

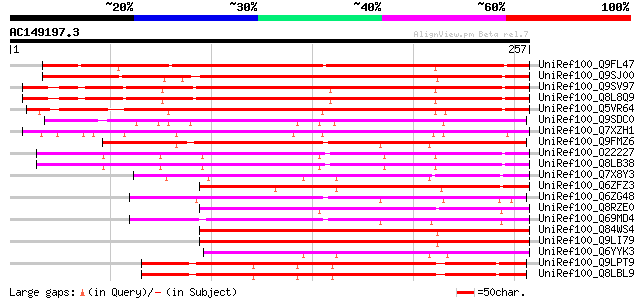
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149197.3 - phase: 0
(257 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FL47 Gb|AAD20416.1 [Arabidopsis thaliana] 280 3e-74
UniRef100_Q9SJ00 Hypothetical protein At2g21990 [Arabidopsis tha... 231 1e-59
UniRef100_Q9SV97 Hypothetical protein AT4g39610 [Arabidopsis tha... 226 4e-58
UniRef100_Q8L8Q9 Hypothetical protein [Arabidopsis thaliana] 224 1e-57
UniRef100_Q5VR64 Hypothetical protein P0702B09.7 [Oryza sativa] 212 7e-54
UniRef100_Q9SDC0 Similar to Arabidopsis thaliana chromosome II B... 184 2e-45
UniRef100_Q7XZH1 Hypothetical protein OSJNBb0033J23.11 [Oryza sa... 177 3e-43
UniRef100_Q9FMZ6 Arabidopsis thaliana genomic DNA, chromosome 5,... 161 2e-38
UniRef100_O22227 Expressed protein [Arabidopsis thaliana] 159 9e-38
UniRef100_Q8LB38 Hypothetical protein [Arabidopsis thaliana] 158 1e-37
UniRef100_Q7X8Y3 OSJNBb0034I13.25 protein [Oryza sativa] 156 4e-37
UniRef100_Q6ZFZ3 Hypothetical protein OJ1311_H06.6-2 [Oryza sativa] 152 6e-36
UniRef100_Q6ZG48 Hypothetical protein OJ1111_E05.14 [Oryza sativa] 149 9e-35
UniRef100_Q8RZE0 Hypothetical protein P0510C12.17 [Oryza sativa] 147 2e-34
UniRef100_Q69MD4 Hypothetical protein OSJNBb0019B14.18-1 [Oryza ... 147 3e-34
UniRef100_Q84WS4 Hypothetical protein At3g25640 [Arabidopsis tha... 144 2e-33
UniRef100_Q9LI79 Gb|AAB84348.1 [Arabidopsis thaliana] 144 2e-33
UniRef100_Q6YYK3 Hypothetical protein OJ1590_E05.28 [Oryza sativa] 142 8e-33
UniRef100_Q9LPT9 T22I11.13 [Arabidopsis thaliana] 142 1e-32
UniRef100_Q8LBL9 Hypothetical protein [Arabidopsis thaliana] 142 1e-32
>UniRef100_Q9FL47 Gb|AAD20416.1 [Arabidopsis thaliana]
Length = 261
Score = 280 bits (715), Expect = 3e-74
Identities = 151/254 (59%), Positives = 182/254 (71%), Gaps = 17/254 (6%)
Query: 17 SPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQ--NSTNKLFGKFRSMFRSFPIIVP 74
+PTT P SP S + TVR I L+ + ++K + + KLF + RS+FRS PI+ P
Sbjct: 12 TPTTMSPLGSPKSKKSTA-TVRPEITLEQPSGRNKTTGSKSTKLFRRVRSVFRSLPIMSP 70
Query: 75 SCKMPTMNGNHRTSETIIHGGIRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGK 134
CK P + G R E +HGG R+TGTLFGYRK RVNLA QE+ + P LLLELAIPTGK
Sbjct: 71 MCKFP-VGGGGRLHENHVHGGTRVTGTLFGYRKTRVNLAVQENPRSLPILLLELAIPTGK 129
Query: 135 LLQDMGMGLNRIALECEKHSSNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQM 194
LLQD+G+GL RIALECEK S +KTKI+DEPIW L+CNGKK GYGVKR PT++DL V+QM
Sbjct: 130 LLQDLGVGLVRIALECEKKPS-EKTKIIDEPIWALYCNGKKSGYGVKRQPTEEDLVVMQM 188
Query: 195 LHSVSVAVGELPSDM-----------SDPQDGELSYMRAHFERVIGSKDSETYYMMMPDG 243
LH+VS+ G LP Q+G+L+YMRAHFERVIGS+DSETYYMM PDG
Sbjct: 189 LHAVSMGAGVLPVSSGAITEQSGGGGGGQQEGDLTYMRAHFERVIGSRDSETYYMMNPDG 248
Query: 244 NSNGPELSVFFVRV 257
NS GPELS+FFVRV
Sbjct: 249 NS-GPELSIFFVRV 261
>UniRef100_Q9SJ00 Hypothetical protein At2g21990 [Arabidopsis thaliana]
Length = 252
Score = 231 bits (589), Expect = 1e-59
Identities = 124/248 (50%), Positives = 169/248 (68%), Gaps = 13/248 (5%)
Query: 17 SPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQNSTNKLFGKFRSMFRSFPIIVPS- 75
S T PSP++ + P R + L + + K+ S K+F FRS+FRSFPII P+
Sbjct: 11 SSTDSYSTPSPSASPSPSPAPRQHVTLLEPSHQHKKKS-KKVFRVFRSVFRSFPIITPAA 69
Query: 76 CKMPTMNGN-----HRTSETIIHGGIRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAI 130
CK+P + G HR+ + G R+TGTLFGYRK RV+L+ QE +C P L++ELA+
Sbjct: 70 CKIPVLPGGSLPDQHRSGSS----GSRVTGTLFGYRKGRVSLSIQESPRCLPSLVVELAM 125
Query: 131 PTGKLLQDMGMGLNRIALECEKHSSNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLY 190
T L +++ G+ RIALE EK +K KI+DEP+WT+F NGKK GYGVKRD T++DL
Sbjct: 126 QTMVLQKELSGGMVRIALETEKRGDKEKIKIMDEPLWTMFSNGKKTGYGVKRDATEEDLN 185
Query: 191 VIQMLHSVSVAVGELPSDMS-DPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPE 249
V+++L VS+ G LP + + D E++YMRA+FERV+GSKDSET+YM+ P+GN NGPE
Sbjct: 186 VMELLRPVSMGAGVLPGNTEFEGPDSEMAYMRAYFERVVGSKDSETFYMLSPEGN-NGPE 244
Query: 250 LSVFFVRV 257
LS+FFVRV
Sbjct: 245 LSIFFVRV 252
>UniRef100_Q9SV97 Hypothetical protein AT4g39610 [Arabidopsis thaliana]
Length = 264
Score = 226 bits (576), Expect = 4e-58
Identities = 130/263 (49%), Positives = 174/263 (65%), Gaps = 23/263 (8%)
Query: 7 SQTTTGQPPPSPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQNSTNKLFGKFRSMF 66
S +++ PP SPT PSP S T P R LQP +SK K+N TN +F R++F
Sbjct: 13 SSSSSSIPPSSPT-----PSPTSTTTPP---RQHPLLQPPSSKKKKNRTN-VFRVLRTVF 63
Query: 67 RSFPIIVP---SCKMPTMNGNHRTSETIIHGGIRITGTLFGYRKARVNLAFQEDSKCHPF 123
RSFPI +CK+P ++ + H RITGTLFGYRK RV+L+ QE+ KC P
Sbjct: 64 RSFPIFTTPSVACKIPVIHPGLGLPDPH-HNTSRITGTLFGYRKGRVSLSIQENPKCLPS 122
Query: 124 LLLELAIPTGKLLQDMGMGLNRIALECEKHSSND--------KTKIVDEPIWTLFCNGKK 175
L++ELA+ T L +++ G+ RIALE EK D KT I++EP+WT++C G+K
Sbjct: 123 LVVELAMQTTTLQKELSTGMVRIALETEKQPRADNNNSKTEKKTDILEEPLWTMYCKGEK 182
Query: 176 MGYGVKRDPTDDDLYVIQMLHSVSVAVGELPSDM-SDPQDGELSYMRAHFERVIGSKDSE 234
GYGVKR+ T++DL V+++L VS+ G LP + S+ DGE++YMRA+FERVIGSKDSE
Sbjct: 183 TGYGVKREATEEDLNVMELLRPVSMGAGVLPGNSESEGPDGEMAYMRAYFERVIGSKDSE 242
Query: 235 TYYMMMPDGNSNGPELSVFFVRV 257
T+YM+ P+GN NGPELS FFVRV
Sbjct: 243 TFYMLSPEGN-NGPELSFFFVRV 264
>UniRef100_Q8L8Q9 Hypothetical protein [Arabidopsis thaliana]
Length = 257
Score = 224 bits (572), Expect = 1e-57
Identities = 129/263 (49%), Positives = 173/263 (65%), Gaps = 23/263 (8%)
Query: 7 SQTTTGQPPPSPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQNSTNKLFGKFRSMF 66
S +++ PP SPT PSP S T P R LQP +SK K+N TN +F R++F
Sbjct: 6 SSSSSSIPPSSPT-----PSPTSTTTPP---RQHPLLQPPSSKKKKNRTN-VFRVLRTVF 56
Query: 67 RSFPIIVP---SCKMPTMNGNHRTSETIIHGGIRITGTLFGYRKARVNLAFQEDSKCHPF 123
RSFPI +CK+P ++ + H RITGTLFGYRK RV+L+ QE+ KC P
Sbjct: 57 RSFPIFTTPSVACKIPVIHPGLGLPDPH-HNTSRITGTLFGYRKGRVSLSIQENPKCLPS 115
Query: 124 LLLELAIPTGKLLQDMGMGLNRIALECEKHSSND--------KTKIVDEPIWTLFCNGKK 175
L++ELA+ T L +++ G+ RIALE EK D KT I++EP+WT++C G+K
Sbjct: 116 LVVELAMQTTTLQKELSTGMVRIALETEKQPRADNNNSKTEKKTDILEEPLWTMYCKGEK 175
Query: 176 MGYGVKRDPTDDDLYVIQMLHSVSVAVGELPSDM-SDPQDGELSYMRAHFERVIGSKDSE 234
GYGVKR+ T++DL V+++L VS+ G LP + S+ DGE++YMRA+FERVIGSKDSE
Sbjct: 176 TGYGVKREATEEDLNVMELLRPVSMGAGVLPGNSESEGPDGEMAYMRAYFERVIGSKDSE 235
Query: 235 TYYMMMPDGNSNGPELSVFFVRV 257
T+YM+ P+GN NGPELS FF RV
Sbjct: 236 TFYMLSPEGN-NGPELSFFFARV 257
>UniRef100_Q5VR64 Hypothetical protein P0702B09.7 [Oryza sativa]
Length = 294
Score = 212 bits (540), Expect = 7e-54
Identities = 121/266 (45%), Positives = 168/266 (62%), Gaps = 29/266 (10%)
Query: 9 TTTGQ-PPPSPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQNSTNKLFGKFRSMFR 67
TT Q PPPSP T P PA T PP+ + A S + R++FR
Sbjct: 41 TTPDQLPPPSPHT----PRPAITLTAPPSKKKRRGGAAARSL-------RAIRAVRALFR 89
Query: 68 SFPIIVPSCK----MPTMNGNHRTSETIIHGGIRITGTLFGYRKARVNLAFQEDSKCHPF 123
S PI+ P+C+ + G R + + G R TGTLFGYRKARV LA QE P
Sbjct: 90 SLPILAPACRFHGAIRAPGGASRAHDGHVSGASRTTGTLFGYRKARVTLAVQETPGSVPI 149
Query: 124 LLLELAIPTGKLLQDMGMGLNRIALECEKH-----SSNDKTKIVDEPIWTLFCNGKKMGY 178
LLLELA+ TG+ +Q+MG R+ALECEK + +T+++DEP+WT + NG+K+GY
Sbjct: 150 LLLELAMQTGRFMQEMGAEHLRVALECEKKPPGAGAGIGRTRLLDEPLWTAYVNGRKIGY 209
Query: 179 GVKRDPTDDDLYVIQMLHSVSVAVGELPSDM------SDPQD-GELSYMRAHFERVIGSK 231
++R+PT+ DL V+Q+L +VSV G LP+D+ ++ QD G+L+YMRA F+RV+GS+
Sbjct: 210 AMRREPTEGDLTVMQLLRTVSVGAGVLPTDVMGGDAGAEVQDAGDLAYMRARFDRVVGSR 269
Query: 232 DSETYYMMMPDGNSNGPELSVFFVRV 257
DSE++YM+ PDGN NGPELS+FF+R+
Sbjct: 270 DSESFYMLNPDGN-NGPELSIFFIRI 294
>UniRef100_Q9SDC0 Similar to Arabidopsis thaliana chromosome II BAC F7D8 genomic
sequence [Oryza sativa]
Length = 588
Score = 184 bits (466), Expect = 2e-45
Identities = 104/273 (38%), Positives = 157/273 (57%), Gaps = 38/273 (13%)
Query: 18 PTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQNSTNKLFGK-------FRSMFRSFP 70
P+T PPP P +T Q K +Q T+K + + R+ FRSFP
Sbjct: 319 PSTPLPPPLPPPTSTAAAAAAAKAKHQ----KQQQQQTSKHWSRPARFVRSVRAAFRSFP 374
Query: 71 II-VPSCK----MPTMNGNHRTS-----ETIIHGGIRITGTLFGYRKARVNLAFQEDSKC 120
I+ PSC+ +P + G H HG R TGTL+G+R+AR+ +AF +
Sbjct: 375 ILPAPSCRGLPSLPHLPGLHHGGAGGAVRNHFHGSTRTTGTLYGHRRARITIAFHDSPGS 434
Query: 121 HPFLLLELAIPTGKLLQDMGM-GLNRIALECEK---------HSSNDKT---KIVDEPIW 167
P LLL++A+PT K +QD+ G+ R+ LEC+K H D +++DEP+W
Sbjct: 435 PPALLLDIAVPTAKFIQDVSAAGMVRVTLECDKQQHQPPPHAHPPGDPLPPRRLLDEPVW 494
Query: 168 TLFCNGKKMGYGVKRDPTDDDLYVIQMLHSVSVAVGELPSDMSDPQ----DGELSYMRAH 223
+ NG+ +GY +R+ T+ D V+++LH++S+ G LP+ +D DGE++YMRAH
Sbjct: 495 SAEVNGESVGYAARREATEADERVMRLLHAMSMGAGVLPAVAADAPTSAADGEVTYMRAH 554
Query: 224 FERVIGSKDSETYYMMMPDGNSNGPELSVFFVR 256
F+RV+GSKD+ETYYM P+G + GPEL++FF+R
Sbjct: 555 FDRVVGSKDAETYYMHNPEGCATGPELTIFFIR 587
>UniRef100_Q7XZH1 Hypothetical protein OSJNBb0033J23.11 [Oryza sativa]
Length = 316
Score = 177 bits (448), Expect = 3e-43
Identities = 114/299 (38%), Positives = 168/299 (56%), Gaps = 48/299 (16%)
Query: 7 SQTTTGQP------PPSPTTGQ-----PPPSPASVNTCPP-TVRMP---INLQPANSKSK 51
S T G P PPSPTT Q P +P PP T R P I L+ +S+++
Sbjct: 18 SPTAGGTPSRLAVAPPSPTTPQCAIPASPHTPGRGRAPPPATPRAPRPEITLRQPSSQAQ 77
Query: 52 QNST----------NKLFGKFRSMFRSFPIIVPSCKMPTM---NGNHRTSETIIHGGIRI 98
Q ++ R++ RS P + P+ P+ ++R + GG R+
Sbjct: 78 QKQKRAPSAAARRPSRALRAIRALLRSLPFVAPAACRPSSALPRRHNRPHDGHAGGGARV 137
Query: 99 TGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDM--GMGLNRIALECEKH--- 153
TGT +G+R+AR+ LA QE P L+LEL +PT KL+Q++ G G R+ALECEK
Sbjct: 138 TGTFYGHRRARITLAVQERPGSLPSLVLELGVPTAKLMQEISTGGGHVRVALECEKRPKK 197
Query: 154 -----SSNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHSVSVAVGELPSD 208
+++E +WT + NG+++GY V+R+ ++ DL V+Q+L +VSV G LP D
Sbjct: 198 LPSAPPEQASVSLLEEAMWTAYVNGRRVGYAVRREASEGDLAVMQLLSTVSVGAGVLPGD 257
Query: 209 -MSDPQ----DGELSYMRAHFERVIGSKDSETYYMMMPDGNS-----NGPELSVFFVRV 257
+++P DGE++YMRA F+RV GSKDSE++YM+ PDG++ G ELS+FFVRV
Sbjct: 258 VLAEPAGAEGDGEVTYMRAGFDRVAGSKDSESFYMVNPDGDAGAGAGGGTELSIFFVRV 316
>UniRef100_Q9FMZ6 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MJB21
[Arabidopsis thaliana]
Length = 238
Score = 161 bits (407), Expect = 2e-38
Identities = 86/214 (40%), Positives = 133/214 (61%), Gaps = 9/214 (4%)
Query: 47 NSKSKQNSTNKLFGKFRSMFRSFPIIVPSCKMPTM-NGNHRTSETIIHGGIRITGTLFGY 105
+SKS + S+ + G MF+ P++ CKM + + HR + TGT+FG+
Sbjct: 28 DSKSSKKSSGSIGGGVLKMFKLIPMLSSGCKMVNLLSRGHRRP---LLKDYATTGTIFGF 84
Query: 106 RKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEKHSSNDKTKIVDEP 165
RK RV LA QED C P ++EL + T L ++M RIALE E +S + K+++E
Sbjct: 85 RKGRVFLAIQEDPHCLPIFIIELPMLTSALQKEMASETVRIALESETKTS--RKKVLEEY 142
Query: 166 IWTLFCNGKKMGYGVKR-DPTDDDLYVIQMLHSVSVAVGELP--SDMSDPQDGELSYMRA 222
+W ++CNG+K+GY ++R + +++++YVI L VS+ G LP + +GE++YMRA
Sbjct: 143 VWGIYCNGRKIGYSIRRKNMSEEEMYVIDALRGVSMGAGVLPCKNQYDQETEGEMTYMRA 202
Query: 223 HFERVIGSKDSETYYMMMPDGNSNGPELSVFFVR 256
F+RVIGSKDSE YM+ P+G+ G ELS++F+R
Sbjct: 203 RFDRVIGSKDSEALYMINPEGSGQGTELSIYFLR 236
>UniRef100_O22227 Expressed protein [Arabidopsis thaliana]
Length = 297
Score = 159 bits (401), Expect = 9e-38
Identities = 104/279 (37%), Positives = 148/279 (52%), Gaps = 38/279 (13%)
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVRMPINLQP---------ANSKSKQNSTNKLFGKFRS 64
P SP SP+S P + L P ++S S ++ K KF
Sbjct: 22 PVTSPARSSHVRSPSSSALIPSIPEHELFLVPCRRCSYVPLSSSSSASHNIGKFHLKFSL 81
Query: 65 MFRSFPIIV--PSCKMPTMNGNHRTSETIIHG------------GIRITGTLFGYRKARV 110
+ RSF I+ P+CKM ++ +S ++ + G R+TGTL+G+++ V
Sbjct: 82 LLRSFINIINIPACKMLSLPSPPSSSSSVSNQLISLVTGGSSSLGRRVTGTLYGHKRGHV 141
Query: 111 NLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEK-HSSNDKTKIVDEPIWTL 169
+ Q + + P LLL+LA+ T L+++M GL RIALECEK H S TK+ EP WT+
Sbjct: 142 TFSVQYNQRSDPVLLLDLAMSTATLVKEMSSGLVRIALECEKRHRSG--TKLFQEPKWTM 199
Query: 170 FCNGKKMGYGVKRDP--TDDDLYVIQMLHSVSVAVGELPSDM---------SDPQDGELS 218
+CNG+K GY V R TD D V+ + V+V G +P+ S + GEL
Sbjct: 200 YCNGRKCGYAVSRGGACTDTDWRVLNTVSRVTVGAGVIPTPKTIDDVSGVGSGTELGELL 259
Query: 219 YMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVRV 257
YMR FERV+GS+DSE +YMM PD N GPELS+F +R+
Sbjct: 260 YMRGKFERVVGSRDSEAFYMMNPDKN-GGPELSIFLLRI 297
>UniRef100_Q8LB38 Hypothetical protein [Arabidopsis thaliana]
Length = 297
Score = 158 bits (399), Expect = 1e-37
Identities = 104/279 (37%), Positives = 147/279 (52%), Gaps = 38/279 (13%)
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVRMPINLQP---------ANSKSKQNSTNKLFGKFRS 64
P SP SP+S P + L P ++S S ++ K KF
Sbjct: 22 PVTSPARSSHVRSPSSSALIPSIPEHELFLVPCRRCSYVPLSSSSSASHNIGKFHLKFSL 81
Query: 65 MFRSFPIIV--PSCKMPTMNGNHRTSETIIHG------------GIRITGTLFGYRKARV 110
+ RSF I+ P+CKM ++ +S ++ + G R+TGTL+G+++ V
Sbjct: 82 LLRSFINIINIPACKMLSLPSPPSSSSSVSNQLISLVTGGSSSLGRRVTGTLYGHKRGHV 141
Query: 111 NLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEK-HSSNDKTKIVDEPIWTL 169
+ Q + P LLL+LA+ T L+++M GL RIALECEK H S TK+ EP WT+
Sbjct: 142 TFSVQYNQSSDPVLLLDLAMSTATLVKEMSSGLVRIALECEKRHRSG--TKLFQEPKWTM 199
Query: 170 FCNGKKMGYGVKRDP--TDDDLYVIQMLHSVSVAVGELPSDM---------SDPQDGELS 218
+CNG+K GY V R TD D V+ + V+V G +P+ S + GEL
Sbjct: 200 YCNGRKCGYAVSRGGACTDTDWRVLNTVSRVTVGAGVIPTPKTIDDVSGVGSGTELGELL 259
Query: 219 YMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVRV 257
YMR FERV+GS+DSE +YMM PD N GPELS+F +R+
Sbjct: 260 YMRGKFERVVGSRDSEAFYMMNPDKN-GGPELSIFLLRI 297
>UniRef100_Q7X8Y3 OSJNBb0034I13.25 protein [Oryza sativa]
Length = 301
Score = 156 bits (395), Expect = 4e-37
Identities = 95/238 (39%), Positives = 138/238 (57%), Gaps = 44/238 (18%)
Query: 62 FRSMFRSFPIIVPSC--------------------KMPTMNGNHRTSETIIHGGIR--IT 99
F +M RS P++ P C M+ ++ T G R +T
Sbjct: 66 FHTMCRSLPVLTPGCGRLQPAGAACRIAAPSRLSPSASLMSKLVASASTGAAGASRRRMT 125
Query: 100 GTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLN-RIALECEKHSSNDK 158
GTLFGYR AR+ L+ Q++ +C P L++ELA+PT LL+D+G RI LE EK +++
Sbjct: 126 GTLFGYRDARIALSLQDNPRCQPTLVVELALPTHALLRDLGTTAGARIVLESEKRAADGG 185
Query: 159 TK-----------------IVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHSVSVA 201
+++E +WT+ CNGKK+GY V+RDPTDDD+ V++ L +VS+
Sbjct: 186 DGAGAGASSRREREQQDGWVLEESMWTMSCNGKKVGYAVRRDPTDDDIAVLETLWAVSMG 245
Query: 202 VGELP--SDMSDPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVRV 257
G LP SDM D +DGE++YMR FE +IGS++SE+ YM+ P G + PEL+VFFVR+
Sbjct: 246 GGVLPGISDM-DGKDGEMAYMRGSFEHIIGSRNSESLYMISPHG-GDCPELAVFFVRL 301
>UniRef100_Q6ZFZ3 Hypothetical protein OJ1311_H06.6-2 [Oryza sativa]
Length = 290
Score = 152 bits (385), Expect = 6e-36
Identities = 80/170 (47%), Positives = 114/170 (67%), Gaps = 8/170 (4%)
Query: 95 GIRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAI-PTGKLLQDMGMGLNRIALECEKH 153
G +TGT+FG R+ RV++A Q D++ P LL+E+A TG L+++M GL R+ALECEK
Sbjct: 122 GSSLTGTIFGRRRGRVHVALQTDTRSPPVLLVEMAAYSTGALVREMSSGLVRLALECEKQ 181
Query: 154 SSNDKTK---IVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHSVSVAVGELPSDMS 210
N K +++EP W +CNG+K G+ V+R+ D+ V+ + VSV G LP D +
Sbjct: 182 PINPGEKRRALLEEPTWRAYCNGRKCGFAVRRECGADEWRVLGAVEPVSVGAGVLPDDAA 241
Query: 211 DP---QDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVRV 257
++G+L YMRA FERV+GS+DSE +YMM PDG S GPELS++ +RV
Sbjct: 242 AAAAAEEGDLMYMRARFERVVGSRDSEAFYMMNPDG-SGGPELSIYLLRV 290
>UniRef100_Q6ZG48 Hypothetical protein OJ1111_E05.14 [Oryza sativa]
Length = 254
Score = 149 bits (375), Expect = 9e-35
Identities = 83/215 (38%), Positives = 127/215 (58%), Gaps = 20/215 (9%)
Query: 60 GKFRSMFRSFPIIVPSCKMPTMNGNHRTSETI-IHGGIRITGTLFGYRKARVNLAFQEDS 118
G MF+ P++ CKM + G H + T TLFG+R+ R++LA ED+
Sbjct: 40 GGIFKMFKLLPMLTTGCKMAALLGRHSGGRAAPLLADHAPTVTLFGHRRGRLSLAIHEDT 99
Query: 119 KCHPFLLLELAIPTGKLLQDMGMGLNRIALECEKHSSNDKTKIVDEPIWTLFCNGKKMGY 178
+ P L+EL + + ++M G ++ALE + S+ + ++++E +W +FCNG+K GY
Sbjct: 100 RAPPAFLIELPMLASAMHREMATGTVKLALESDTRSA--RRRLLEEYVWAVFCNGRKAGY 157
Query: 179 GVKR-DPTDDDLYVIQMLHSVSVAVGELPSDMSDPQ-----DGELSYMRAHFERVIGSKD 232
++R D +DDD +V+++L VS+ G LP +D + DGEL+YMRA ERV+GSKD
Sbjct: 158 AIRRKDASDDDRHVLRLLRGVSMGAGVLPPPPADRRGGAGPDGELTYMRARVERVVGSKD 217
Query: 233 SETYYMMMP-DGNSNG----------PELSVFFVR 256
SE +YM+ P DG+ NG PELS+F VR
Sbjct: 218 SEAFYMINPDDGSDNGGAAGRDRECAPELSIFLVR 252
>UniRef100_Q8RZE0 Hypothetical protein P0510C12.17 [Oryza sativa]
Length = 307
Score = 147 bits (372), Expect = 2e-34
Identities = 82/193 (42%), Positives = 113/193 (58%), Gaps = 31/193 (16%)
Query: 95 GIRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEK-- 152
G R+TGTL+G+R+ V+LAFQ D + P LLLELA PT L+++M GL RIALECE+
Sbjct: 116 GARLTGTLYGHRRGHVHLAFQVDPRACPALLLELAAPTASLVREMASGLVRIALECERAK 175
Query: 153 ----------------------HSSNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLY 190
SS K+V+E +W +CNG+ GY V+R+ D
Sbjct: 176 GGGACAFPTAAAAPSSSSSSSSSSSAGGRKLVEETVWRAYCNGRSCGYAVRRECGAADWR 235
Query: 191 VIQMLHSVSVAVGELPSDMSDPQDGELSYMRAHFERVIGSKDSETYYMMMPD------GN 244
V++ L VS+ G +P+ +G++ YMRA FERV+GS+DSE +YMM PD N
Sbjct: 236 VLRALEPVSMGAGVIPAACGG-GEGDVMYMRARFERVVGSRDSEAFYMMNPDCGGSGSNN 294
Query: 245 SNGPELSVFFVRV 257
+ GPELSV+ +RV
Sbjct: 295 NGGPELSVYLLRV 307
>UniRef100_Q69MD4 Hypothetical protein OSJNBb0019B14.18-1 [Oryza sativa]
Length = 244
Score = 147 bits (371), Expect = 3e-34
Identities = 79/214 (36%), Positives = 129/214 (59%), Gaps = 21/214 (9%)
Query: 60 GKFRSMFRSFPIIVPSCKMPTMNGNHRTSETIIHGGIRITGTLFGYRKARVNLAFQEDSK 119
G MF+ P++ CKM + G H + H T TLFG+R+ RV+LA ED++
Sbjct: 35 GGIFKMFKLMPMLTSGCKMVALLGRHNRALLADHA---TTVTLFGHRRGRVSLAIHEDTR 91
Query: 120 CHPFLLLELAIPTGKLLQDMGMGLNRIALECEKHSSNDKTKIVDEPIWTLFCNGKKMGYG 179
P L+EL + T L +++ G+ ++ALE + S+ + ++V+E +W ++CNG+K GY
Sbjct: 92 APPVFLIELPMLTSALHKEISSGVVKLALESDTRSA--RRRLVEEYVWAVYCNGRKAGYS 149
Query: 180 VKR-DPTDDDLYVIQMLHSVSVAVGELPS------DMSDPQDGELSYMRAHFERVIGSKD 232
++R + +DD+ +V+++L VS+ G LP+ + DGEL+Y+RA ERV+GSKD
Sbjct: 150 IRRKEASDDERHVLRLLRGVSMGAGVLPAAPEKEGGVPAGPDGELTYVRARVERVVGSKD 209
Query: 233 SETYYMMMPD---------GNSNGPELSVFFVRV 257
SE +YM+ P+ G+ + PELS+F VR+
Sbjct: 210 SEAFYMINPNEGGVGGDSAGDGSAPELSIFLVRM 243
>UniRef100_Q84WS4 Hypothetical protein At3g25640 [Arabidopsis thaliana]
Length = 267
Score = 144 bits (364), Expect = 2e-33
Identities = 72/173 (41%), Positives = 108/173 (61%), Gaps = 10/173 (5%)
Query: 95 GIRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEKHS 154
G R+ GTLFG R+ V A Q+D P +L++L PT L+++M GL RIALE +
Sbjct: 95 GFRVVGTLFGNRRGHVYFAVQDDPTRLPAVLIQLPTPTSVLVREMASGLVRIALETAAYK 154
Query: 155 SNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHSVSVAVGELPSDMS---- 210
++ K K+++E W +CNGKK GY +++ + + V++ + +++ G LP+ +
Sbjct: 155 TDSKKKLLEESTWRTYCNGKKCGYAARKECGEAEWKVLKAVGPITMGAGVLPATTTTVDE 214
Query: 211 ------DPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVRV 257
+ GEL YMRA FERV+GS+DSE +YMM PD +S GPELSV+F+RV
Sbjct: 215 EGNGAVGSEKGELMYMRARFERVVGSRDSEAFYMMNPDVSSGGPELSVYFLRV 267
>UniRef100_Q9LI79 Gb|AAB84348.1 [Arabidopsis thaliana]
Length = 293
Score = 144 bits (364), Expect = 2e-33
Identities = 72/173 (41%), Positives = 108/173 (61%), Gaps = 10/173 (5%)
Query: 95 GIRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEKHS 154
G R+ GTLFG R+ V A Q+D P +L++L PT L+++M GL RIALE +
Sbjct: 121 GFRVVGTLFGNRRGHVYFAVQDDPTRLPAVLIQLPTPTSVLVREMASGLVRIALETAAYK 180
Query: 155 SNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHSVSVAVGELPSDMS---- 210
++ K K+++E W +CNGKK GY +++ + + V++ + +++ G LP+ +
Sbjct: 181 TDSKKKLLEESTWRTYCNGKKCGYAARKECGEAEWKVLKAVGPITMGAGVLPATTTTVDE 240
Query: 211 ------DPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVRV 257
+ GEL YMRA FERV+GS+DSE +YMM PD +S GPELSV+F+RV
Sbjct: 241 EGNGAVGSEKGELMYMRARFERVVGSRDSEAFYMMNPDVSSGGPELSVYFLRV 293
>UniRef100_Q6YYK3 Hypothetical protein OJ1590_E05.28 [Oryza sativa]
Length = 315
Score = 142 bits (358), Expect = 8e-33
Identities = 78/184 (42%), Positives = 109/184 (58%), Gaps = 23/184 (12%)
Query: 97 RITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLN-RIALECEKHSS 155
R+TGTLFG+RK RV LA QE +C P L++ELAI T LL+++ RI LE E+ +
Sbjct: 132 RVTGTLFGHRKGRVALALQETPRCLPTLVIELAIQTNALLRELANPAGARIVLETERRAP 191
Query: 156 NDKTK---------IVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHSVSVAVGELP 206
+ ++D WT+FCNG+K G V+R+ TDDDL V++ L VS+ G LP
Sbjct: 192 STDAAAGKHRRAPPLLDVAAWTMFCNGRKTGLAVRREATDDDLAVLETLRPVSMGAGVLP 251
Query: 207 -------SDMSDPQDG------ELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVF 253
S P+ G E++YMR F+ +GS+DSE+ YM+ P G GPEL++F
Sbjct: 252 ASNRSSSSSSQSPEKGAAAADDEVAYMRGCFDHFVGSRDSESLYMIAPQGGGTGPELAIF 311
Query: 254 FVRV 257
FVR+
Sbjct: 312 FVRL 315
>UniRef100_Q9LPT9 T22I11.13 [Arabidopsis thaliana]
Length = 242
Score = 142 bits (357), Expect = 1e-32
Identities = 78/195 (40%), Positives = 123/195 (63%), Gaps = 12/195 (6%)
Query: 66 FRSFPIIVPSCKMPTMNGNHRTSETIIHGGIRITGTLFGYRKARVNLAFQEDSK-CHPFL 124
+++ + V S + + +H +S ++GT FG+R+ RV+ Q+ + P L
Sbjct: 55 YQNDTLSVSSTSSSSSSSDHSSSS---QSSSTVSGTFFGHRRGRVSFCLQDAAVGSSPLL 111
Query: 125 LLELAIPTGKLLQDMGM-GLNRIALECEKHSSNDK--TKIVDEPIWTLFCNGKKMGYGVK 181
LLELA+PT L ++M G+ RIALEC++ S++ + I D P+W++FCNG+KMG+ V+
Sbjct: 112 LLELAVPTAALAKEMDEEGVLRIALECDRRRSSNSRSSSIFDVPVWSMFCNGRKMGFAVR 171
Query: 182 RDPTDDDLYVIQMLHSVSVAVGELPSDMSDPQDGELSYMRAHFERVIGSKDSETYYMMMP 241
R T++D ++M+ SVSV G +PS+ D ++ Y+RA FERV GS DSE+++MM P
Sbjct: 172 RKVTENDAVFLRMMQSVSVGAGVVPSEEED----QMLYLRARFERVTGSSDSESFHMMNP 227
Query: 242 DGNSNGPELSVFFVR 256
G S G ELS+F +R
Sbjct: 228 -GGSYGQELSIFLLR 241
>UniRef100_Q8LBL9 Hypothetical protein [Arabidopsis thaliana]
Length = 245
Score = 142 bits (357), Expect = 1e-32
Identities = 78/195 (40%), Positives = 123/195 (63%), Gaps = 12/195 (6%)
Query: 66 FRSFPIIVPSCKMPTMNGNHRTSETIIHGGIRITGTLFGYRKARVNLAFQEDSK-CHPFL 124
+++ + V S + + +H +S ++GT FG+R+ RV+ Q+ + P L
Sbjct: 58 YQNDTLSVSSTSSSSSSSDHSSSS---QSSSTVSGTFFGHRRGRVSFCLQDATVGSSPLL 114
Query: 125 LLELAIPTGKLLQDMGM-GLNRIALECEKHSSNDK--TKIVDEPIWTLFCNGKKMGYGVK 181
LLELA+PT L ++M G+ RIALEC++ S++ + I D P+W++FCNG+KMG+ V+
Sbjct: 115 LLELAVPTAALAKEMDEEGVLRIALECDRRRSSNSRSSSIFDVPVWSMFCNGRKMGFAVR 174
Query: 182 RDPTDDDLYVIQMLHSVSVAVGELPSDMSDPQDGELSYMRAHFERVIGSKDSETYYMMMP 241
R T++D ++M+ SVSV G +PS+ D ++ Y+RA FERV GS DSE+++MM P
Sbjct: 175 RKVTENDAVFLRMMQSVSVGAGVVPSEEED----QMLYLRARFERVTGSSDSESFHMMNP 230
Query: 242 DGNSNGPELSVFFVR 256
G S G ELS+F +R
Sbjct: 231 -GGSYGQELSIFLLR 244
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 475,491,332
Number of Sequences: 2790947
Number of extensions: 21095317
Number of successful extensions: 138112
Number of sequences better than 10.0: 255
Number of HSP's better than 10.0 without gapping: 114
Number of HSP's successfully gapped in prelim test: 147
Number of HSP's that attempted gapping in prelim test: 135113
Number of HSP's gapped (non-prelim): 1941
length of query: 257
length of database: 848,049,833
effective HSP length: 125
effective length of query: 132
effective length of database: 499,181,458
effective search space: 65891952456
effective search space used: 65891952456
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 73 (32.7 bits)
Medicago: description of AC149197.3


