

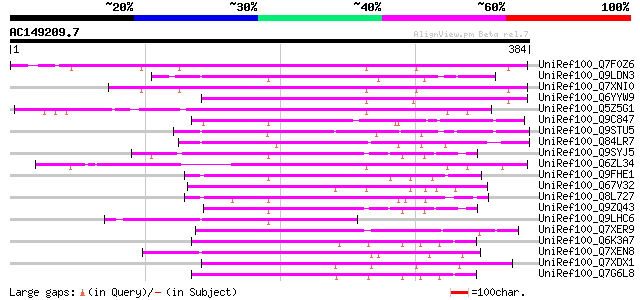
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149209.7 - phase: 0
(384 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7F0Z6 Myb family protein-like [Oryza sativa] 125 2e-27
UniRef100_Q9LDN3 Gb|AAF34841.1 [Arabidopsis thaliana] 107 7e-22
UniRef100_Q7XNI0 OSJNBb0032D24.13 protein [Oryza sativa] 106 1e-21
UniRef100_Q6YYW9 Ribosomal protein-like [Oryza sativa] 102 1e-20
UniRef100_Q5Z5G1 Myb protein-like [Oryza sativa] 102 1e-20
UniRef100_Q9C847 Hypothetical protein F8D11.7 [Arabidopsis thali... 102 2e-20
UniRef100_Q9STU5 Hypothetical protein T23J7.10 [Arabidopsis thal... 101 4e-20
UniRef100_Q84LR7 Hypothetical protein [Arabidopsis thaliana] 91 4e-17
UniRef100_Q9SYJ5 Hypothetical protein T7B11.25 [Arabidopsis thal... 89 3e-16
UniRef100_Q6ZL34 Hypothetical protein OJ1699_E05.47 [Oryza sativa] 88 4e-16
UniRef100_Q9FHE1 Glutathione transferase-like [Arabidopsis thali... 88 5e-16
UniRef100_Q67V32 NAM-like protein [Oryza sativa] 86 1e-15
UniRef100_Q8L727 Glutathione transferase [Arabidopsis thaliana] 86 2e-15
UniRef100_Q9ZQ43 Hypothetical protein At2g22710 [Arabidopsis tha... 84 9e-15
UniRef100_Q9LHC6 Gb|AAF34841.1 [Arabidopsis thaliana] 83 1e-14
UniRef100_Q7XER9 Hypothetical protein [Oryza sativa] 82 2e-14
UniRef100_Q6K3A7 Cytokinin inducibl protein-like [Oryza sativa] 80 1e-13
UniRef100_Q7XEN8 Putative NAM-like protein [Oryza sativa] 80 1e-13
UniRef100_Q7XDX1 Hypothetical protein [Oryza sativa] 80 1e-13
UniRef100_Q7G6L8 Hypothetical protein OSJNBa0032N04.18 [Oryza sa... 79 2e-13
>UniRef100_Q7F0Z6 Myb family protein-like [Oryza sativa]
Length = 417
Score = 125 bits (314), Expect = 2e-27
Identities = 109/416 (26%), Positives = 187/416 (44%), Gaps = 41/416 (9%)
Query: 1 MDPTNNQSNTQNSSDYPFSYQNQFPNSHPQNPNQFPNQHPQNPN--QFPNQNPQNMSNFG 58
M+ N S +Q SS QN P P +Q +Q PQ+P+ Q N Q + FG
Sbjct: 9 MNLLNQGSPSQQSS------QNSPPTQFPSTFSQ--SQFPQSPHFTQASPPNFQTFNPFG 60
Query: 59 FASNFNHPSFVPNNFNPYFRSMMGYSSQTPPFNGYMPM---VNENFQSVGEYPDFSSQIN 115
+N++ P NF + + S F G+ P ++ Q VG S+
Sbjct: 61 PPANYHLYGSSPPNFQGFQQQASWLQSAPISFQGFRPQESWMHSPNQVVGSASSHGSESA 120
Query: 116 RGGMTRDNE-----VAPTPEDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQT 170
R E + + +++ +R + +W E+NL L+S W+ S+ G ++
Sbjct: 121 SQCPARQEENNLVNIEESSDNSQETGRRGTRVNWTEEENLRLLSSWLNNSLDSINGNDKK 180
Query: 171 SESYWGKIAEYCNEHCSFDS-PRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSED 229
E YW +A N + S ++ R +V C+ + + K I K+ GAY A+R SG+S+D
Sbjct: 181 GEYYWRDVAAEFNGNASSNNRKRTVVQCKTHWGGVKKDIAKFCGAYSRARRTWSSGFSDD 240
Query: 230 DVFKKAQELYGCGKNVQ-FTLMEEWRALRDQPRY-----------GSQVGGNVDSGSSGS 277
+ +KA LY N + FTL WR L+DQP++ +++ + SS +
Sbjct: 241 MIMEKAHALYKSENNDKTFTLEYMWRELKDQPKWQRILEEDSKNKRTKISESGAYTSSSN 300
Query: 278 KRSYEDSVGSSARPMDRDAAKK----KGKKKSKGETLEKVEKEWVQFKELKEQEIEQLKE 333
+ + E++ RP + AK KGKK + ++ +++V F E + E + +
Sbjct: 301 QETEEETSRKEKRPEGQKKAKAKLKGKGKKPAPSPLGDQPSQDFVLFNEAVKLRAEAVLK 360
Query: 334 LTLVKQQKNKLLQEKTQAKKMKMYLKLRDEEHLD------DRKNELLGKLERELFE 383
+ + +E+T+ +K + YLKL D++ + R +L KL EL E
Sbjct: 361 SAEATTKSAEAKKEQTRMEKYQTYLKLLDKDTANFSDAKLKRHEAVLEKLATELAE 416
>UniRef100_Q9LDN3 Gb|AAF34841.1 [Arabidopsis thaliana]
Length = 287
Score = 107 bits (266), Expect = 7e-22
Identities = 76/279 (27%), Positives = 131/279 (46%), Gaps = 35/279 (12%)
Query: 106 EYPDFSSQINRGGMTRDNEVAPTPEDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVV 165
E P FSSQ++ G +++ + ++ PK +++ W ++++VL+S W+ V+
Sbjct: 10 EVPAFSSQLSEEG----SDLEGSEDEVKPKQSISRK-KWTAKEDIVLVSAWLNTSKDPVI 64
Query: 166 GRNQTSESYWGKIAEYCNEHCSFDS--PRDIVACRNRFNYMNKLINKWVGAYDSAKRMQG 223
G +Q +S+W +IA Y S D R+ C++R+ +NK + K+VG Y + +
Sbjct: 65 GNDQQGQSFWKRIAAYVAASPSLDGLPKREHAKCKHRWGKVNKSVTKFVGCYKTTTTHKT 124
Query: 224 SGWSEDDVFKKAQELYGCGKNVQFTLMEEWRALRDQPRYGSQVGGNVD---------SGS 274
SG SEDDV K A E+Y FTL WR LR ++ D G+
Sbjct: 125 SGQSEDDVMKLAYEIYFNDTKKNFTLDHAWRELRYDQKWCEATSRKGDKNAKRKKCGDGN 184
Query: 275 SGSKRSYEDSVGSSARPMDRDAAKKKGKKKS--------------KGETLEKVEKEWVQF 320
+ S+ + + +RP AAK K +K + G+++E + W
Sbjct: 185 ASSQPIHVEDDSVMSRPPGVKAAKAKARKSATIKEGKKPATVKDDSGQSVEHFQNLW--- 241
Query: 321 KELKEQEIEQLKELTLVKQQKNKLLQEKTQAKKMKMYLK 359
ELKE++ ++ KE QQ +LL + + ++LK
Sbjct: 242 -ELKEKDWDR-KEKQSKHQQLERLLSKTEPLSDIDLFLK 278
>UniRef100_Q7XNI0 OSJNBb0032D24.13 protein [Oryza sativa]
Length = 399
Score = 106 bits (264), Expect = 1e-21
Identities = 86/341 (25%), Positives = 153/341 (44%), Gaps = 31/341 (9%)
Query: 74 NPYFRSMMGYSSQTPPFNGYMPM---VNENFQSVGEYPDFSSQINRGGMTRDNE-----V 125
N + + S F G+ P ++ Q VG S+ R E +
Sbjct: 58 NGFLQQASWLQSAPISFQGFRPQESWMHSPNQVVGSASSHGSKSASQCPARQEENNLVNI 117
Query: 126 APTPEDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEH 185
+ +++ +R +W E+NL L+S W+ S+ G ++ E YW +A N +
Sbjct: 118 EESSDNSQETGRRGTHVNWTEEENLRLLSSWLNNSLDSINGNDKKGEYYWRDVAAEFNGN 177
Query: 186 CSFDS-PRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKN 244
S ++ R +V C+ + + K I K+ GAY A+R SG+S+D + +KA LY N
Sbjct: 178 ASSNNRKRTVVQCKTHWGGVKKDIAKFCGAYSRARRTWSSGFSDDMIMEKAHALYKSENN 237
Query: 245 VQ-FTLMEEWRALRDQPRY-----------GSQVGGNVDSGSSGSKRSYEDSVGSSARPM 292
+ FTL WR L+DQP++ +++ + SS ++ + E++ RP
Sbjct: 238 DKTFTLEYMWRELKDQPKWRRILEEDSKNKRTKISESGAYTSSSNQETEEETSRKEKRPE 297
Query: 293 DRDAAKK----KGKKKSKGETLEKVEKEWVQFKELKEQEIEQLKELTLVKQQKNKLLQEK 348
+ AK KGKK + ++ +++V F E + E + + + + +E+
Sbjct: 298 GQKKAKAKLKGKGKKPAPSPLGDQPSQDFVLFNEAVKLRAEAVLKSAEATTKSAEAKKEQ 357
Query: 349 TQAKKMKMYLKLRDEEHLD------DRKNELLGKLERELFE 383
T+ +K + YLKL D++ + R +L KL EL E
Sbjct: 358 TRMEKYQTYLKLLDKDTANFSDAKLKRHEAVLEKLATELAE 398
>UniRef100_Q6YYW9 Ribosomal protein-like [Oryza sativa]
Length = 303
Score = 102 bits (255), Expect = 1e-20
Identities = 74/264 (28%), Positives = 129/264 (48%), Gaps = 23/264 (8%)
Query: 143 SWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDS-PRDIVACRNRF 201
+W E+NL L+S W+ S+ G ++ E YW +A N + S ++ R +V C+ +
Sbjct: 39 NWTEEENLRLLSSWLNNSLDSINGNDKKGEYYWRDVAAEFNGNTSSNNRKRTVVQCKTHW 98
Query: 202 NYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQ-FTLMEEWRALRDQP 260
+ K I K+ GAY A+R SG+S+D + +KA LY N + FTL WR L+DQP
Sbjct: 99 GGVKKDIAKFCGAYSRARRTWSSGFSDDMIMEKAHALYKSENNDKTFTLEYMWRELKDQP 158
Query: 261 RY-----------GSQVGGNVDSGSSGSKRSYEDSVGSSARPMDRDAA----KKKGKKKS 305
++ +++ + SS ++ + E++ RP + A K KGKK +
Sbjct: 159 KWRRILEEDSKNKRTKISESGAYTSSSNQETEEETSRKEKRPEGQKKAKVKLKGKGKKHA 218
Query: 306 KGETLEKVEKEWVQFKELKEQEIEQLKELTLVKQQKNKLLQEKTQAKKMKMYLKLRDEEH 365
++ +++V E + E + + T + + +E+T+ +K + YLKL D++
Sbjct: 219 PSPLGDQPSQDFVLLNEAVKLRAEAVLKSTEATTKSAEAKKEQTRMEKYQTYLKLLDKDT 278
Query: 366 LD------DRKNELLGKLERELFE 383
+ R +L KL EL E
Sbjct: 279 ANFNDVKLKRHEAILEKLATELAE 302
>UniRef100_Q5Z5G1 Myb protein-like [Oryza sativa]
Length = 409
Score = 102 bits (255), Expect = 1e-20
Identities = 95/385 (24%), Positives = 164/385 (41%), Gaps = 43/385 (11%)
Query: 4 TNNQSNTQNSSDYPFSYQNQF-----PNSHPQNP--NQFPNQHP------QNPNQFPNQN 50
TN+++ S+ +P SY F N HP P N P +HP Q +
Sbjct: 21 TNSEAQNSLSTQFPTSYPQNFRPSFLQNFHPFGPPSNYQPYRHPPIFQGAQQQEYYGQPT 80
Query: 51 PQNMSNFGFASNFNHPSFVPNNFNPYFRSMMGYSSQTPPFNGYMPMVNENFQSVGEYPDF 110
P ++ F N H S F RS G T N S G
Sbjct: 81 PGSLEGFQLQENLVHSSNQAFGFAAN-RSQFGMQYSTSI------RATANTSSHGSASPC 133
Query: 111 SSQINRGGMTRDNEVAPTPEDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQT 170
++ N + E + D++ + +R + +W + N+ L+S W+ + G ++
Sbjct: 134 HTRHNEKEVVEVEEAS----DSSEEGRRGTRINWTEDDNIRLMSSWLNNSVDPIKGNDKK 189
Query: 171 SESYWGKIAEYCNEHC-SFDSPRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSED 229
SE YW +A N + S + R+ CR ++ + + + K+ G Y A+ SG+S+D
Sbjct: 190 SEQYWKAVAREFNSNMPSNGNKRNPKQCRTHWDNVKRDVTKFCGFYSKARTTFTSGYSDD 249
Query: 230 DVFKKAQELYGCGKNVQ-FTLMEEWRALRDQPRY-----------GSQVGGNVDSGSSGS 277
+ +KA+E Y N + FTL W+ L+DQP++ S++ + SS +
Sbjct: 250 MIMEKAREWYKKHNNQKPFTLEYMWKDLKDQPKWRRVLEESSHNKRSKISESGAYTSSSN 309
Query: 278 KRSYEDSVGSSARPMDRDAAKKKGKKKSKGETL-EKVEKEWVQFKE---LKEQEIEQLKE 333
+ + E++ RP + AAK++ K K L +K + V F E K + + E
Sbjct: 310 QDTEEETERKEKRPEGQKAAKQRQKGKGAPSPLGDKPSQNMVLFHEAITTKAAALLKAAE 369
Query: 334 LTLV--KQQKNKLLQEKTQAKKMKM 356
TL+ + +K K + +K + K K+
Sbjct: 370 ATLIGAEAKKEKAIAKKREGKGRKI 394
>UniRef100_Q9C847 Hypothetical protein F8D11.7 [Arabidopsis thaliana]
Length = 579
Score = 102 bits (253), Expect = 2e-20
Identities = 81/257 (31%), Positives = 127/257 (48%), Gaps = 18/257 (7%)
Query: 135 KSKRNQQP--SWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDS-- 190
K K+ ++P +W++ +++VLISGW+ VVG Q ++W +IA Y N
Sbjct: 329 KGKKAKKPRRNWSSTEDVVLISGWLNTSKDPVVGNEQKGAAFWERIAVYYNSSPKLKGVE 388
Query: 191 PRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQFTLM 250
R + C+ R++ +N +NK+VG+Y +A + Q SG ++DDV A +++ +FT
Sbjct: 389 KRGHICCKQRWSKVNDAVNKFVGSYLAASKQQTSGQNDDDVVSLAHQIFSKDYGCKFTCE 448
Query: 251 EEWRALRDQPRYGSQVGGNVDSGSSGSKRSYEDS--VG---SSARPMDRDAAKKKGKKKS 305
WR + RY + G + ++ DS VG ARP+ AAK K K
Sbjct: 449 HAWR----ERRYDQKWIAQSTHGKAKRRKCEADSEPVGVEDKEARPIGVKAAKAAAKAKG 504
Query: 306 KGETLEKVEKEWVQFKELKE-QEIEQLKELTLVKQQKNKLLQEKTQAKKMKMYLKLRDEE 364
K + L VE E + LKE Q I ++KE ++K ++++K K L L E
Sbjct: 505 KAK-LSPVEGE--ETNALKEIQSIWEIKEKDHAAKEKLIIIKDKKNRTKFFERL-LGKTE 560
Query: 365 HLDDRKNELLGKLEREL 381
L D + EL KL EL
Sbjct: 561 PLSDIEIELKNKLINEL 577
>UniRef100_Q9STU5 Hypothetical protein T23J7.10 [Arabidopsis thaliana]
Length = 302
Score = 101 bits (251), Expect = 4e-20
Identities = 76/282 (26%), Positives = 139/282 (48%), Gaps = 28/282 (9%)
Query: 122 DNEVAPTPEDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEY 181
D++V+P TPK KR ++ W+ ++LVL+S W+ +V+G Q ++W +IA Y
Sbjct: 29 DDDVSPNQAAHTPKVKRERR-KWSAGEDLVLVSAWLNTSKDAVIGNEQKGYAFWSRIAAY 87
Query: 182 CNEHCSFD--SPRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELY 239
+ R+ + R+ +N+ + K+VG+Y++A + + SG ++DDV A E+Y
Sbjct: 88 YGASPKLNGVEKRETGHIKQRWTKINEGVGKFVGSYEAATKQKSSGQNDDDVVALAHEIY 147
Query: 240 GCGKNVQFTLMEEWRALRDQPRYGSQVGGNV---------DSGSSGSKRSYEDSVGSSAR 290
++ +FTL WR LR + ++ S D S+ +++E S R
Sbjct: 148 N-SEHGKFTLEHAWRVLRFEQKWLSAPSTKATVMSKRRKSDKASTSQPQTHEAEEAMS-R 205
Query: 291 PMDRDAAKKKGKK--------KSKGETLEKVEKEWVQFKELKEQEIEQLKELTLVKQQKN 342
P+ AAK K KK + KG + +++ W E+K+++ E L++ +++++
Sbjct: 206 PIGVKAAKAKAKKAVSKTTTVEDKGNVMLEIQSIW----EIKQKDWE-LRQKDREQEKED 260
Query: 343 KLLQEKTQAKKMKMYLKLRDEEHLDDRKNELLGKLERELFEN 384
QE+ K+ L +E L D + L KL E+ N
Sbjct: 261 FEKQERLSRTKLLESL-FTKKEPLTDIEVALKNKLINEMLSN 301
>UniRef100_Q84LR7 Hypothetical protein [Arabidopsis thaliana]
Length = 392
Score = 91.3 bits (225), Expect = 4e-17
Identities = 77/282 (27%), Positives = 136/282 (47%), Gaps = 36/282 (12%)
Query: 126 APTPEDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEH 185
+P+ E+ PK KR + W+ ++++VL+S W+ +V+G Q + ++W +IA Y +
Sbjct: 87 SPSLEEHVPKVKRERM-KWSAKEDMVLVSSWLNTSKDAVIGNEQKANTFWSRIAAYYDAS 145
Query: 186 CSFDSPRDIVA--CRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGK 243
+ R + + R+ +N + K+VG+Y++A R + SG +++DV A E++
Sbjct: 146 PQLNGLRKRMQGNIKQRWAKINDGVCKFVGSYEAASREKSSGQNDNDVISLAHEIFNNDY 205
Query: 244 NVQFTLMEEWRALRDQPRYGSQVGGNVDSGSSGSKRSYEDSVG---------SSARPMDR 294
+F L WR LR ++ SQ +V S ++ + S + +RP+
Sbjct: 206 GYKFPLEHAWRVLRHDQKWCSQ--ASVMSKRRKCDKAAQPSTSQPPSHGVEEAMSRPIGV 263
Query: 295 DAAKKKGKK--------KSKGETLEKVEKEW-VQFK--ELKEQEIEQLKELTLVKQQKNK 343
AAK K KK + KG + +++ W ++ K EL++++ EQ KE K + +K
Sbjct: 264 KAAKAKAKKTVTKTTTVEDKGNAMLEIQSIWEIKQKDWELRQKDREQEKEDFEKKDRLSK 323
Query: 344 -LLQEKTQAKKMKMYLKLRDEEHLDDRKNELLGKLERELFEN 384
L E AKK E L D + L KL L E+
Sbjct: 324 TTLLESLIAKK----------EPLTDNEVTLKNKLIDFLLEH 355
>UniRef100_Q9SYJ5 Hypothetical protein T7B11.25 [Arabidopsis thaliana]
Length = 302
Score = 88.6 bits (218), Expect = 3e-16
Identities = 78/279 (27%), Positives = 126/279 (44%), Gaps = 36/279 (12%)
Query: 91 NGYMPMVNENFQS----VGEYPDFSSQINRGGMTRDNEVAPTPEDTTPKSKRNQQPSWNT 146
+G+M +++ +S +Y SS+I G R E + E+ + W
Sbjct: 11 SGFMDLLHSQQESHNNETNQYEVGSSEIPVFGTQRCQE---SHEERYASRVAIARKKWAA 67
Query: 147 EQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDS--PRDIVACRNRFNYM 204
++++VLIS W+ VVG Q + ++W +IA Y + D R C+ R+ M
Sbjct: 68 KEDIVLISAWLNTSKDPVVGNEQKAPAFWKRIASYVAANPDLDGVPKRASAQCKQRWAKM 127
Query: 205 NKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQFTLMEEWRALRDQPRYGS 264
N+L+ K+VG Y + + SG +E+DV A EL+ +F+L WR LR ++
Sbjct: 128 NELVMKFVGCYATTTNQKASGQTENDVMLFANELFENDMKKKFSLDHAWRLLRHDQKW-- 185
Query: 265 QVGGNVDSGSSGSKRSYEDSVGSSA---------------RPMDRDAAKKKGKKKS--KG 307
+ N G +KR + VG+ A RP AAK K KK KG
Sbjct: 186 -LISNAPKGKGIAKRR-KVRVGTQAASSQPIDLEDDDVMRRPPGVKAAKAKAKKTPTVKG 243
Query: 308 ETLEKVEKEWVQFKELKEQEIEQLKELTLVKQQKNKLLQ 346
E + VE +LKE++ ++ KQ KN++L+
Sbjct: 244 EEVNSVE-HLTSLWDLKERDWDRKD-----KQSKNQMLE 276
>UniRef100_Q6ZL34 Hypothetical protein OJ1699_E05.47 [Oryza sativa]
Length = 361
Score = 88.2 bits (217), Expect = 4e-16
Identities = 102/395 (25%), Positives = 162/395 (40%), Gaps = 69/395 (17%)
Query: 20 YQNQFPNSHPQ-NPNQFPNQHPQNP--NQFPNQNPQNMSNFGFASNFNHPSFVPNNFNPY 76
++ F N Q + +Q N QN QFP PQN F NF HP P+N+ PY
Sbjct: 4 FKGNFSNLLNQGSSSQATNSEAQNSLSTQFPTSYPQNFRP-SFLQNF-HPFGPPSNYQPY 61
Query: 77 FRSMMGYSSQTPPFNGY-MPMVNENFQSVGEYPDFSSQINRGGMTRDNEVAPTPEDTTPK 135
+ +Q + G P E FQ +Q R V P
Sbjct: 62 RHPPIFQGAQQQEYYGQPTPGSLEGFQLQENLVHSFNQAFGFAANRSQFVDP-------- 113
Query: 136 SKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHC-SFDSPRDI 194
+ G ++ SE YW +A N + S + R+
Sbjct: 114 ----------------------------IKGNDKKSEQYWKAVAREFNSNMPSNGNKRNP 145
Query: 195 VACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQ-FTLMEEW 253
CR ++ + + + K+ G Y A+ SG+S+D + +KA E Y N + FTL W
Sbjct: 146 KQCRTHWDNVKRDVTKFCGFYSKARTTFTSGYSDDMIMEKACEWYKKHNNQKPFTLEYMW 205
Query: 254 RALRDQPRY-----------GSQVGGNVDSGSSGSKRSYEDSVGSSARPMDRDAAKKKGK 302
+ L+DQP++ +++ + SS ++ + E++ RP + AAK++ K
Sbjct: 206 KDLKDQPKWRRVLEESSHNKRNKISESGAYTSSSNQDTEEETERKEKRPEGQKAAKQRQK 265
Query: 303 KKSKGETL-EKVEKEWVQFKE---LKEQEIEQLKELTLV----KQQKNKLLQEKTQAKKM 354
K L +K + V F E K + + E TL+ K++K +EK +A+K
Sbjct: 266 GKGAPSPLGDKPSQNMVLFHEAITTKAAALLKAAEATLIGAEAKKEKAIAKKEKARAEKY 325
Query: 355 KMYLKLRDE------EHLDDRKNELLGKLERELFE 383
+MYLKL ++ E R +L +L REL E
Sbjct: 326 QMYLKLMEKDTSTFSEAKLKRHENVLDQLARELAE 360
>UniRef100_Q9FHE1 Glutathione transferase-like [Arabidopsis thaliana]
Length = 590
Score = 87.8 bits (216), Expect = 5e-16
Identities = 66/251 (26%), Positives = 116/251 (45%), Gaps = 35/251 (13%)
Query: 130 EDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFD 189
++TT + K ++ W+ ++ +LIS W+ +V + ++W +I Y N S
Sbjct: 261 QNTTDRRKHRRK--WSRAEDAILISAWLNTSKDPIVDNEHKACAFWKRIGAYFNNSASLA 318
Query: 190 S--PRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQF 247
+ R+ C+ R++ +N + K+VG YD A + SG SEDDVF+ A ++Y F
Sbjct: 319 NLPKREPSHCKQRWSKLNDKVCKFVGCYDQALNQRSSGQSEDDVFQVAYQVYTNNYKSNF 378
Query: 248 TLMEEWRALRDQPRYGSQVGGNVDSGSSGSKRS--------YEDSVGSSARPMDRD---- 295
TL WR LR ++ S G SKR+ Y S + P+ D
Sbjct: 379 TLEHAWRELRHSKKWCSLYPFENSKGGGSSKRTKLNNGDRVYSSSSNPESVPIALDEEEQ 438
Query: 296 ---------AAKKKGKKKSKGETLE--------KVEKEWVQFKELKEQEIEQLKELTLVK 338
++K+K KK + T+E ++E WV +E EQ +++ + ++
Sbjct: 439 VMDLPLGVKSSKQKEKKVATIITIEEREADSGSRLENLWVLDEE--EQVMDRPLGVKSLE 496
Query: 339 QQKNKLLQEKT 349
Q++NK+ + T
Sbjct: 497 QKENKVAPKPT 507
>UniRef100_Q67V32 NAM-like protein [Oryza sativa]
Length = 376
Score = 86.3 bits (212), Expect = 1e-15
Identities = 65/254 (25%), Positives = 123/254 (47%), Gaps = 32/254 (12%)
Query: 132 TTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDSP 191
T K + + ++ T+++++L+S W+ G + G +Q+ +YW +I EY + + F+S
Sbjct: 100 TAAKISQGRTKNFTTQEDILLVSAWLNVGMDPIQGVDQSQGTYWARIHEYFHANKEFEST 159
Query: 192 RDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELY--GCGKNVQFTL 249
R + NR++ + +N + G + SG DD A EL+ K+ +F L
Sbjct: 160 RSESSLLNRWSAIQHDVNIFCGCMSRIEARNQSGSRVDDKIANACELFKEEDKKHRKFNL 219
Query: 250 MEEWRALRDQPRY--GSQVGGNVDSGSSGSKRSYEDSVGSSARPMDRD--------AAKK 299
M W L+D+P++ + G S+ +++ +S +S P D D + +
Sbjct: 220 MHCWNILKDKPKWMDNRKKVGCAKKPSNKKQKTVANSSPTSVEPADLDVYCSDAQPSVRP 279
Query: 300 KGKKKSK-----GETLEKVE------KEWVQFKELKEQEI---------EQLKELTLVKQ 339
GKK +K G T+E V+ KE +ELK++E+ E+ K +++
Sbjct: 280 DGKKAAKQKLRQGRTIEAVDYLMEKKKEADVVRELKKEEMCKKAFALQEERCKRAFALQE 339
Query: 340 QKNKLLQEKTQAKK 353
++NKL +EK + +K
Sbjct: 340 ERNKLEREKFEFQK 353
>UniRef100_Q8L727 Glutathione transferase [Arabidopsis thaliana]
Length = 591
Score = 85.5 bits (210), Expect = 2e-15
Identities = 74/258 (28%), Positives = 111/258 (42%), Gaps = 43/258 (16%)
Query: 130 EDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCS--VVGRNQTSESYWGKIAEYCNEHCS 187
+DTT + R ++ W+ +++LIS W+ VV Q + ++W +I + + S
Sbjct: 261 QDTTDRKARRRK--WSPPDDVILISAWLNTSKDRKVVVYDEQQAHTFWKRIGAHVSNSAS 318
Query: 188 FDS--PRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNV 245
+ R+ CR R+ +N + K+VG YD A + SG SEDDVF+ A +LY
Sbjct: 319 LANLPKREWNHCRQRWRKINDYVCKFVGCYDQALNQRASGQSEDDVFQVAYQLYYNNYMS 378
Query: 246 QFTLMEEWRALRDQPRYGSQVGGNVDSGSSGSKRSYEDSVG---SSARP----------- 291
F L WR LR ++ S G SKR+ + G SS P
Sbjct: 379 NFKLEHAWRELRHNKKWCSTYTSENSKGGGSSKRTKLNGGGVYSSSCNPESVPIALDGEE 438
Query: 292 --MDRDAAKKKGKKKSK------------GETLEKVEKEWVQFKELKEQEIEQLKELTL- 336
MDR K K+K K ++ ++E WV +E EQ+ +L L
Sbjct: 439 QVMDRPLGVKSSKQKEKKVATKTMLEEREADSRSRLENLWVLDEE------EQVMDLPLG 492
Query: 337 VKQQKNKLLQEKTQAKKM 354
VK K K + K K M
Sbjct: 493 VKSSKQK--ERKVATKTM 508
>UniRef100_Q9ZQ43 Hypothetical protein At2g22710 [Arabidopsis thaliana]
Length = 300
Score = 83.6 bits (205), Expect = 9e-15
Identities = 66/222 (29%), Positives = 102/222 (45%), Gaps = 29/222 (13%)
Query: 144 WNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDS--PRDIVACRNRF 201
W ++++VLIS W+ VVG Q + ++W +IA Y D R C+ R+
Sbjct: 63 WAAKEDIVLISAWLNTSKDPVVGNEQKAPAFWKRIASYVAASPDLDGVPKRASAQCKQRW 122
Query: 202 NYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQFTLMEEWRALRDQPR 261
MN+L+ K+VG Y + + SG +E+DV A EL+ +F+L WR LR +
Sbjct: 123 AKMNELVMKFVGCYATTTNQKASGQTENDVMLFANELFKNDMKKKFSLDHAWRLLRHDQK 182
Query: 262 YGSQVGGNVDSGSSGSKRSYEDSVGSSA---------------RPMDRDAAKKKGKKKS- 305
+ + N G +KR + VG+ A RP AAK K K
Sbjct: 183 W---LISNAPKGKGIAKRR-KVRVGTQAASSQPIDLEDDDVMRRPPGVKAAKAKAKMTPT 238
Query: 306 -KGETLEKVEKEWVQFKELKEQEIEQLKELTLVKQQKNKLLQ 346
KGE + VE +LKE++ ++ KQ KN++L+
Sbjct: 239 VKGEEVNSVE-HLTSLWDLKEKDWDRKD-----KQSKNQMLE 274
>UniRef100_Q9LHC6 Gb|AAF34841.1 [Arabidopsis thaliana]
Length = 211
Score = 83.2 bits (204), Expect = 1e-14
Identities = 53/190 (27%), Positives = 92/190 (47%), Gaps = 8/190 (4%)
Query: 71 NNFNPYFRSMMGYSSQTPPFNGYMPMVNENFQSVGEYPDFSSQI-NRGGMTRDNEVAPTP 129
++ NP+++S S + +N E P F++Q+ G + + T
Sbjct: 2 DDMNPFYQS----SGLIDLLHSQQEDINRFASGSSEVPAFNTQMFEEGSKDKVTLMGCTQ 57
Query: 130 EDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFD 189
++ PK +Q+ W ++++VL+ W+ V+G +Q +S+ +IA Y + D
Sbjct: 58 DEVKPKVGLSQK-KWFPKEDIVLVRAWLNTNKDHVIGNDQQCQSFLKQIASYVDISPQLD 116
Query: 190 S--PRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQF 247
R+ C+ R++ +NK + K+VG Y +A + SG SEDDV K A E+Y +F
Sbjct: 117 DLPKREHAKCKQRWSKVNKSVTKFVGCYKTATTHKTSGQSEDDVMKLAYEIYYNDTKKKF 176
Query: 248 TLMEEWRALR 257
L WR LR
Sbjct: 177 NLEHTWRKLR 186
>UniRef100_Q7XER9 Hypothetical protein [Oryza sativa]
Length = 300
Score = 82.4 bits (202), Expect = 2e-14
Identities = 69/247 (27%), Positives = 108/247 (42%), Gaps = 15/247 (6%)
Query: 138 RNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDS-PRDIVA 196
R + +W E+NL L+S WI Y T V G ++ SE YW +A+ N + R +
Sbjct: 61 RRTRINWTEEENLHLLSYWIHYSTDPVKGIDRKSEFYWKAVADVFNTSAPTNGHKRSVKQ 120
Query: 197 CRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGC-GKNVQFTLMEEWRA 255
+ + + + I K+ G Y K SG S+D V A ++ K+ FTL WR
Sbjct: 121 LKTHWGDVKRDITKFCGVYGRLKATWKSGQSDDMVMNSAHAIFEKENKDKPFTLEYMWRE 180
Query: 256 LRDQPRYGSQVGGNVDSGSSGSKRSYEDSVGSSARPMDRDAAKKKGKKKSKGETLEKVEK 315
++D P++ + SG+KR+ E+++ RP + AK K K K +
Sbjct: 181 VKDLPKW-----CRIVQEDSGNKRTKEETISKEKRPDGQKKAKAGLKGKGKDAAPSPLGN 235
Query: 316 EWVQFKELKEQEIEQLKELTLVKQQKNKLLQ------EKTQAKKMKMYLKLRDEEHLDDR 369
+ Q + E LK ++K K K Q EK + + + K R E LD
Sbjct: 236 QPSQ-NMILYHEAMSLKATAMIKSAKEKKYQTYLKLLEKDTSNFSEAHHK-RHEGVLDQL 293
Query: 370 KNELLGK 376
EL G+
Sbjct: 294 AKELAGE 300
>UniRef100_Q6K3A7 Cytokinin inducibl protein-like [Oryza sativa]
Length = 320
Score = 79.7 bits (195), Expect = 1e-13
Identities = 60/247 (24%), Positives = 111/247 (44%), Gaps = 37/247 (14%)
Query: 135 KSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDSPRDI 194
K + + ++N ++++ L W + ++G Q ++YW +IAE+ + + F+S R+
Sbjct: 27 KGRAKRSSNFNRKEDIQLCISWQSISSDPIIGNEQPGKAYWQRIAEHYHANRDFESDRNT 86
Query: 195 VACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCG--KNVQFTLMEE 252
+ +R+ + K ++K+ G Y+ +R SG ++ +A+ LY KN F
Sbjct: 87 FSLEHRWGNIQKEVSKFQGCYEQIERRHPSGIPHQELVLEAEALYSSNAPKNRAFQFNHC 146
Query: 253 WRALRDQPRYGS-----------------QVGGNVDSGSSGSKRSYEDSVGSSA--RPMD 293
W LR+ P++ + G + G K + D SA RPM
Sbjct: 147 WLKLRNSPKFQTLKSHKRPRSRKSSTPIESAGEEDEEGDDARKSTTPDLSQPSAKKRPMG 206
Query: 294 RDAAKKKGK--------KKSKGETLEKVEKE-------WVQFKELKEQEIEQLKELTLVK 338
R AK+K K K++ + L+ EKE W + KE++E ++ E LV
Sbjct: 207 RKEAKEKLKNGGEDGPYKEAMKDLLDAKEKEAKVKEERWKETKEIQEHKL-LFAERKLVW 265
Query: 339 QQKNKLL 345
Q+ K++
Sbjct: 266 DQEQKIM 272
>UniRef100_Q7XEN8 Putative NAM-like protein [Oryza sativa]
Length = 370
Score = 79.7 bits (195), Expect = 1e-13
Identities = 63/273 (23%), Positives = 120/273 (43%), Gaps = 25/273 (9%)
Query: 99 ENFQSVGEYPDFSSQINRGGMTRDNEVAPTPEDTTPKSKRNQQPSWNTEQNLVLISGWIK 158
EN+ +P Q M + A + P +KR++ ++NT ++ +L+S W+
Sbjct: 57 ENWDPSQNFPPTGQQPQHSPMVGEQPSAEVGGSSRPNNKRSK--NFNTAEDQMLVSAWLN 114
Query: 159 YGTCSVVGRNQTSESYWGKIAEYCNEHCSFDSPRDIVACRNRFNYMNKLINKWVGAYDSA 218
++ G Q +SYW +I +Y +E+ +F S ++ + N + + + +NK+ G Y+
Sbjct: 115 TTLDAITGVEQHRDSYWARIHQYFHENKNFPSDQNQNSLNNCWGTIQEQVNKFCGYYEQI 174
Query: 219 KRMQGSGWSEDDVFKKAQELYGCGKNVQFTLMEEWRALRDQPRYGSQV-----GGNVD-- 271
SG D +A LY + F+LM W L P++ ++ +VD
Sbjct: 175 LNGPQSGMVVQDYITQAFTLYKSAEKKTFSLMHCWEILHHHPKWNDRLFQKRQKAHVDPL 234
Query: 272 ---SGSSGSKRSYEDSVGSSARPMDRDAAKKKGKKK-SKGET----------LEKVEKEW 317
S S+ S + +++ P+ R KK K K +G T + + W
Sbjct: 235 VRPSASTNSSEFHYSPDINTSDPLVRPPGKKVEKAKCPRGNTSSCSSESSPVVIALNNMW 294
Query: 318 VQFKELKEQEIEQLKE--LTLVKQQKNKLLQEK 348
+ KE+ Q E+ + + ++ +K +L EK
Sbjct: 295 SEKKEMSAQSREERNDRLVEVISLEKERLKVEK 327
>UniRef100_Q7XDX1 Hypothetical protein [Oryza sativa]
Length = 348
Score = 79.7 bits (195), Expect = 1e-13
Identities = 52/246 (21%), Positives = 114/246 (46%), Gaps = 16/246 (6%)
Query: 143 SWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDSPRDIVACRNRFN 202
+W ++++L L+S W+ + + G + +ESYWG + E N + R++ ++R+
Sbjct: 86 TWASDEDLRLVSAWLYHSNDPINGNGKKNESYWGDVVELYNSTTPINRKREVKHLKDRWQ 145
Query: 203 YMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELY--GCGKNVQFTLMEEWRALRDQP 260
+ + + + ++ A + SG+S+D + A + Y FT++ W+ LRD+P
Sbjct: 146 RIKRWVGFFCASWKKAALVYTSGYSDDQLRDFANQFYVDDYPNEGPFTVLHCWKVLRDEP 205
Query: 261 RYGSQV----GGNVDSGSSGSKRSYEDSVGSSARPMDRDAAKK------KGKKKSKGETL 310
++ + + + S GS + +G RP+ R+ AKK KGK K ++L
Sbjct: 206 KWHAVLEELEKPHKRSLDDGSDTLSQKDIGEKERPIGRNEAKKQRNGKGKGKGKDDDDSL 265
Query: 311 EKVEKEWVQFKELKEQEIEQL----KELTLVKQQKNKLLQEKTQAKKMKMYLKLRDEEHL 366
+ K+++ + + E+ ++ K + +L +E + + + + +
Sbjct: 266 REDMKKYMDVQAAASKRHEEFLGTQHRISDAKVEVARLRREAVLTESYQKMMSMDTSQMT 325
Query: 367 DDRKNE 372
D+ K E
Sbjct: 326 DEMKAE 331
>UniRef100_Q7G6L8 Hypothetical protein OSJNBa0032N04.18 [Oryza sativa]
Length = 330
Score = 79.3 bits (194), Expect = 2e-13
Identities = 61/246 (24%), Positives = 111/246 (44%), Gaps = 36/246 (14%)
Query: 135 KSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDSPRDI 194
K + ++ ++++ L W + ++G Q ++YW +IAE+ + + F S R+
Sbjct: 35 KGSAKRSSNYTRKEDIQLCISWQSISSDPIIGNEQPGKAYWQRIAEHYHANHDFKSDRNA 94
Query: 195 VACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYG--CGKNVQFTLMEE 252
+ +R+ + K ++K+ G Y+ +R SG ++ +A+ LY KN F
Sbjct: 95 NSLEHRWGNIQKEVSKFQGCYNQTERRHPSGIPHQELVLEAEALYSSTAPKNRAFQFNHC 154
Query: 253 WRALRDQPRYGS----------------QVGGNVDSGSSGSKRSYEDSVGSSA--RPMDR 294
W LR+ P++ + + G D G SK + D SA RP+DR
Sbjct: 155 WLKLRNSPKFQTLESHKRPRSRKSSTPIERAGEEDEGDDASKSTAPDLSQPSAKKRPIDR 214
Query: 295 DAAKKKGK--------KKSKGETLEKVEKE-------WVQFKELKEQEIEQLKELTLVKQ 339
AK+K K K++ + L+ EKE W KE++E+++ E LV
Sbjct: 215 KQAKEKLKNGGEDGPYKEAMKDLLDAKEKEAKLKEERWKGTKEIQERKL-LFAERKLVWD 273
Query: 340 QKNKLL 345
Q+ K++
Sbjct: 274 QEQKIM 279
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.311 0.129 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 734,890,303
Number of Sequences: 2790947
Number of extensions: 34504739
Number of successful extensions: 174124
Number of sequences better than 10.0: 1974
Number of HSP's better than 10.0 without gapping: 395
Number of HSP's successfully gapped in prelim test: 1712
Number of HSP's that attempted gapping in prelim test: 153861
Number of HSP's gapped (non-prelim): 11409
length of query: 384
length of database: 848,049,833
effective HSP length: 129
effective length of query: 255
effective length of database: 488,017,670
effective search space: 124444505850
effective search space used: 124444505850
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC149209.7


