

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149197.11 - phase: 0
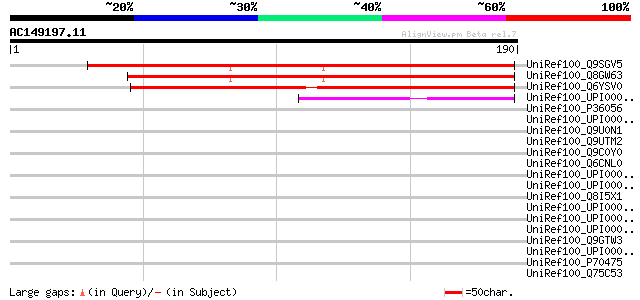
(190 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SGV5 F1N19.17 [Arabidopsis thaliana] 169 3e-41
UniRef100_Q8GW63 Hypothetical protein At1g64600/F1N19_16 [Arabid... 168 6e-41
UniRef100_Q6YSV0 Hypothetical protein P0680C01.9 [Oryza sativa] 165 7e-40
UniRef100_UPI000042FB3F UPI000042FB3F UniRef100 entry 48 1e-04
UniRef100_P36056 Hypothetical 72.2 kDa protein in RPS27A-GPM1 in... 44 0.002
UniRef100_UPI000046861F UPI000046861F UniRef100 entry 44 0.003
UniRef100_Q9U0N1 Glutamic acid-rich protein [Plasmodium falciparum] 43 0.004
UniRef100_Q9UTM2 Cox1101 protein [Schizosaccharomyces pombe] 43 0.004
UniRef100_Q9C0Y0 Cox11-b protein [Schizosaccharomyces pombe] 43 0.004
UniRef100_Q6CNL0 Similar to sp|P36056 Saccharomyces cerevisiae Y... 43 0.005
UniRef100_UPI00002C7A2B UPI00002C7A2B UniRef100 entry 42 0.006
UniRef100_UPI000023DFFA UPI000023DFFA UniRef100 entry 42 0.008
UniRef100_Q8I5X1 Hypothetical protein [Plasmodium falciparum] 42 0.008
UniRef100_UPI0000466BA5 UPI0000466BA5 UniRef100 entry 41 0.019
UniRef100_UPI00003C12C8 UPI00003C12C8 UniRef100 entry 41 0.019
UniRef100_UPI000023E705 UPI000023E705 UniRef100 entry 41 0.019
UniRef100_Q9GTW3 Glutamic acid-rich protein [Plasmodium falciparum] 41 0.019
UniRef100_UPI000018232A UPI000018232A UniRef100 entry 40 0.024
UniRef100_P70475 Neural zinc finger factor-1 [Rattus norvegicus] 40 0.024
UniRef100_Q75C53 ACR064Wp [Ashbya gossypii] 40 0.024
>UniRef100_Q9SGV5 F1N19.17 [Arabidopsis thaliana]
Length = 555
Score = 169 bits (429), Expect = 3e-41
Identities = 87/165 (52%), Positives = 110/165 (65%), Gaps = 5/165 (3%)
Query: 30 HNVILIFVSSLIPLTREPWPLDGITFDTLKEQQAKRNPEDLEIDYEDWLKLQ--EADDDA 87
H +L L RE WPLDG+ +TLKE++A + PEDLEIDYED++K Q E
Sbjct: 361 HETLLRIKLGLCLQFRELWPLDGMKLETLKERRANKKPEDLEIDYEDFIKSQVVEVPYID 420
Query: 88 PREVDAIRRYESDGLETDGDGEDDNEEVK---ETEEETEIADLGGGWGRIVFMPIRRGKQ 144
PR D+ E++ + DG G D++EE K E EEE+E A +GGGWGRI+F P R+GKQ
Sbjct: 421 PRAYDSDTMDENEEEQEDGGGTDEDEEDKIEEEIEEESERASVGGGWGRIIFPPFRKGKQ 480
Query: 145 VTMNVCRSIKRDASEGEFARMVVTKSKNPALHRQAKRSIWGDLWP 189
VT+++C K D SEG F R V+TKSKNP LH QAK+S WGDLWP
Sbjct: 481 VTLDMCVPTKEDGSEGAFERRVITKSKNPDLHLQAKKSFWGDLWP 525
>UniRef100_Q8GW63 Hypothetical protein At1g64600/F1N19_16 [Arabidopsis thaliana]
Length = 537
Score = 168 bits (426), Expect = 6e-41
Identities = 84/150 (56%), Positives = 106/150 (70%), Gaps = 5/150 (3%)
Query: 45 REPWPLDGITFDTLKEQQAKRNPEDLEIDYEDWLKLQ--EADDDAPREVDAIRRYESDGL 102
RE WPLDG+ +TLKE++A + PEDLEIDYED++K Q E PR D+ E++
Sbjct: 358 RELWPLDGMKLETLKERRANKKPEDLEIDYEDFIKSQVVEVPYIDPRAYDSDTMDENEEE 417
Query: 103 ETDGDGEDDNEEVK---ETEEETEIADLGGGWGRIVFMPIRRGKQVTMNVCRSIKRDASE 159
+ DG G D++EE K E EEE+E A +GGGWGRI+F P R+GKQVT+++C K D SE
Sbjct: 418 QEDGGGTDEDEEDKIEEEIEEESERASVGGGWGRIIFPPFRKGKQVTLDMCVPTKEDGSE 477
Query: 160 GEFARMVVTKSKNPALHRQAKRSIWGDLWP 189
G F R V+TKSKNP LH QAK+S WGDLWP
Sbjct: 478 GAFERRVITKSKNPDLHLQAKKSFWGDLWP 507
>UniRef100_Q6YSV0 Hypothetical protein P0680C01.9 [Oryza sativa]
Length = 711
Score = 165 bits (417), Expect = 7e-40
Identities = 78/144 (54%), Positives = 104/144 (72%), Gaps = 4/144 (2%)
Query: 46 EPWPLDGITFDTLKEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETD 105
E WPLDG+ F+TLKE+ AKRNPEDL IDY++ +E ++ D + Y+SD E
Sbjct: 571 EAWPLDGMKFETLKERHAKRNPEDLIIDYDEQFPSEEDEEAPVNAEDCLVPYDSDAQELG 630
Query: 106 GDGEDDNEEVKETEEETEIADLGGGWGRIVFMPIRRGKQVTMNVCRSIKRDASEGEFARM 165
E + +E EE++ ADLGGGWGRI++ PIRRG+QV ++VCR+ KRDASEG F R+
Sbjct: 631 LFHETE----EEFEEQSVRADLGGGWGRIIYSPIRRGRQVQLDVCRATKRDASEGAFERV 686
Query: 166 VVTKSKNPALHRQAKRSIWGDLWP 189
V+T+SKNP +H QA+RS+WGDLWP
Sbjct: 687 VITQSKNPTMHHQARRSLWGDLWP 710
>UniRef100_UPI000042FB3F UPI000042FB3F UniRef100 entry
Length = 1030
Score = 48.1 bits (113), Expect = 1e-04
Identities = 25/81 (30%), Positives = 40/81 (48%), Gaps = 6/81 (7%)
Query: 109 EDDNEEVKETEEETEIADLGGGWGRIVFMPIRRGKQVTMNVCRSIKRDASEGEFARMVVT 168
E+ + V E E + W R+V P++R VTM+ C ++G R+ T
Sbjct: 509 EETSPSVNSEELEENLRKEAYSWPRMVAPPMKRKGHVTMDTC------CADGNIQRLTYT 562
Query: 169 KSKNPALHRQAKRSIWGDLWP 189
KS + + A++S WGDL+P
Sbjct: 563 KSHSKQSYHDARKSSWGDLFP 583
>UniRef100_P36056 Hypothetical 72.2 kDa protein in RPS27A-GPM1 intergenic region
[Saccharomyces cerevisiae]
Length = 628
Score = 43.9 bits (102), Expect = 0.002
Identities = 32/127 (25%), Positives = 56/127 (43%), Gaps = 23/127 (18%)
Query: 64 KRNPEDLEIDYEDWLKLQEA--DDDAPREVDAIRRYESDGLETDGDGEDDNEEVKETEEE 121
+RN D EI +L + + D++ +E+ +R +G G DD +
Sbjct: 440 RRNGRDYEILNYSYLIFERSHKDENTLKEIKKLRNENVNGKYDIGSLGDDTQN------- 492
Query: 122 TEIADLGGGWGRIVFMPIRRGKQVTMNVCRSIKRDASEGEFARMVVTKSKNPALHRQAKR 181
W RI+ P++R V M++C A GE + V++S + ++ A++
Sbjct: 493 --------SWPRIINDPVKRKGHVMMDLC------APSGELEKWTVSRSFSKQIYHDARK 538
Query: 182 SIWGDLW 188
S GDLW
Sbjct: 539 SKKGDLW 545
>UniRef100_UPI000046861F UPI000046861F UniRef100 entry
Length = 755
Score = 43.5 bits (101), Expect = 0.003
Identities = 25/73 (34%), Positives = 43/73 (58%), Gaps = 3/73 (4%)
Query: 53 ITFDTLKEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGE--D 110
I D +E++ + ED EI+ ++ + +E DD+ EV+A E + ++ DG+ E D
Sbjct: 38 IEVDEEEEEEEDDDEEDEEIEVDEEEEEEEEDDEEDEEVEADEEEEEE-IDDDGEEEADD 96
Query: 111 DNEEVKETEEETE 123
D EE++E EEE +
Sbjct: 97 DEEEIEEDEEEVD 109
Score = 39.3 bits (90), Expect = 0.054
Identities = 23/74 (31%), Positives = 42/74 (56%), Gaps = 5/74 (6%)
Query: 53 ITFDTLKEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGEDDN 112
I D +E++ + + ED E++ ++ + +E DDD E D + + +E D + DD+
Sbjct: 57 IEVDEEEEEEEEDDEEDEEVEADEEEE-EEIDDDGEEEADD----DEEEIEEDEEEVDDD 111
Query: 113 EEVKETEEETEIAD 126
EE++E EEE + D
Sbjct: 112 EEIEEDEEEMDDDD 125
Score = 38.1 bits (87), Expect = 0.12
Identities = 26/70 (37%), Positives = 36/70 (51%), Gaps = 3/70 (4%)
Query: 60 EQQAKRNPEDLEIDYEDWLKLQEADD---DAPREVDAIRRYESDGLETDGDGEDDNEEVK 116
+++ K + ED E D ED ++E D+ D E + I E + E D D ED+ EV
Sbjct: 1 DEEDKEDDEDDEDDEEDEEDVEEDDEEEEDDDEEDEEIEVDEEEEEEEDDDEEDEEIEVD 60
Query: 117 ETEEETEIAD 126
E EEE E D
Sbjct: 61 EEEEEEEEDD 70
Score = 36.6 bits (83), Expect = 0.35
Identities = 23/73 (31%), Positives = 37/73 (50%), Gaps = 6/73 (8%)
Query: 56 DTLKEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGEDDNEEV 115
D +E+ E++E+D E+ + +E DD+ E++ E E + D E+D E
Sbjct: 24 DDEEEEDDDEEDEEIEVDEEE--EEEEDDDEEDEEIEVDEEEE----EEEEDDEEDEEVE 77
Query: 116 KETEEETEIADLG 128
+ EEE EI D G
Sbjct: 78 ADEEEEEEIDDDG 90
Score = 33.9 bits (76), Expect = 2.3
Identities = 23/70 (32%), Positives = 37/70 (52%), Gaps = 5/70 (7%)
Query: 60 EQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGEDDNEEVKETE 119
E++ + ED EI+ ++ + +E DD+ E++ E E + D E+D EEV+ E
Sbjct: 26 EEEEDDDEEDEEIEVDEEEEEEEDDDEEDEEIEVDEEEE----EEEEDDEED-EEVEADE 80
Query: 120 EETEIADLGG 129
EE E D G
Sbjct: 81 EEEEEIDDDG 90
>UniRef100_Q9U0N1 Glutamic acid-rich protein [Plasmodium falciparum]
Length = 673
Score = 43.1 bits (100), Expect = 0.004
Identities = 20/68 (29%), Positives = 39/68 (56%)
Query: 59 KEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGEDDNEEVKET 118
+E++ + E+ E + E+ +++E +DDA + D E D E D D E+D+++ +E
Sbjct: 579 EEEEEEEEEEEEEEEEEEEDEVEEDEDDAEEDEDDAEEDEDDAEEDDDDAEEDDDDAEED 638
Query: 119 EEETEIAD 126
++E E D
Sbjct: 639 DDEDEDED 646
Score = 42.0 bits (97), Expect = 0.008
Identities = 22/65 (33%), Positives = 36/65 (54%), Gaps = 3/65 (4%)
Query: 59 KEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGEDDNEEVKET 118
+E + + + +D E D +D +E +DDA + D + D E D + ED++EE +E
Sbjct: 596 EEDEVEEDEDDAEEDEDD---AEEDEDDAEEDDDDAEEDDDDAEEDDDEDEDEDEEEEED 652
Query: 119 EEETE 123
EEE E
Sbjct: 653 EEEEE 657
Score = 39.7 bits (91), Expect = 0.041
Identities = 19/65 (29%), Positives = 37/65 (56%), Gaps = 3/65 (4%)
Query: 59 KEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGEDDNEEVKET 118
+E++ + +++E D +D +E +DDA + D + D E D D E+D++E ++
Sbjct: 589 EEEEEEEEEDEVEEDEDD---AEEDEDDAEEDEDDAEEDDDDAEEDDDDAEEDDDEDEDE 645
Query: 119 EEETE 123
+EE E
Sbjct: 646 DEEEE 650
Score = 35.8 bits (81), Expect = 0.60
Identities = 19/62 (30%), Positives = 34/62 (54%), Gaps = 3/62 (4%)
Query: 60 EQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGEDDNEEVKETE 119
E A+ + +D E D +D +E DDDA + D + + + D + E+D EE +E+E
Sbjct: 604 EDDAEEDEDDAEEDEDD---AEEDDDDAEEDDDDAEEDDDEDEDEDEEEEEDEEEEEESE 660
Query: 120 EE 121
++
Sbjct: 661 KK 662
Score = 35.4 bits (80), Expect = 0.78
Identities = 16/67 (23%), Positives = 35/67 (51%)
Query: 60 EQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGEDDNEEVKETE 119
E+ + E+ E + E+ + +E +D+ + D E D E + D E+D+++ +E +
Sbjct: 573 EEDEEEEEEEEEEEEEEEEEEEEEEDEVEEDEDDAEEDEDDAEEDEDDAEEDDDDAEEDD 632
Query: 120 EETEIAD 126
++ E D
Sbjct: 633 DDAEEDD 639
>UniRef100_Q9UTM2 Cox1101 protein [Schizosaccharomyces pombe]
Length = 695
Score = 43.1 bits (100), Expect = 0.004
Identities = 21/59 (35%), Positives = 33/59 (55%), Gaps = 6/59 (10%)
Query: 131 WGRIVFMPIRRGKQVTMNVCRSIKRDASEGEFARMVVTKSKNPALHRQAKRSIWGDLWP 189
W RI+ P++R V ++VC S + R +V KS+ +R A++S WGDL+P
Sbjct: 353 WPRIIRPPLKRDGHVIIDVCDS------DARLRRNIVPKSQGKLAYRLARKSAWGDLFP 405
>UniRef100_Q9C0Y0 Cox11-b protein [Schizosaccharomyces pombe]
Length = 732
Score = 43.1 bits (100), Expect = 0.004
Identities = 21/59 (35%), Positives = 33/59 (55%), Gaps = 6/59 (10%)
Query: 131 WGRIVFMPIRRGKQVTMNVCRSIKRDASEGEFARMVVTKSKNPALHRQAKRSIWGDLWP 189
W RI+ P++R V ++VC S + R +V KS+ +R A++S WGDL+P
Sbjct: 390 WPRIIRPPLKRDGHVIIDVCDS------DARLRRNIVPKSQGKLAYRLARKSAWGDLFP 442
>UniRef100_Q6CNL0 Similar to sp|P36056 Saccharomyces cerevisiae YKL155c CAP1
singleton [Kluyveromyces lactis]
Length = 729
Score = 42.7 bits (99), Expect = 0.005
Identities = 25/81 (30%), Positives = 42/81 (50%), Gaps = 10/81 (12%)
Query: 112 NEEVKETEEETEIADLG----GGWGRIVFMPIRRGKQVTMNVCRSIKRDASEGEFARMVV 167
N E +E + + +I LG W RIV P +R V +++C A G+F + +V
Sbjct: 562 NREREENKIKYDIGSLGDETANTWPRIVKQPTKRKGHVILDLC------APSGKFEKWIV 615
Query: 168 TKSKNPALHRQAKRSIWGDLW 188
KS + ++ A+++ GDLW
Sbjct: 616 PKSLDKQIYHDARKTQKGDLW 636
>UniRef100_UPI00002C7A2B UPI00002C7A2B UniRef100 entry
Length = 258
Score = 42.4 bits (98), Expect = 0.006
Identities = 28/71 (39%), Positives = 39/71 (54%), Gaps = 6/71 (8%)
Query: 56 DTLKEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDG---EDDN 112
++ + +Q+K + + EID EDWL QE +DD D YE D E + D EDD
Sbjct: 56 ESSEPEQSKESTK--EIDNEDWLD-QEYEDDEEEYEDDEEEYEDDEEEYEDDEEEYEDDE 112
Query: 113 EEVKETEEETE 123
EE ++ EEE E
Sbjct: 113 EEYEDDEEEYE 123
Score = 35.4 bits (80), Expect = 0.78
Identities = 27/84 (32%), Positives = 37/84 (43%), Gaps = 5/84 (5%)
Query: 43 LTREPWPLDGITFDTLKEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGL 102
+ E W D + + + ED E +YED +E +DD D YE D
Sbjct: 70 IDNEDWLDQEYEDDEEEYEDDEEEYEDDEEEYED--DEEEYEDDEEEYEDDEEEYEDDEE 127
Query: 103 ETDGDGE---DDNEEVKETEEETE 123
E + D E DD EE ++ EEE E
Sbjct: 128 EYEDDEEEYEDDEEEYEDDEEEYE 151
Score = 32.3 bits (72), Expect = 6.6
Identities = 23/59 (38%), Positives = 30/59 (49%), Gaps = 4/59 (6%)
Query: 68 EDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGED--DNEEVKETEEETEI 124
ED E +YED +E +DD D YE D E + D E+ D+EE E +EE I
Sbjct: 102 EDDEEEYED--DEEEYEDDEEEYEDDEEEYEDDEEEYEDDEEEYEDDEEEYEDDEENMI 158
>UniRef100_UPI000023DFFA UPI000023DFFA UniRef100 entry
Length = 989
Score = 42.0 bits (97), Expect = 0.008
Identities = 28/96 (29%), Positives = 46/96 (47%), Gaps = 8/96 (8%)
Query: 93 AIRRYESDGLETDGDGEDDNEEVKETEEETEIADLGGGWGRIVFMPIRRGKQVTMNVCRS 152
AIRR S E G++ + E E +++ R++ P++R VT+++C
Sbjct: 797 AIRRGVSK--EKPVTGKEAANQAFEGYENSDVKPDMQSLPRMIMPPLKRKGHVTLDLC-- 852
Query: 153 IKRDASEGEFARMVVTKSKNPALHRQAKRSIWGDLW 188
EG R V+KS + + A++S WGDLW
Sbjct: 853 ----TPEGRVERWTVSKSFSKLAYHDARKSKWGDLW 884
>UniRef100_Q8I5X1 Hypothetical protein [Plasmodium falciparum]
Length = 395
Score = 42.0 bits (97), Expect = 0.008
Identities = 25/98 (25%), Positives = 53/98 (53%), Gaps = 1/98 (1%)
Query: 30 HNVILIFVSSLIPLTREPWPLDGITFDTLKEQQAKRNPEDLEIDYEDWLKLQEADDD-AP 88
H++ I S +R+ ++ T+++ +E + + E+ E + E+ + +E +++
Sbjct: 122 HSIDTISDISFTQTSRKSLEIESNTYESYREVEKEDIEEEEEEEKEEEYEEEEEEEEYEE 181
Query: 89 REVDAIRRYESDGLETDGDGEDDNEEVKETEEETEIAD 126
E + YE +GL+T+ + E+DN+EV+ EE E D
Sbjct: 182 EEEEEEEEYEEEGLKTEEEKEEDNKEVEPEEELKEEDD 219
Score = 34.7 bits (78), Expect = 1.3
Identities = 20/68 (29%), Positives = 38/68 (55%), Gaps = 2/68 (2%)
Query: 59 KEQQAKRNPEDLEIDYED-WLKLQEADDDAPREVDAIRRY-ESDGLETDGDGEDDNEEVK 116
+E++ + E+ E +YE+ LK +E ++ +EV+ E D E + + E +NE+ K
Sbjct: 175 EEEEYEEEEEEEEEEYEEEGLKTEEEKEEDNKEVEPEEELKEEDDKEVEPEEEKENEQKK 234
Query: 117 ETEEETEI 124
E +EE +
Sbjct: 235 EEQEENNL 242
>UniRef100_UPI0000466BA5 UPI0000466BA5 UniRef100 entry
Length = 532
Score = 40.8 bits (94), Expect = 0.019
Identities = 24/74 (32%), Positives = 36/74 (48%), Gaps = 21/74 (28%)
Query: 131 WGRIVFMPIRRGKQVTMNVCRSIKRDASEGEFARMVVTKSKNPAL--------------H 176
W R+V I+ GK V ++VC S F R+VVTKS + +
Sbjct: 410 WPRVVMPTIKAGKHVLLDVC-------SPYNFERLVVTKSSSLISNLKTKTGTILKGYGY 462
Query: 177 RQAKRSIWGDLWPF 190
++A++ +WGDLW F
Sbjct: 463 KKARKILWGDLWRF 476
>UniRef100_UPI00003C12C8 UPI00003C12C8 UniRef100 entry
Length = 977
Score = 40.8 bits (94), Expect = 0.019
Identities = 16/59 (27%), Positives = 33/59 (55%), Gaps = 6/59 (10%)
Query: 131 WGRIVFMPIRRGKQVTMNVCRSIKRDASEGEFARMVVTKSKNPALHRQAKRSIWGDLWP 189
W R++ P+++G VT++ C + G R ++K+ ++ A+++ WGDL+P
Sbjct: 734 WPRLIRSPLKKGGHVTLDAC------CATGNIERFTISKACGKQAYQDARKAKWGDLFP 786
>UniRef100_UPI000023E705 UPI000023E705 UniRef100 entry
Length = 144
Score = 40.8 bits (94), Expect = 0.019
Identities = 30/97 (30%), Positives = 43/97 (43%), Gaps = 14/97 (14%)
Query: 57 TLKEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAI--------RRYESDGLETDGDG 108
T E++ ED E + E E DDD +E DA + E+D E +GD
Sbjct: 31 TADEEEQSEEDEDAEEEEE------EDDDDEDKEADAKVEPPVKKRKTAEADDQELNGDA 84
Query: 109 EDDNEEVKETEEETEIADLGGGWGRIVFMPIRRGKQV 145
EDD EE +E +EE + + G +P R + V
Sbjct: 85 EDDEEEEEEEKEENDDEEDAEGDDEAADVPARTKEAV 121
>UniRef100_Q9GTW3 Glutamic acid-rich protein [Plasmodium falciparum]
Length = 682
Score = 40.8 bits (94), Expect = 0.019
Identities = 20/65 (30%), Positives = 36/65 (54%), Gaps = 3/65 (4%)
Query: 59 KEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGEDDNEEVKET 118
+E + + + +D E D +D +E +DDA + D E D E D D E+D++E ++
Sbjct: 595 EEDEVEEDEDDAEEDEDD---AEEDEDDAGEDEDDAEEDEDDAEEDDDDAEEDDDEEEDV 651
Query: 119 EEETE 123
E+E +
Sbjct: 652 EDEED 656
Score = 38.9 bits (89), Expect = 0.070
Identities = 19/68 (27%), Positives = 38/68 (54%), Gaps = 2/68 (2%)
Query: 59 KEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGEDDNEEVKET 118
+E++ + E+ E + ED +++E +DDA + D E D E + D E+D ++ +E
Sbjct: 580 EEEEEEEEEEEEEEEEED--EVEEDEDDAEEDEDDAEEDEDDAGEDEDDAEEDEDDAEED 637
Query: 119 EEETEIAD 126
+++ E D
Sbjct: 638 DDDAEEDD 645
Score = 38.5 bits (88), Expect = 0.092
Identities = 18/65 (27%), Positives = 37/65 (56%), Gaps = 3/65 (4%)
Query: 59 KEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGEDDNEEVKET 118
+E++ + +++E D +D +E +DDA + D E D E + D E+D+++ +E
Sbjct: 588 EEEEEEEEEDEVEEDEDD---AEEDEDDAEEDEDDAGEDEDDAEEDEDDAEEDDDDAEED 644
Query: 119 EEETE 123
++E E
Sbjct: 645 DDEEE 649
Score = 35.8 bits (81), Expect = 0.60
Identities = 23/66 (34%), Positives = 33/66 (49%), Gaps = 5/66 (7%)
Query: 60 EQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGED--DNEEVKE 117
E A+ + +D E D +D E +DDA + D + D E D + ED D E+ +E
Sbjct: 603 EDDAEEDEDDAEEDEDD---AGEDEDDAEEDEDDAEEDDDDAEEDDDEEEDVEDEEDEEE 659
Query: 118 TEEETE 123
EEE E
Sbjct: 660 DEEEEE 665
Score = 35.0 bits (79), Expect = 1.0
Identities = 16/65 (24%), Positives = 36/65 (54%), Gaps = 1/65 (1%)
Query: 59 KEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGEDDNEEVKET 118
+ ++ + + E++E D E+ + +E +++ E D + E D E + D E+D ++ E
Sbjct: 565 ESKEVQEDEEEVEEDEEEEEE-EEEEEEEEEEEDEVEEDEDDAEEDEDDAEEDEDDAGED 623
Query: 119 EEETE 123
E++ E
Sbjct: 624 EDDAE 628
>UniRef100_UPI000018232A UPI000018232A UniRef100 entry
Length = 1182
Score = 40.4 bits (93), Expect = 0.024
Identities = 24/73 (32%), Positives = 39/73 (52%), Gaps = 6/73 (8%)
Query: 51 DGITFDTLKEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGED 110
D F ++Q + ED E+D ED +++E DD+ + D + D +E + D +D
Sbjct: 106 DDEEFSEDNDEQGDDDDED-EVDREDEEEIEEEDDEDDEDDD-----DGDDVEEEEDDDD 159
Query: 111 DNEEVKETEEETE 123
+ EE +E EEE E
Sbjct: 160 EEEEEEEEEEENE 172
>UniRef100_P70475 Neural zinc finger factor-1 [Rattus norvegicus]
Length = 1187
Score = 40.4 bits (93), Expect = 0.024
Identities = 24/73 (32%), Positives = 39/73 (52%), Gaps = 6/73 (8%)
Query: 51 DGITFDTLKEQQAKRNPEDLEIDYEDWLKLQEADDDAPREVDAIRRYESDGLETDGDGED 110
D F ++Q + ED E+D ED +++E DD+ + D + D +E + D +D
Sbjct: 106 DDEEFSEDNDEQGDDDDED-EVDREDEEEIEEEDDEDDEDDD-----DGDDVEEEEDDDD 159
Query: 111 DNEEVKETEEETE 123
+ EE +E EEE E
Sbjct: 160 EEEEEEEEEEENE 172
>UniRef100_Q75C53 ACR064Wp [Ashbya gossypii]
Length = 709
Score = 40.4 bits (93), Expect = 0.024
Identities = 20/70 (28%), Positives = 37/70 (52%), Gaps = 6/70 (8%)
Query: 119 EEETEIADLGGGWGRIVFMPIRRGKQVTMNVCRSIKRDASEGEFARMVVTKSKNPALHRQ 178
E + +A+ W R++ P +R V +++C A G+F + VV+KS + +
Sbjct: 552 ESGSTVANASETWPRVIRQPTKRKGHVMLDLC------APSGQFEKWVVSKSFDKQAYHD 605
Query: 179 AKRSIWGDLW 188
A+++ GDLW
Sbjct: 606 ARKAQKGDLW 615
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.138 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 330,495,027
Number of Sequences: 2790947
Number of extensions: 14172419
Number of successful extensions: 91060
Number of sequences better than 10.0: 668
Number of HSP's better than 10.0 without gapping: 216
Number of HSP's successfully gapped in prelim test: 485
Number of HSP's that attempted gapping in prelim test: 84324
Number of HSP's gapped (non-prelim): 3702
length of query: 190
length of database: 848,049,833
effective HSP length: 120
effective length of query: 70
effective length of database: 513,136,193
effective search space: 35919533510
effective search space used: 35919533510
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 71 (32.0 bits)
Medicago: description of AC149197.11


