKG--NCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNE 201
E EE+G + E+ EE E + + E ++E E+ E
Sbjct: 700 KGEDEEEEEGEEEEGEDEEEEGEEEEEEEEEEEEEEEEEEEKEEGEEEEGEDEEEGEEEE 759
Query: 202 EEEENDKKNKVMKWALPGLQFQQPVKDGDEQSE 234
EEEE + + K + G + ++ V+D +E E
Sbjct: 760 EEEEEEGEEKEGEDEEEGEEEEEEVEDEEEVGE 792
Score = 41.6 bits (96), Expect = 0.045
Identities = 34/119 (28%), Positives = 56/119 (46%), Gaps = 6/119 (5%)
Query: 92 ESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEE-NE 150
E +EG E+E GE++ +EE E E+ E + EE + +EEE +EE E
Sbjct: 687 EEEEGEEEEEEEEKGEDEEEEEGEEEEGEDEEEEGEEEEEEEEEEEEEEEEEEEKEEGEE 746
Query: 151 EKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKK 209
E+G + E+ EE E+ + + E ++ E+E EEEEE +++
Sbjct: 747 EEGEDEEEGEEEEEEEEEEGEEKEGEDEE-----EGEEEEEEVEDEEEVGEEEEEEEEE 800
>UniRef100_UPI000046C3AD UPI000046C3AD UniRef100 entry
Length = 2030
Score = 50.8 bits (120), Expect = 7e-05
Identities = 33/130 (25%), Positives = 67/130 (51%), Gaps = 18/130 (13%)
Query: 94 QEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKG 153
QE ++EN E + +E EV + N + +E +I +E +++ENEE
Sbjct: 817 QENEVKQENEIKQENEVKQENEVKQENEVKQENEV-----KQENEIKQENEIKQENEE-- 869
Query: 154 NCSSMVKRRREKVMEEARNSVPEN--------GKVMHLVKAFERLLSIKKEKEKNEEEEE 205
+ +KR++E ++E R S+ N ++ ++ E + + K E+E+ EEEE+
Sbjct: 870 ---NEIKRQKEMLLETVRKSISNNCESYKKDLNMFLNKIEHNELMKNEKDEEEEEEEEED 926
Query: 206 NDKKNKVMKW 215
+++N+ ++W
Sbjct: 927 EEEENEEIEW 936
Score = 40.0 bits (92), Expect = 0.13
Identities = 40/169 (23%), Positives = 63/169 (36%), Gaps = 21/169 (12%)
Query: 12 ENLNPNEFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIRQRKFVVAKKKKNDQSPRKTVLC 71
+N+NP N+ +K L N +N + ++ + + ND+S
Sbjct: 1533 KNINPQNVVNKAGKEKEL-----NVINKEGQNENSNDEIKEESIKDESSNDES------- 1580
Query: 72 KCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHD 131
DD SK CE + E +E EN D + D E ++ I + +
Sbjct: 1581 -----DDASKS-CEENDEMSEKEEDKQRDENEKDNKSDEDNELEHEQKKKEINDETRVKE 1634
Query: 132 IGNEERKIDEEEGVE---EENEEKGNCSSMVKRRREKVMEEARNSVPEN 177
I EE + + E E EE E + + V+ E E RN EN
Sbjct: 1635 IEKEEESMSDAESQENSKEETESEESSEEEVETESEGDDETERNKQKEN 1683
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.339 0.149 0.490
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 737,146,495
Number of Sequences: 2790947
Number of extensions: 32828733
Number of successful extensions: 335849
Number of sequences better than 10.0: 3185
Number of HSP's better than 10.0 without gapping: 323
Number of HSP's successfully gapped in prelim test: 3094
Number of HSP's that attempted gapping in prelim test: 284279
Number of HSP's gapped (non-prelim): 17450
length of query: 439
length of database: 848,049,833
effective HSP length: 130
effective length of query: 309
effective length of database: 485,226,723
effective search space: 149935057407
effective search space used: 149935057407
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC149305.8


