

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149207.4 - phase: 0
(168 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
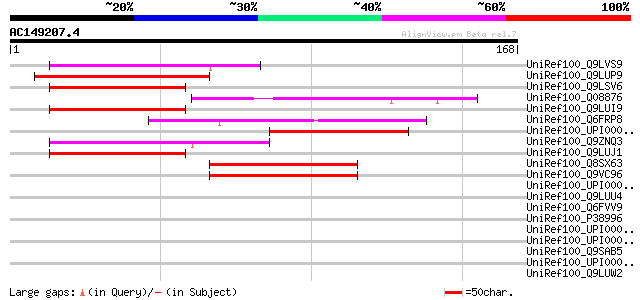
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LVS9 Emb|CAB62106.1 [Arabidopsis thaliana] 49 4e-05
UniRef100_Q9LUP9 Gb|AAB60771.1 [Arabidopsis thaliana] 47 1e-04
UniRef100_Q9LSV6 Emb|CAB62106.1 [Arabidopsis thaliana] 47 2e-04
UniRef100_Q08876 Suppressor of hairy wing protein [Drosophila vi... 47 2e-04
UniRef100_Q9LUI9 Gb|AAD25583.1 [Arabidopsis thaliana] 46 3e-04
UniRef100_Q6FRP8 Similar to sp|P11747 Saccharomyces cerevisiae Y... 46 4e-04
UniRef100_UPI000042E91A UPI000042E91A UniRef100 entry 45 5e-04
UniRef100_Q9ZNQ3 Hypothetical protein At2g27520 [Arabidopsis tha... 45 7e-04
UniRef100_Q9LUJ1 Gb|AAD25583.1 [Arabidopsis thaliana] 45 7e-04
UniRef100_Q8SX63 LD40380p [Drosophila melanogaster] 45 7e-04
UniRef100_Q9VC96 CG31132-PA [Drosophila melanogaster] 45 7e-04
UniRef100_UPI00003C15E6 UPI00003C15E6 UniRef100 entry 45 0.001
UniRef100_Q9LUU4 Gb|AAD25686.1 [Arabidopsis thaliana] 45 0.001
UniRef100_Q6FVV9 Candida glabrata strain CBS138 chromosome D com... 45 0.001
UniRef100_P38996 Nuclear polyadenylated RNA-binding protein 3 [S... 45 0.001
UniRef100_UPI000023F348 UPI000023F348 UniRef100 entry 44 0.001
UniRef100_UPI000021B71D UPI000021B71D UniRef100 entry 44 0.001
UniRef100_Q9SAB5 F25C20.23 [Arabidopsis thaliana] 44 0.001
UniRef100_UPI0000318879 UPI0000318879 UniRef100 entry 44 0.002
UniRef100_Q9LUW2 Gb|AAD25583.1 [Arabidopsis thaliana] 44 0.002
>UniRef100_Q9LVS9 Emb|CAB62106.1 [Arabidopsis thaliana]
Length = 416
Score = 49.3 bits (116), Expect = 4e-05
Identities = 29/71 (40%), Positives = 41/71 (56%), Gaps = 1/71 (1%)
Query: 14 LPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLADITDL-LSTATEH 72
LP +L+ EIL RV + + QLRS CK W +L + +F +KHL K+ D +L LS +
Sbjct: 46 LPGDLLEEILCRVPATSLKQLRSTCKQWNNLFNNGRFTRKHLDKAPKDFQNLMLSDSRVF 105
Query: 73 RKSFVSHQIPN 83
S H IP+
Sbjct: 106 SMSVSFHGIPS 116
>UniRef100_Q9LUP9 Gb|AAB60771.1 [Arabidopsis thaliana]
Length = 374
Score = 47.4 bits (111), Expect = 1e-04
Identities = 20/58 (34%), Positives = 39/58 (66%)
Query: 9 SPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLADITDLL 66
SP ++L +L+ +IL+RV + + ++LRS CK W +++ D +F++KH + + D+L
Sbjct: 7 SPMSVLTEDLVEDILSRVPATSLVRLRSTCKQWNAILNDRRFIKKHFDTAEKEYLDML 64
>UniRef100_Q9LSV6 Emb|CAB62106.1 [Arabidopsis thaliana]
Length = 361
Score = 47.0 bits (110), Expect = 2e-04
Identities = 21/45 (46%), Positives = 29/45 (63%)
Query: 14 LPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKS 58
LP +L++EIL RV + + +LRS CK W L D +F KH HK+
Sbjct: 6 LPEDLLVEILCRVPATSLKRLRSTCKLWNHLYNDKRFKSKHCHKA 50
>UniRef100_Q08876 Suppressor of hairy wing protein [Drosophila virilis]
Length = 899
Score = 46.6 bits (109), Expect = 2e-04
Identities = 35/106 (33%), Positives = 54/106 (50%), Gaps = 17/106 (16%)
Query: 61 DITDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELD 120
D +L + A E+ + FV + +E++DDDDDD+DE V E A +R+ N L E+
Sbjct: 163 DEVELGAGAKENGEEFVVSGV------DEDDDDDDDDEDEGVVEGGAKRRSGNNELKEMV 216
Query: 121 NLLVG--------VRSIKRELEIIKVDT---LAMKDRMKCLESFLK 155
+ G V+S+K+ LE + D+ L D +K LE K
Sbjct: 217 EHVCGKCYKTFRRVKSLKKHLEFCRYDSGYHLRKADMLKNLEKIEK 262
>UniRef100_Q9LUI9 Gb|AAD25583.1 [Arabidopsis thaliana]
Length = 379
Score = 46.2 bits (108), Expect = 3e-04
Identities = 19/45 (42%), Positives = 33/45 (73%)
Query: 14 LPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKS 58
LP++L+ +IL+RV + + +LRS C+ W +L+ D +F +KH HK+
Sbjct: 5 LPLDLVEKILSRVPATSLKRLRSTCRRWNALLKDRRFTEKHFHKA 49
>UniRef100_Q6FRP8 Similar to sp|P11747 Saccharomyces cerevisiae YML098w FUN81
[Candida glabrata]
Length = 163
Score = 45.8 bits (107), Expect = 4e-04
Identities = 32/95 (33%), Positives = 50/95 (51%), Gaps = 4/95 (4%)
Query: 47 DDQFVQKHLHKSLADITDLLST---ATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVD 103
D +FV + LA +L+ST TE +K F ++ N + +E+EDDDDD+DD+E D
Sbjct: 68 DFKFVLRKDPIKLARAEELISTNALITEAKKQFNENENQNLKRYKEDEDDDDDEDDDE-D 126
Query: 104 EEEAAKRARMNRLAELDNLLVGVRSIKRELEIIKV 138
++E +K E V V+ K+ + KV
Sbjct: 127 DDEDSKPKEAPVTVETQESTVPVKEKKKRTKKAKV 161
>UniRef100_UPI000042E91A UPI000042E91A UniRef100 entry
Length = 219
Score = 45.4 bits (106), Expect = 5e-04
Identities = 21/46 (45%), Positives = 30/46 (64%)
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLLVGVRSIKRE 132
EEEEEDDDDDDD ++ EEE+ +AR + L+E L +K++
Sbjct: 44 EEEEEDDDDDDDSDDAPEEESTSKARSDILSEQKRLQQVEAELKKQ 89
>UniRef100_Q9ZNQ3 Hypothetical protein At2g27520 [Arabidopsis thaliana]
Length = 347
Score = 45.1 bits (105), Expect = 7e-04
Identities = 25/74 (33%), Positives = 42/74 (55%), Gaps = 1/74 (1%)
Query: 14 LPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSL-ADITDLLSTATEH 72
LP +L+ EIL+R+ + + +LR CK W +L D +F+ K HK+ D+ +LS +
Sbjct: 6 LPWDLVDEILSRLPATSLGRLRFTCKRWNALFKDPEFITKQFHKAAKQDLVLMLSNFGVY 65
Query: 73 RKSFVSHQIPNQLQ 86
S +IPN ++
Sbjct: 66 SMSTNLKEIPNNIE 79
>UniRef100_Q9LUJ1 Gb|AAD25583.1 [Arabidopsis thaliana]
Length = 339
Score = 45.1 bits (105), Expect = 7e-04
Identities = 19/45 (42%), Positives = 32/45 (70%)
Query: 14 LPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKS 58
LP++L+ EIL+RV + + +LRS C+ W +L+ D +F +KH K+
Sbjct: 5 LPLDLVEEILSRVPATSLKRLRSTCRQWNALLKDRRFTEKHFRKA 49
>UniRef100_Q8SX63 LD40380p [Drosophila melanogaster]
Length = 1765
Score = 45.1 bits (105), Expect = 7e-04
Identities = 19/49 (38%), Positives = 31/49 (62%)
Query: 67 STATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRARMNR 115
ST + + + ++P E+E+++DDDDDD++ DEEEA+ R R R
Sbjct: 1048 STRRNQKPTAAARRLPVANSSEDEDEEDDDDDDDDDDEEEASTRGRSRR 1096
>UniRef100_Q9VC96 CG31132-PA [Drosophila melanogaster]
Length = 2232
Score = 45.1 bits (105), Expect = 7e-04
Identities = 19/49 (38%), Positives = 31/49 (62%)
Query: 67 STATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRARMNR 115
ST + + + ++P E+E+++DDDDDD++ DEEEA+ R R R
Sbjct: 1515 STRRNQKPTAAARRLPVANSSEDEDEEDDDDDDDDDDEEEASTRGRSRR 1563
>UniRef100_UPI00003C15E6 UPI00003C15E6 UniRef100 entry
Length = 161
Score = 44.7 bits (104), Expect = 0.001
Identities = 24/57 (42%), Positives = 35/57 (61%), Gaps = 9/57 (15%)
Query: 84 QLQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLLVGVRSIKRELEIIKVDT 140
Q EE+++DDDDDDDD+E +EEE + + LAEL I+ E E+ ++DT
Sbjct: 63 QRSEEDDDDDDDDDDDDEEEEEEDDYNIQDDELAEL---------IEDEEELAEIDT 110
>UniRef100_Q9LUU4 Gb|AAD25686.1 [Arabidopsis thaliana]
Length = 135
Score = 44.7 bits (104), Expect = 0.001
Identities = 37/115 (32%), Positives = 57/115 (49%), Gaps = 7/115 (6%)
Query: 11 TTI--LPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLADITDLLST 68
TTI LP EL EIL+RV + + + +++CK W +L D +FV+K+L KS ++ LL
Sbjct: 3 TTIPNLPDELESEILSRVPAKSLAKWKTICKRWYALFRDPRFVKKNLCKSAREVM-LLMN 61
Query: 69 ATEHRKSFVSHQIPNQLQEEEEEDDD----DDDDDEEVDEEEAAKRARMNRLAEL 119
H S H I ++ + E D D +V +E R+ A+L
Sbjct: 62 DRVHSISVNRHGIDDRFEPSMEFSGKLRSLIDSKDVKVSKEANLARSLNTMYAKL 116
>UniRef100_Q6FVV9 Candida glabrata strain CBS138 chromosome D complete sequence
[Candida glabrata]
Length = 123
Score = 44.7 bits (104), Expect = 0.001
Identities = 34/115 (29%), Positives = 53/115 (45%), Gaps = 11/115 (9%)
Query: 59 LADITDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAE 118
++ ITDLL E V+ E+EEE+D+D+D+DE+ DEEE + R E
Sbjct: 2 ISSITDLLEELKETIVPVVAEAAEEDPVEDEEEEDEDEDEDEDEDEEEGVDQLDALR-EE 60
Query: 119 LDNLLVGVRSIKRELEIIKVDTLAMKD--------RMKCLESF--LKIYLKSATS 163
N G + +E ++ T +D + C+E F L+ YL S +
Sbjct: 61 CKNTEEGKHLVHHYMECVERVTKQQEDPEYENLDYKEDCVEEFFHLQHYLDSCAA 115
>UniRef100_P38996 Nuclear polyadenylated RNA-binding protein 3 [Saccharomyces
cerevisiae]
Length = 802
Score = 44.7 bits (104), Expect = 0.001
Identities = 30/73 (41%), Positives = 38/73 (51%), Gaps = 8/73 (10%)
Query: 83 NQLQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLLVGVRSIKRELEIIKVDTLA 142
N+ +EEEEEDDDDDDDD++ DEEE + DN VG S + E D
Sbjct: 107 NEEEEEEEEDDDDDDDDDDDDEEEEEEEEEEGN----DNSSVGSDSAAEDGE----DEED 158
Query: 143 MKDRMKCLESFLK 155
KD+ K E L+
Sbjct: 159 KKDKTKDKEVELR 171
Score = 37.7 bits (86), Expect = 0.11
Identities = 15/36 (41%), Positives = 26/36 (71%)
Query: 71 EHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEE 106
EH++ + + +EEEEE++DDDDDD++ D++E
Sbjct: 93 EHQQKGGNDDDDDDNEEEEEEEEDDDDDDDDDDDDE 128
Score = 32.0 bits (71), Expect = 6.2
Identities = 16/32 (50%), Positives = 21/32 (65%), Gaps = 5/32 (15%)
Query: 80 QIPNQLQEEEEED-----DDDDDDDEEVDEEE 106
++ N +EEEEE +DDDDDD E +EEE
Sbjct: 82 EVINTEEEEEEEHQQKGGNDDDDDDNEEEEEE 113
>UniRef100_UPI000023F348 UPI000023F348 UniRef100 entry
Length = 1168
Score = 44.3 bits (103), Expect = 0.001
Identities = 24/78 (30%), Positives = 43/78 (54%), Gaps = 2/78 (2%)
Query: 42 KSLVVDDQFVQK--HLHKSLADITDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDD 99
K +VV D V+ L +I D + T ++ ++ EE+ EDDD+DDDD
Sbjct: 1071 KQIVVPDTIVRPIVPLRPQSEEINDSMEDVTLTKEPVDEYKGLRSQLEEDNEDDDNDDDD 1130
Query: 100 EEVDEEEAAKRARMNRLA 117
++ D+++ R+R++R+A
Sbjct: 1131 DDDDDQDVVVRSRIDRVA 1148
>UniRef100_UPI000021B71D UPI000021B71D UniRef100 entry
Length = 1357
Score = 44.3 bits (103), Expect = 0.001
Identities = 19/32 (59%), Positives = 24/32 (74%)
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRARMNRLAE 118
EEEE+D+DDDD+DEE DEEE R + RL +
Sbjct: 60 EEEEDDEDDDDEDEEDDEEEEEPRLKYTRLTQ 91
>UniRef100_Q9SAB5 F25C20.23 [Arabidopsis thaliana]
Length = 363
Score = 44.3 bits (103), Expect = 0.001
Identities = 20/45 (44%), Positives = 32/45 (70%)
Query: 11 TTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHL 55
T L +L+ EIL+RV + + ++LRS CK W++L+ + +FV KHL
Sbjct: 3 TMDLSSDLVEEILSRVPARSLVRLRSTCKQWEALIAEPRFVNKHL 47
>UniRef100_UPI0000318879 UPI0000318879 UniRef100 entry
Length = 1439
Score = 43.9 bits (102), Expect = 0.002
Identities = 36/134 (26%), Positives = 64/134 (46%), Gaps = 26/134 (19%)
Query: 16 VELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLADITDLLS-------- 67
VE+M++IL + S +R VC+ + + D +LA I D+L
Sbjct: 706 VEVMVDILLSLLSRPSRHIRQVCRTVFASICADV-----TPAALAAILDVLDPEQDDEES 760
Query: 68 ---TATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEV---DEEEAAK-------RARMN 114
T+ + + +++EEE+E++D+D+DDE + DEEE + R +
Sbjct: 761 SAVVVTDDNGTLGKGEEDEEMEEEEDEEEDEDEDDEALEDDDEEELEEEPVDQNFRLELM 820
Query: 115 RLAELDNLLVGVRS 128
++ + N LVGV+S
Sbjct: 821 KVLQQRNALVGVKS 834
>UniRef100_Q9LUW2 Gb|AAD25583.1 [Arabidopsis thaliana]
Length = 380
Score = 43.9 bits (102), Expect = 0.002
Identities = 20/43 (46%), Positives = 28/43 (64%)
Query: 14 LPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLH 56
LP++L+ EIL+RV + + +LRS CK W +L F QKH H
Sbjct: 6 LPLDLVEEILSRVSATSLKRLRSTCKQWNTLFKKRSFSQKHFH 48
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.129 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 252,966,297
Number of Sequences: 2790947
Number of extensions: 10393255
Number of successful extensions: 331235
Number of sequences better than 10.0: 4598
Number of HSP's better than 10.0 without gapping: 3631
Number of HSP's successfully gapped in prelim test: 1116
Number of HSP's that attempted gapping in prelim test: 233339
Number of HSP's gapped (non-prelim): 40619
length of query: 168
length of database: 848,049,833
effective HSP length: 118
effective length of query: 50
effective length of database: 518,718,087
effective search space: 25935904350
effective search space used: 25935904350
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 70 (31.6 bits)
Medicago: description of AC149207.4


