

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149305.6 - phase: 0 /pseudo/partial
(543 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
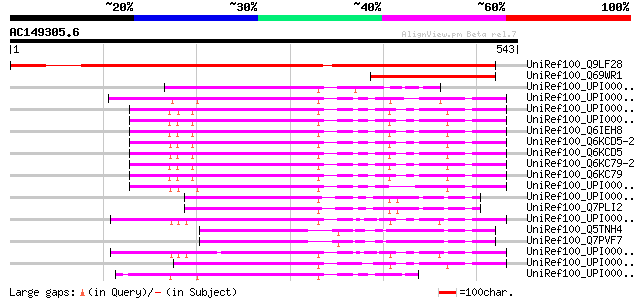
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LF28 Hypothetical protein T20K14_150 [Arabidopsis th... 593 e-168
UniRef100_Q69WR1 Putative IDN3 protein isoform A [Oryza sativa] 167 6e-40
UniRef100_UPI0000431B8B UPI0000431B8B UniRef100 entry 127 1e-27
UniRef100_UPI000043614F UPI000043614F UniRef100 entry 124 5e-27
UniRef100_UPI00001CED5F UPI00001CED5F UniRef100 entry 120 1e-25
UniRef100_UPI000006E798 UPI000006E798 UniRef100 entry 119 2e-25
UniRef100_Q6IEH8 Transcriptional regulator [Homo sapiens] 119 2e-25
UniRef100_Q6KCD5-2 Splice isoform 2 of Q6KCD5 [Mus musculus] 119 2e-25
UniRef100_Q6KCD5 Nipped-B-like protein [Mus musculus] 119 2e-25
UniRef100_Q6KC79-2 Splice isoform 2 of Q6KC79 [Homo sapiens] 119 2e-25
UniRef100_Q6KC79 Nipped-B-like protein [Homo sapiens] 119 2e-25
UniRef100_UPI00003B0050 UPI00003B0050 UniRef100 entry 117 1e-24
UniRef100_UPI000044FA01 UPI000044FA01 UniRef100 entry 116 1e-24
UniRef100_Q7PLI2 Nipped-B protein [Drosophila melanogaster] 116 1e-24
UniRef100_UPI0000439DE5 UPI0000439DE5 UniRef100 entry 116 2e-24
UniRef100_Q5TNH4 ENSANGP00000028248 [Anopheles gambiae str. PEST] 113 2e-23
UniRef100_Q7PVF7 ENSANGP00000010656 [Anopheles gambiae str. PEST] 113 2e-23
UniRef100_UPI00004366F2 UPI00004366F2 UniRef100 entry 112 4e-23
UniRef100_UPI0000337DCB UPI0000337DCB UniRef100 entry 112 4e-23
UniRef100_UPI0000360ED0 UPI0000360ED0 UniRef100 entry 110 1e-22
>UniRef100_Q9LF28 Hypothetical protein T20K14_150 [Arabidopsis thaliana]
Length = 1755
Score = 593 bits (1530), Expect = e-168
Identities = 308/521 (59%), Positives = 376/521 (72%), Gaps = 45/521 (8%)
Query: 1 MAIDILGTIAARLKRDAVICSQEKFWVLQDLLSEDAAPQHYPKDTCCVCLGGRVENLFKC 60
MAI++LGTIAARLKRDAV+CS+++FW L + SE + Q
Sbjct: 596 MAIELLGTIAARLKRDAVLCSKDRFWTLLESDSEISVDQ--------------------- 634
Query: 61 SGCDRLFHADCLDVKENEVPNRNWYCLMCICSKQLLVLQSYCNSQRKDDAKKNRKVS-KD 119
E ++ +RNW+C +C+C +QLLVLQSYC + K K + S ++
Sbjct: 635 ---------------ELDISSRNWHCPLCVCKRQLLVLQSYCKTDTKGTGKLESEESIEN 679
Query: 120 DSTFSNHEIVQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKPIYYFARMKSR 179
S + E+VQQ+LLNY QDV SADD+H FICWFYLC WYK+ PK Q K YY AR+K++
Sbjct: 680 PSMITKTEVVQQMLLNYLQDVGSADDVHTFICWFYLCLWYKDVPKSQNKFKYYIARLKAK 739
Query: 180 TIVRDSGSVSSMLTRDSIKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRA 239
+I+R+SG+ +S LTRD+IK+ITLALG NSSF RGFDKI + LL SL+EN+P IRAKALRA
Sbjct: 740 SIIRNSGATTSFLTRDAIKQITLALGMNSSFSRGFDKILNMLLASLRENAPNIRAKALRA 799
Query: 240 VSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEKITE 299
VSIIVEADPEVL DK VQ +VEGRFCD+AISVREAALELVGRHIASHPDVG KYFEK+ E
Sbjct: 800 VSIIVEADPEVLCDKRVQLAVEGRFCDSAISVREAALELVGRHIASHPDVGIKYFEKVAE 859
Query: 300 RIKDTGVSVRKRAIKIIRDMCCSDGNFLRVYKSLHRDNFTCY***IEYPAYSASDLQDLV 359
RIKDTGVSVRKRAIKIIRDMC S+ NF + + + S +QDLV
Sbjct: 860 RIKDTGVSVRKRAIKIIRDMCTSNPNFSEFTSACAEI--------LSRISDDESSVQDLV 911
Query: 360 CKTFYEFWFEEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNNQLLVTVIKRCLTL 419
CKTFYEFWFEEP TQ D S++PLE+ KKT+Q+V +L R PN QLLVT+IKR L L
Sbjct: 912 CKTFYEFWFEEPPGHHTQFASDASSIPLELEKKTKQMVGLLSRTPNQQLLVTIIKRALAL 971
Query: 420 DFLPQSAKASGVNPVSLVTVRKRCELMCKCLLEKILQVDEMNSNELEKHALPYVLVLHAF 479
DF PQ+AKA+G+NPV+L +VR+RCELMCKCLLEKILQV+EM+ E E LPYVLVLHAF
Sbjct: 972 DFFPQAAKAAGINPVALASVRRRCELMCKCLLEKILQVEEMSREEGEVQVLPYVLVLHAF 1031
Query: 480 CLVDPTLCAPASNPSQFVLTLQPYLKTQVKTNLKTYILGSL 520
CLVDP LC PAS+P++FV+TLQPYLK+Q + +L S+
Sbjct: 1032 CLVDPGLCTPASDPTKFVITLQPYLKSQADSRTGAQLLESI 1072
>UniRef100_Q69WR1 Putative IDN3 protein isoform A [Oryza sativa]
Length = 764
Score = 167 bits (424), Expect = 6e-40
Identities = 81/134 (60%), Positives = 105/134 (77%)
Query: 387 LEVAKKTEQIVEMLKRLPNNQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCELM 446
+E+A KTEQIV+ML+++PN+ L+T++KR L LDFLPQSAKA+G+N + ++RKRCEL+
Sbjct: 1 MEIAVKTEQIVDMLRKMPNHLPLITIVKRNLALDFLPQSAKATGINSSFMASLRKRCELI 60
Query: 447 CKCLLEKILQVDEMNSNELEKHALPYVLVLHAFCLVDPTLCAPASNPSQFVLTLQPYLKT 506
CK LLE+ILQV+E ++E E HALPYVL L AFC+VDPTLC PA+ P QFV TLQPYLK
Sbjct: 61 CKRLLERILQVEEGAASETEVHALPYVLALQAFCIVDPTLCTPATQPFQFVETLQPYLKK 120
Query: 507 QVKTNLKTYILGSL 520
QV +L S+
Sbjct: 121 QVDNKSTAQLLESI 134
>UniRef100_UPI0000431B8B UPI0000431B8B UniRef100 entry
Length = 1345
Score = 127 bits (318), Expect = 1e-27
Identities = 91/304 (29%), Positives = 152/304 (49%), Gaps = 32/304 (10%)
Query: 166 QQKPIYYFARMKSRTIVRDSGSVSSMLTRDSIKKITLALGQNSSFCRGFDKIFHTLLVSL 225
++K Y +R++ + + + + +S + I+ L F + FD+ +L L
Sbjct: 1066 EEKKKYIISRIRPYSTGTKPDILQTYIDYNSAELISQYLASKRPFSQSFDRYLKQILHVL 1125
Query: 226 KENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREAALELVGRHIAS 285
E+S IR KA++ +++IVEADP VL +Q V+ F D A SVREAA++LVG+ + S
Sbjct: 1126 TESSIAIRTKAMKCLTMIVEADPSVLARVDMQLGVKHSFLDHATSVREAAVDLVGKFVLS 1185
Query: 286 HPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----YKSLHRDNFTCY 341
P++ KY++ ++ RI DTGVSVRKR IKI++D+C +F ++ K + R N
Sbjct: 1186 RPELIDKYYDMLSARILDTGVSVRKRVIKILKDICMECPDFPKIPEICVKMIRRVN---- 1241
Query: 342 ***IEYPAYSASDLQDLVCKTFYEFWF----EEPSTPQTQVFKDGSTVPLEVAKKTEQIV 397
++ LV + F WF E P+ + + + VA + +
Sbjct: 1242 ---------DEEGIRKLVMEVFQNMWFTPVRERPTLDSESLLRKVMNITDVVAASKDMGL 1292
Query: 398 EMLKRLPNNQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCELMCKCLLEKILQV 457
E + QLLV++ K D S K P +L+T C+ + CL+E +L++
Sbjct: 1293 EWFE-----QLLVSLFKPKEDKD---DSTKMQTEPPKALLTA---CKQIVDCLIENVLRL 1341
Query: 458 DEMN 461
+E N
Sbjct: 1342 EETN 1345
>UniRef100_UPI000043614F UPI000043614F UniRef100 entry
Length = 1301
Score = 124 bits (312), Expect = 5e-27
Identities = 119/475 (25%), Positives = 196/475 (41%), Gaps = 93/475 (19%)
Query: 107 KDDAKKNRKVSKDDSTFSNHEIVQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQ 166
K D + ++ ++S + +Q+ LLNY + D F FYL WY++
Sbjct: 108 KMDQRSINRILGENSGSDEIQQLQKALLNYLDENVETDPFLLFARKFYLAQWYRDTSTET 167
Query: 167 QKPIYYF--------------------------ARMKS-RTIVRDSGSVSSMLTRDSIKK 199
+K + AR K R++++ + S S L +S
Sbjct: 168 EKAMKSQRDDDSSDGPHHAKDVETTSEILQKAEARKKFLRSVIKTTASKFSSLRVNSDTV 227
Query: 200 -------ITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLG 252
I L F + FD +L L E++ +R KA++ +S +V DP +L
Sbjct: 228 DYEDSCLIVRYLASMRPFAQSFDIYLTQILRVLGESAIAVRTKAMKCLSEVVAVDPSILA 287
Query: 253 DKFVQSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRA 312
+Q V GR D + SVREAA+EL+GR + S P + +Y++ + ERI DTG+SVRKR
Sbjct: 288 RLDMQRGVHGRLMDNSTSVREAAVELLGRFVLSRPQLTEQYYDMLIERILDTGISVRKRV 347
Query: 313 IKIIRDMCCSDGNFLRV----YKSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWF 368
IKI+RD+C F +V K + R N ++ LV +TF + WF
Sbjct: 348 IKILRDICLEQPTFNKVTEMCVKMIRRVN-------------DEEGIKKLVNETFQKLWF 394
Query: 369 EEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNN------QLLVTVIKRCLTLDFL 422
TP K+ T +K I +++ ++ QLL ++K +
Sbjct: 395 ----TPTPNHDKEAMT------RKILNITDVVAACRDSGYDWFEQLLQNLLKSEEDASYK 444
Query: 423 PQSAKASGVNPVSLVTVRKRCELMCKCLLEKILQVDEM-----NSNELEKHALPYVLVLH 477
P RK C + L+E IL+ +E N + + L+
Sbjct: 445 P---------------ARKACAQLVDSLVEHILKYEESLADCENKGLTSNRLVACITTLY 489
Query: 478 AFCLVDPTLCAPASNPSQFVLTLQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
F + P L + +T+QPYL T+ T ++ ++ L + L++
Sbjct: 490 LFSKIRPHLMV------KHAMTMQPYLTTKCNTQSDFMVICNVAKILELVVPLMD 538
>UniRef100_UPI00001CED5F UPI00001CED5F UniRef100 entry
Length = 2642
Score = 120 bits (300), Expect = 1e-25
Identities = 116/453 (25%), Positives = 189/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKP------------------- 169
+Q+ LL+Y + T D F FY+ W+++ +K
Sbjct: 1630 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSDGAHHAKEI 1689
Query: 170 -----IYYFARMKSR---TIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
I + A + R +I++ + S S L D I L F + F
Sbjct: 1690 ETTGQIMHRAESRKRFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1749
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1750 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1809
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1810 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1869
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1870 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1906
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1907 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1951
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1952 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 2005
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 2006 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 2038
>UniRef100_UPI000006E798 UPI000006E798 UniRef100 entry
Length = 2265
Score = 119 bits (299), Expect = 2e-25
Identities = 116/453 (25%), Positives = 189/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKP------------------- 169
+Q+ LL+Y + T D F FY+ W+++ +K
Sbjct: 1112 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSEGTHHAKEI 1171
Query: 170 -----IYYFARMKS---RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
I + A + R+I++ + S S L D I L F + F
Sbjct: 1172 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1231
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1232 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1291
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1292 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1351
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1352 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1388
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1389 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1433
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1434 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 1487
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 1488 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 1520
>UniRef100_Q6IEH8 Transcriptional regulator [Homo sapiens]
Length = 2804
Score = 119 bits (299), Expect = 2e-25
Identities = 116/453 (25%), Positives = 189/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKP------------------- 169
+Q+ LL+Y + T D F FY+ W+++ +K
Sbjct: 1651 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSEGTHHAKEI 1710
Query: 170 -----IYYFARMKS---RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
I + A + R+I++ + S S L D I L F + F
Sbjct: 1711 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1770
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1771 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1830
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1831 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1890
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1891 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1927
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1928 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1972
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1973 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 2026
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 2027 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 2059
>UniRef100_Q6KCD5-2 Splice isoform 2 of Q6KCD5 [Mus musculus]
Length = 2691
Score = 119 bits (299), Expect = 2e-25
Identities = 114/453 (25%), Positives = 191/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKPI------------YYFARM 176
+Q+ LL+Y + T D F FY+ W+++ +K + ++ +
Sbjct: 1645 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSDATHHAKEL 1704
Query: 177 KS---------------RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
++ R+I++ + S S L D I L F + F
Sbjct: 1705 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1764
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1765 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1824
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1825 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1884
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1885 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1921
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1922 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1966
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1967 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 2020
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 2021 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 2053
>UniRef100_Q6KCD5 Nipped-B-like protein [Mus musculus]
Length = 2798
Score = 119 bits (299), Expect = 2e-25
Identities = 114/453 (25%), Positives = 191/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKPI------------YYFARM 176
+Q+ LL+Y + T D F FY+ W+++ +K + ++ +
Sbjct: 1645 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSDATHHAKEL 1704
Query: 177 KS---------------RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
++ R+I++ + S S L D I L F + F
Sbjct: 1705 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1764
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1765 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1824
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1825 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1884
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1885 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1921
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1922 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1966
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1967 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 2020
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 2021 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 2053
>UniRef100_Q6KC79-2 Splice isoform 2 of Q6KC79 [Homo sapiens]
Length = 2697
Score = 119 bits (299), Expect = 2e-25
Identities = 116/453 (25%), Positives = 189/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKP------------------- 169
+Q+ LL+Y + T D F FY+ W+++ +K
Sbjct: 1651 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSEGTHHAKEI 1710
Query: 170 -----IYYFARMKS---RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
I + A + R+I++ + S S L D I L F + F
Sbjct: 1711 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1770
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1771 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1830
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1831 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1890
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1891 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1927
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1928 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1972
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1973 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 2026
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 2027 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 2059
>UniRef100_Q6KC79 Nipped-B-like protein [Homo sapiens]
Length = 2804
Score = 119 bits (299), Expect = 2e-25
Identities = 116/453 (25%), Positives = 189/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKP------------------- 169
+Q+ LL+Y + T D F FY+ W+++ +K
Sbjct: 1651 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSEGTHHAKEI 1710
Query: 170 -----IYYFARMKS---RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
I + A + R+I++ + S S L D I L F + F
Sbjct: 1711 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1770
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1771 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1830
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1831 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1890
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1891 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1927
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1928 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1972
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1973 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 2026
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 2027 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 2059
>UniRef100_UPI00003B0050 UPI00003B0050 UniRef100 entry
Length = 1994
Score = 117 bits (292), Expect = 1e-24
Identities = 113/447 (25%), Positives = 183/447 (40%), Gaps = 99/447 (22%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKPI------------------ 170
+Q+ LL+Y + T D F FY+ W+++ +K I
Sbjct: 853 LQKALLDYLDENTETDASLVFSRKFYIAQWFRDTTMETEKAIKSQKDEDSSEGTHHAKDV 912
Query: 171 ----YYFARMKSR-----TIVRDSGSVSSMLT--RDSIKK-----ITLALGQNSSFCRGF 214
R +SR +I++ + S S L D++ I L F + F
Sbjct: 913 ETTGQIMHRAESRKKFLRSIIKTAPSQFSTLKIYSDTVDYEDACLIVRYLASMRPFAQSF 972
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 973 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARPDMQRGVHGRLMDNSTSVREA 1032
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1033 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1092
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1093 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHHDKEAMT------ 1129
Query: 391 KKTEQIVEMLKRLPNNQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCELMCKCL 450
+K I ++L + + S V+K C + L
Sbjct: 1130 RKILNITDVLLKSEED---------------------------ASYKPVKKACTQLVDNL 1162
Query: 451 LEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLTLQPYLK 505
+E IL+ +E S+ K + + L F + P L + +T+QPYL
Sbjct: 1163 VEHILKYEESLSDSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMTMQPYLA 1216
Query: 506 TQVKTNLKTYILGSLQCYYALCIELLE 532
T+ T ++ ++ L + L+E
Sbjct: 1217 TKCSTQNDFMVICNVAKILELVVPLME 1243
>UniRef100_UPI000044FA01 UPI000044FA01 UniRef100 entry
Length = 2069
Score = 116 bits (291), Expect = 1e-24
Identities = 87/331 (26%), Positives = 164/331 (49%), Gaps = 50/331 (15%)
Query: 188 VSSMLTRDSIKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRAVSIIVEAD 247
+ + + ++ + I L F + FD +++ + E S +R +A++ ++ IVE D
Sbjct: 1093 IKTYIDYNNAQLIAQYLATKRPFSQSFDGCLKKIILVVNEPSIAVRTRAMKCLANIVEVD 1152
Query: 248 PEVLGDKFVQSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVS 307
P VL K +Q V +F DTAISVREAA++LVG+ + S+ D+ +Y++ ++ RI DTGVS
Sbjct: 1153 PLVLKRKDMQMGVNQKFLDTAISVREAAVDLVGKFVLSNQDLIDQYYDMLSTRILDTGVS 1212
Query: 308 VRKRAIKIIRDMCCSDGNFLRV----YKSLHRDNFTCY***IEYPAYSASDLQDLVCKTF 363
VRKR IKI+RD+C +F ++ K + R + +Q LV + F
Sbjct: 1213 VRKRVIKILRDICLEYPDFSKIPEICVKMIRR-------------VHDEEGIQKLVTEVF 1259
Query: 364 YEFWFEEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPN------NQLLVTVIK--- 414
+ WF TP T+ K G + +K I++++ + LL+++ K
Sbjct: 1260 MKMWF----TPCTKNDKIG------IQRKINHIIDVVNTAHDTGTTWLEGLLMSIFKPRD 1309
Query: 415 -RCLTLDFLPQSAKASGVNPVSLVTVRKRCELMCKCLLEKILQVDEMNSNELEKHALPYV 473
+ + + K + P+ +V C+ + L+++++++++ +++ + L +
Sbjct: 1310 NMLRSEGCVQEFIKKNSEPPMDIVIA---CQQLADGLVDRLIELEDTDNSRM----LGCI 1362
Query: 474 LVLHAFCLVDPTLCAPASNPSQFVLTLQPYL 504
LH V P L + +T++PYL
Sbjct: 1363 TTLHLLAKVRPQLLV------KHAITIEPYL 1387
>UniRef100_Q7PLI2 Nipped-B protein [Drosophila melanogaster]
Length = 2077
Score = 116 bits (291), Expect = 1e-24
Identities = 87/331 (26%), Positives = 164/331 (49%), Gaps = 50/331 (15%)
Query: 188 VSSMLTRDSIKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRAVSIIVEAD 247
+ + + ++ + I L F + FD +++ + E S +R +A++ ++ IVE D
Sbjct: 1101 IKTYIDYNNAQLIAQYLATKRPFSQSFDGCLKKIILVVNEPSIAVRTRAMKCLANIVEVD 1160
Query: 248 PEVLGDKFVQSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVS 307
P VL K +Q V +F DTAISVREAA++LVG+ + S+ D+ +Y++ ++ RI DTGVS
Sbjct: 1161 PLVLKRKDMQMGVNQKFLDTAISVREAAVDLVGKFVLSNQDLIDQYYDMLSTRILDTGVS 1220
Query: 308 VRKRAIKIIRDMCCSDGNFLRV----YKSLHRDNFTCY***IEYPAYSASDLQDLVCKTF 363
VRKR IKI+RD+C +F ++ K + R + +Q LV + F
Sbjct: 1221 VRKRVIKILRDICLEYPDFSKIPEICVKMIRR-------------VHDEEGIQKLVTEVF 1267
Query: 364 YEFWFEEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPN------NQLLVTVIK--- 414
+ WF TP T+ K G + +K I++++ + LL+++ K
Sbjct: 1268 MKMWF----TPCTKNDKIG------IQRKINHIIDVVNTAHDTGTTWLEGLLMSIFKPRD 1317
Query: 415 -RCLTLDFLPQSAKASGVNPVSLVTVRKRCELMCKCLLEKILQVDEMNSNELEKHALPYV 473
+ + + K + P+ +V C+ + L+++++++++ +++ + L +
Sbjct: 1318 NMLRSEGCVQEFIKKNSEPPMDIVIA---CQQLADGLVDRLIELEDTDNSRM----LGCI 1370
Query: 474 LVLHAFCLVDPTLCAPASNPSQFVLTLQPYL 504
LH V P L + +T++PYL
Sbjct: 1371 TTLHLLAKVRPQLLV------KHAITIEPYL 1395
>UniRef100_UPI0000439DE5 UPI0000439DE5 UniRef100 entry
Length = 1466
Score = 116 bits (290), Expect = 2e-24
Identities = 115/473 (24%), Positives = 200/473 (41%), Gaps = 94/473 (19%)
Query: 109 DAKKNRKVSKDDSTFSNHEIVQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQK 168
D K ++ ++++ + +Q+ LL+Y + D F FY+ W+++ +K
Sbjct: 844 DQKAIERIIRENTEGDETQRLQKALLDYMDENAETDPALAFARKFYIAQWFRDCTTETEK 903
Query: 169 PIY-----------------------YFARMKSR-----TIVRDSGSV-------SSMLT 193
+ R ++R ++V+ + + S +
Sbjct: 904 AMRSQNQKEDDSDGAQHAKELQATGDIMQRAETRKKFLHSVVKSTPNQFTTLRMNSDTVD 963
Query: 194 RDSIKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGD 253
D I L F + FD +L L E++ +R KA++ +S +V DP +L
Sbjct: 964 YDDACLIVRYLASTRPFSQSFDIYLTQILRVLGESAIAVRTKAMKCLSEVVAVDPSILAR 1023
Query: 254 KFVQSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAI 313
+Q V GR D + SVREAA+EL+GR + S P + +Y++ + ERI DTG+SVRKR I
Sbjct: 1024 SDMQRGVHGRLMDNSTSVREAAVELLGRFVLSRPQLTEQYYDMLIERILDTGISVRKRVI 1083
Query: 314 KIIRDMCCSDGNFLRV----YKSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFE 369
KI+RD+C NF ++ K + R N ++ LV +TF + WF
Sbjct: 1084 KILRDICLEQPNFSKITEMCVKMIRRVN-------------DEEGIKKLVNETFQKLWF- 1129
Query: 370 EPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNN---QLLVTVIKRCLTLDFLPQSA 426
TP D T+ ++ T+ +V K + QLL ++K S
Sbjct: 1130 -TPTPN----HDKETMNRKILNITD-VVSACKDTGYDWFEQLLQNLLK----------SE 1173
Query: 427 KASGVNPVSLVTVRKRCELMCKCLLEKILQVDE-------MNSNELEKHALPYVLVLHAF 479
+ S P RK C + L+E IL+ +E +NS L + + L+ F
Sbjct: 1174 EDSSYKP-----TRKACVQLVDNLVEHILKYEEALAEHKSVNSTRL----VACITTLYLF 1224
Query: 480 CLVDPTLCAPASNPSQFVLTLQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+ L + +T+QPYL T+ + ++ ++ L + L++
Sbjct: 1225 SKIRAQLMV------KHAMTMQPYLTTKCSSQSDFMVICNVAKILELVVPLMD 1271
>UniRef100_Q5TNH4 ENSANGP00000028248 [Anopheles gambiae str. PEST]
Length = 1425
Score = 113 bits (282), Expect = 2e-23
Identities = 89/334 (26%), Positives = 152/334 (44%), Gaps = 63/334 (18%)
Query: 204 LGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGR 263
L SF + +DK +++ ++E IR +A++ ++ IVE D VL K +Q V+ +
Sbjct: 659 LASKRSFSQSYDKYLQKIILVVREPVVAIRTRAMKCLANIVEVDQLVLARKDMQMGVQQK 718
Query: 264 FCDTAISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSD 323
DTAISVREAA++LVG++I S P++ +Y+E I++RI DTGVSVRKR IKI+RD+C
Sbjct: 719 LLDTAISVREAAVDLVGKYILSDPELIDQYYEMISQRILDTGVSVRKRVIKILRDIC--- 775
Query: 324 GNFLRVYKSLHRDNFTCY***IEYPAY---------------SASDLQDLVCKTFYEFWF 368
IEYPA+ +Q LV F WF
Sbjct: 776 ---------------------IEYPAHEKIPDICVKMIRRVNDEEGIQKLVMDVFMTMWF 814
Query: 369 EEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNNQLLVTVIKRCLTLDFLPQSAKA 428
P + D +K QI++++ +++ L F P+ +K
Sbjct: 815 -TPCNDNDKAAMD---------RKITQIIDVV--CSSHETGTQGFDALLKTIFEPKESKD 862
Query: 429 SGVNPVSLV--TVRKRCELMCKCLLEKILQVDEMNSNELEKHALPYVLVLHAFCLVDPTL 486
+ T+ K C+ + L++ ++++ + L + + LH F + P L
Sbjct: 863 DNKKLKKEIPKTLIKACQQIVDGLVDATMRLEGAENTRL----VGCITALHLFAKIQPQL 918
Query: 487 CAPASNPSQFVLTLQPYLKTQVKTNLKTYILGSL 520
++L+PYL + + + + + S+
Sbjct: 919 LV------NHAMSLEPYLNMRCQNQIISKFISSI 946
>UniRef100_Q7PVF7 ENSANGP00000010656 [Anopheles gambiae str. PEST]
Length = 1518
Score = 113 bits (282), Expect = 2e-23
Identities = 89/334 (26%), Positives = 152/334 (44%), Gaps = 63/334 (18%)
Query: 204 LGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGR 263
L SF + +DK +++ ++E IR +A++ ++ IVE D VL K +Q V+ +
Sbjct: 659 LASKRSFSQSYDKYLQKIILVVREPVVAIRTRAMKCLANIVEVDQLVLARKDMQMGVQQK 718
Query: 264 FCDTAISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSD 323
DTAISVREAA++LVG++I S P++ +Y+E I++RI DTGVSVRKR IKI+RD+C
Sbjct: 719 LLDTAISVREAAVDLVGKYILSDPELIDQYYEMISQRILDTGVSVRKRVIKILRDIC--- 775
Query: 324 GNFLRVYKSLHRDNFTCY***IEYPAY---------------SASDLQDLVCKTFYEFWF 368
IEYPA+ +Q LV F WF
Sbjct: 776 ---------------------IEYPAHEKIPDICVKMIRRVNDEEGIQKLVMDVFMTMWF 814
Query: 369 EEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNNQLLVTVIKRCLTLDFLPQSAKA 428
P + D +K QI++++ +++ L F P+ +K
Sbjct: 815 -TPCNDNDKAAMD---------RKITQIIDVV--CSSHETGTQGFDALLKTIFEPKESKD 862
Query: 429 SGVNPVSLV--TVRKRCELMCKCLLEKILQVDEMNSNELEKHALPYVLVLHAFCLVDPTL 486
+ T+ K C+ + L++ ++++ + L + + LH F + P L
Sbjct: 863 DNKKLKKEIPKTLIKACQQIVDGLVDATMRLEGAENTRL----VGCITALHLFAKIQPQL 918
Query: 487 CAPASNPSQFVLTLQPYLKTQVKTNLKTYILGSL 520
++L+PYL + + + + + S+
Sbjct: 919 LV------NHAMSLEPYLNMRCQNQIISKFISSI 946
>UniRef100_UPI00004366F2 UPI00004366F2 UniRef100 entry
Length = 1833
Score = 112 bits (279), Expect = 4e-23
Identities = 114/473 (24%), Positives = 202/473 (42%), Gaps = 97/473 (20%)
Query: 109 DAKKNRKVSKDDSTFSNHEIVQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQK 168
D K ++ ++++ + +Q+ LL+Y + D F FY+ W+++ +K
Sbjct: 665 DQKAIERIIRENTEGDETQRLQKALLDYMDENAETDPALAFARKFYIAQWFRDCTTETEK 724
Query: 169 PIY-----------------------YFARMKSR-----TIVRDSGSV-------SSMLT 193
+ R ++R ++V+ + + S +
Sbjct: 725 AMRSQNQKEDDSDGAQHAKELQATGDIMQRAETRKKFLHSVVKSTPNQFTTLRMNSDTVD 784
Query: 194 RDSIKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGD 253
D I L + F + FD+ + +L E++ +R KA++ +S +V DP +L
Sbjct: 785 YDDACLIVRYLASSRPFSQSFDRGGNRVL---GESAIAVRTKAMKCLSEVVAVDPSILAR 841
Query: 254 KFVQSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAI 313
+Q V GR D + SVREAA+EL+GR + S P + +Y++ + ERI DTG+SVRKR I
Sbjct: 842 SDMQRGVHGRLMDNSTSVREAAVELLGRFVLSRPQLTEQYYDMLIERILDTGISVRKRVI 901
Query: 314 KIIRDMCCSDGNFLRV----YKSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFE 369
KI+RD+C NF ++ K + R N ++ LV +TF + WF
Sbjct: 902 KILRDICLEQPNFSKITEMCVKMIRRVN-------------DEEGIKKLVNETFQKLWF- 947
Query: 370 EPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNN---QLLVTVIKRCLTLDFLPQSA 426
TP D T+ ++ T+ +V K + QLL ++K S
Sbjct: 948 -TPTPN----HDKETMNRKILNITD-VVSACKDTGYDWFEQLLQNLLK----------SE 991
Query: 427 KASGVNPVSLVTVRKRCELMCKCLLEKILQVDE-------MNSNELEKHALPYVLVLHAF 479
+ S P RK C + L+E IL+ +E +NS L + + L+ F
Sbjct: 992 EDSSYKP-----TRKACVQLVDNLVEHILKYEEALAEHKSVNSTRL----VACITTLYLF 1042
Query: 480 CLVDPTLCAPASNPSQFVLTLQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+ L + +T+QPYL T+ + ++ ++ L + L++
Sbjct: 1043 SKIRAQLMV------KHAMTMQPYLTTKCSSQSDFMVICNVAKILELVVPLMD 1089
>UniRef100_UPI0000337DCB UPI0000337DCB UniRef100 entry
Length = 1794
Score = 112 bits (279), Expect = 4e-23
Identities = 98/373 (26%), Positives = 165/373 (43%), Gaps = 60/373 (16%)
Query: 176 MKSRTIVRDS-GSVSSMLTRDSIKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRA 234
MK+ ++ S GS S + + I L F + FD +L L E++ +R
Sbjct: 779 MKASSVRSHSQGSNSETVEYEDSCLIVRYLASMRPFAQSFDIYLSQILRVLGESAIAVRT 838
Query: 235 KALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYF 294
KA++ +S +V D +L +Q V GR D + SVREAA+ELVGR + S P++ +Y+
Sbjct: 839 KAMKCLSEVVAVDSSILARVDMQRGVHGRLMDNSTSVREAAVELVGRFVLSRPELIEQYY 898
Query: 295 EKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----YKSLHRDNFTCY***IEYPAY 350
+ + ERI DTG+SVRKR IKI+RD+C F ++ K + R N
Sbjct: 899 DMLIERILDTGISVRKRVIKILRDICLEQPGFRKITEMCVKMIRRVN------------- 945
Query: 351 SASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNN---- 406
++ LV +TF + WF T+ + +K I +++ ++
Sbjct: 946 DEEGIKKLVNETFQKLWFTPTPGQDTEA----------MTRKILNITDVVLACKDSGYDW 995
Query: 407 --QLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCELMCKCLLEKILQVDEMNSNE 464
QLL ++K S + + P +K C + L+E IL+ ++ ++
Sbjct: 996 FEQLLQNLLK----------SEEQASYKP-----TKKACVQLVDNLVEHILKYEDSLADC 1040
Query: 465 LEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLTLQPYLKTQVKTNLKTYILGS 519
EK + LH F + L + +T+QPYL T+ T ++ +
Sbjct: 1041 EEKGFNSDRLVSCTTTLHLFSKIRAQLMV------KHAMTIQPYLTTKCNTQNDFMVICN 1094
Query: 520 LQCYYALCIELLE 532
+ L + L+E
Sbjct: 1095 VAKILELVVPLME 1107
>UniRef100_UPI0000360ED0 UPI0000360ED0 UniRef100 entry
Length = 1268
Score = 110 bits (275), Expect = 1e-22
Identities = 93/364 (25%), Positives = 156/364 (42%), Gaps = 69/364 (18%)
Query: 114 RKVSKDDSTFSNHEIVQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKPI--- 170
R + ++D T + +Q+ LL+Y +D D F FY+ W+++ +K +
Sbjct: 329 RSIDRNDET----QQLQKALLDYLEDSADTDASLVFARKFYIAQWFRDTTTEAEKSMRNQ 384
Query: 171 -----------YYFARMKSRTIVRDSGSVSSMLTRDSIKK-------------------- 199
+ +++ + R+ IK
Sbjct: 385 NQKDDDSSDGPQHAKEIETTGEIMQRAEKRKKFLRNIIKTTPAHFATLKMNSDTVDYEDS 444
Query: 200 --ITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQ 257
I L F + FD +L L E++ +R KA++ +S +V DP +L +Q
Sbjct: 445 CLIVRYLASMRPFAQSFDIYLTQILRVLGESAIAVRTKAMKCLSEVVAVDPSILARSDMQ 504
Query: 258 SSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIR 317
V GR D + SVREAA+EL+GR + S P + +Y++ + ERI DTG+SVRKR IKI+R
Sbjct: 505 RGVHGRLMDNSTSVREAAVELLGRFVLSRPQLTEQYYDMLIERILDTGISVRKRVIKILR 564
Query: 318 DMCCSDGNFLRV----YKSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPST 373
D+C F ++ K + R N ++ LV +TF + WF T
Sbjct: 565 DICLEQPAFSKITEMCVKMIRRVN-------------DEEGIKKLVNETFQKLWF----T 607
Query: 374 PQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNNQLLVTVIKRCLTLDFLPQSAKASGVNP 433
P K+ T +K I +++ + QL+ +++ L + A++ GVN
Sbjct: 608 PTPAHDKETMT------RKILNITDVVAKKACVQLVDNLVEHILKYE--ESLAESKGVNS 659
Query: 434 VSLV 437
LV
Sbjct: 660 TRLV 663
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.327 0.139 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 868,232,122
Number of Sequences: 2790947
Number of extensions: 34739165
Number of successful extensions: 107953
Number of sequences better than 10.0: 778
Number of HSP's better than 10.0 without gapping: 422
Number of HSP's successfully gapped in prelim test: 356
Number of HSP's that attempted gapping in prelim test: 106861
Number of HSP's gapped (non-prelim): 1392
length of query: 543
length of database: 848,049,833
effective HSP length: 132
effective length of query: 411
effective length of database: 479,644,829
effective search space: 197134024719
effective search space used: 197134024719
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC149305.6


