

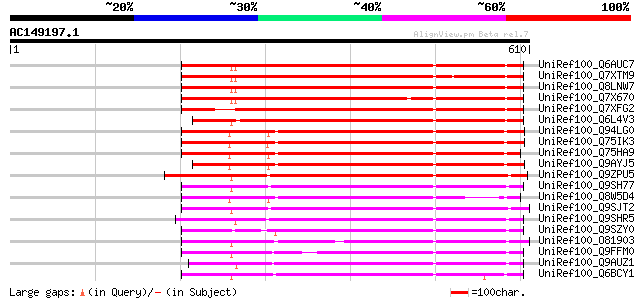
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149197.1 + phase: 0 /pseudo
(610 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6AUC7 Putative polyprotein [Oryza sativa] 443 e-123
UniRef100_Q7XTM9 OSJNBa0033G05.13 protein [Oryza sativa] 441 e-122
UniRef100_Q8LNW7 Putative polyprotein [Oryza sativa] 435 e-120
UniRef100_Q7X670 OSJNBa0093P23.9 protein [Oryza sativa] 433 e-120
UniRef100_Q7XFG2 Putative retroelement [Oryza sativa] 423 e-117
UniRef100_Q6L4V3 Putative polyprotein [Oryza sativa] 419 e-115
UniRef100_Q94LG0 Putative retroelement pol polyprotein [Oryza sa... 395 e-108
UniRef100_Q75IK3 Putative polyprotein [Oryza sativa] 395 e-108
UniRef100_Q75HA9 Putative polyprotein [Oryza sativa] 393 e-108
UniRef100_Q9AYJ5 Putative pol polyprotein [Oryza sativa] 370 e-101
UniRef100_Q9ZPU5 Putative retroelement pol polyprotein [Arabidop... 350 9e-95
UniRef100_Q9SH77 Putative retroelement pol polyprotein [Arabidop... 314 6e-84
UniRef100_Q8W5D4 Putative retrotransposon-related protein [Oryza... 312 2e-83
UniRef100_Q9SJT2 Putative retroelement pol polyprotein [Arabidop... 308 2e-82
UniRef100_Q9SHR5 F28L22.3 protein [Arabidopsis thaliana] 307 5e-82
UniRef100_Q9SZY0 Putative retrotransposon [Arabidopsis thaliana] 304 4e-81
UniRef100_O81903 Putative transposable element [Arabidopsis thal... 304 6e-81
UniRef100_Q9FFM0 Copia-like retrotransposable element [Arabidops... 302 2e-80
UniRef100_Q9AUZ1 Polyprotein, putative [Arabidopsis thaliana] 301 3e-80
UniRef100_Q6BCY1 Gag-Pol [Ipomoea batatas] 298 3e-79
>UniRef100_Q6AUC7 Putative polyprotein [Oryza sativa]
Length = 1241
Score = 443 bits (1140), Expect = e-123
Identities = 221/422 (52%), Positives = 293/422 (69%), Gaps = 23/422 (5%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN-- 259
+LDT +HMCP RD F T+E V G VLMG+D C+VA IG V K DG +RTL +
Sbjct: 210 ILDTACTYHMCPNRDWFATYEAVQGGTVLMGDDTPCEVAGIGTVQIKMFDGCIRTLLDVR 269
Query: 260 ------ESLVC---------KYSAEGGVLMVSKGYLVLLKAS-RIDSLYVLQGIVVTGS- 302
SL+ KYS G+L V+KG LV++KA + +LY L+G + G+
Sbjct: 270 HIPNLKRSLISLCTLDRKGYKYSGGDGILKVTKGSLVVMKADIKYANLYHLRGTTILGNV 329
Query: 303 AAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVS 362
AAVS S+ D T LWHM LGHM+E G+ LSK+G L Q I KL+FC+H +F K K+V
Sbjct: 330 AAVSDSLSNSDATNLWHMRLGHMSEIGLAELSKRGLLDGQSIGKLKFCEHCIFGKHKRVK 389
Query: 363 FSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTF 422
F+T+TH T+GILDY+HSDLWGP++ TS+GG RYMMTI+DD+ RKVW YFL++K + F F
Sbjct: 390 FNTSTHTTEGILDYVHSDLWGPARKTSFGGARYMMTIVDDYSRKVWPYFLKHKYQAFDVF 449
Query: 423 KKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMI 482
K+W+ +VE QT + VK L TDN +E CS F +C + GI RH T+P PQQNGVAERM
Sbjct: 450 KEWKTMVERQTERKVKILRTDNGMELCSKIFKSYCKSEGIVRHYTVPHTPQQNGVAERMN 509
Query: 483 RTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSN 542
RT++ +ARCMLSNA L + W EA STAC+L+NRSP A+D K P ++WSG+ +YS+
Sbjct: 510 RTIISKARCMLSNASL--PKQFWAEAVSTACYLINRSPSYAIDKKTPIEVWSGSPANYSD 567
Query: 543 LRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDA 602
LR+FGC AYA V++GKL PR ++CIFL Y S KGY+LWC P+++K+++SR+V F+E
Sbjct: 568 LRVFGCTAYAHVDNGKLEPRVIKCIFLGYLSGVKGYKLWC--PETKKVVISRNVVFHESI 625
Query: 603 LL 604
+L
Sbjct: 626 ML 627
>UniRef100_Q7XTM9 OSJNBa0033G05.13 protein [Oryza sativa]
Length = 1181
Score = 441 bits (1134), Expect = e-122
Identities = 222/422 (52%), Positives = 295/422 (69%), Gaps = 24/422 (5%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN-- 259
+LDT +HMCP RD F T+E V G VLMG+D C+VA IG V K DG +RTL++
Sbjct: 301 ILDTACTYHMCPNRDWFATYEVVQGGTVLMGDDTPCEVAGIGTVQIKMFDGCIRTLSDVR 360
Query: 260 ------ESLVC---------KYSAEGGVLMVSKGYLVLLKAS-RIDSLYVLQGIVVTGSA 303
SL+ KYS G+L V+KG LV++KAS + +LY LQG + G+
Sbjct: 361 HIPNLKRSLISLCTLDRKGYKYSGGDGILKVTKGSLVVMKASIKSANLYHLQGTTILGNV 420
Query: 304 A-VSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVS 362
A VS S+ D T LWHM LGHM+E G+ LSK+G L Q I KL+FC+H +F K K+V
Sbjct: 421 ATVSDSLSNSDATNLWHMRLGHMSEIGLAELSKRGLLDGQSISKLKFCEHCIFGKHKRVK 480
Query: 363 FSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTF 422
F+T+TH T+GILDY+HSDLWGP++ TS+GG RYMMTI+DD+ RKVW YFL++K + F F
Sbjct: 481 FNTSTHTTEGILDYVHSDLWGPARKTSFGGARYMMTIVDDYSRKVWPYFLKHKYQAFNVF 540
Query: 423 KKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMI 482
K+W+ +VE QT + VK L TDN +EFCS F +C + GI RH T+P PQQNGVAERM
Sbjct: 541 KEWKTMVERQTERKVKILRTDNGMEFCSKIFKSYCKSEGIVRHYTVPHTPQQNGVAERMN 600
Query: 483 RTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSN 542
R ++ +ARCMLSNAGL + W EA STAC+L+NRSP S + K P ++WSG+ +YS+
Sbjct: 601 RIIISKARCMLSNAGL--PKQFWAEAVSTACYLINRSP-SYANKKTPIEVWSGSPANYSD 657
Query: 543 LRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDA 602
L++FGC AYA V++GKL PRA++CIFL Y S KGY+LWC P+++K+++SR+V F+E
Sbjct: 658 LKVFGCTAYAHVDNGKLEPRAIKCIFLGYPSSVKGYKLWC--PETKKVVISRNVVFHESI 715
Query: 603 LL 604
+L
Sbjct: 716 ML 717
>UniRef100_Q8LNW7 Putative polyprotein [Oryza sativa]
Length = 1280
Score = 435 bits (1119), Expect = e-120
Identities = 218/422 (51%), Positives = 290/422 (68%), Gaps = 23/422 (5%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN-- 259
+LDT + +HMCP RD F T+E + G VLMG+D C+VA IG V K DG +RTL++
Sbjct: 336 ILDTAWTYHMCPNRDWFATYEALQGGTVLMGDDTPCEVAGIGTVQIKMFDGYIRTLSDVR 395
Query: 260 ------ESLVC---------KYSAEGGVLMVSKGYLVLLKAS-RIDSLYVLQGIVVTGS- 302
SL+ KYS G+L V+KG LV++KA + +LY L+G + G+
Sbjct: 396 HIPNLKRSLISLCTLDRKGYKYSGGDGILKVTKGSLVVMKADIKSANLYHLRGTTILGNV 455
Query: 303 AAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVS 362
AAVS S+ D T LWHM LGHM+E G+ LSK+ L Q I KL+FC+H +F K K+V
Sbjct: 456 AAVSDSLSNSDATNLWHMRLGHMSEIGLAELSKRELLDGQSIGKLKFCEHCIFGKHKRVK 515
Query: 363 FSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTF 422
F+T+TH T+GILDY+HSDLWGP+ TS+GG RYMMTI+DD+ RKVW YFL++K + F F
Sbjct: 516 FNTSTHTTEGILDYVHSDLWGPACKTSFGGARYMMTIVDDYSRKVWPYFLKHKYQAFDVF 575
Query: 423 KKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMI 482
K+W+ +VE QT K VK L TDN +EFCS F +C + GI H T+P PQQNGVAERM
Sbjct: 576 KEWKTMVERQTEKKVKILRTDNGMEFCSKIFKSYCKSEGIVHHYTVPHTPQQNGVAERMN 635
Query: 483 RTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSN 542
++ +ARCMLSNA L + W EA ST C+L+NRSP A D K P ++WSG+ +YS+
Sbjct: 636 MAIISKARCMLSNADL--PKQFWAEAVSTTCYLINRSPSYATDKKTPIEVWSGSPANYSD 693
Query: 543 LRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDA 602
LR+FGC AYA V++GKL PRA++CIFL Y S KGY+LWC P+++K+++SR+V F+E
Sbjct: 694 LRVFGCTAYAHVDNGKLEPRAIKCIFLGYPSGVKGYKLWC--PETKKVVISRNVVFHESV 751
Query: 603 LL 604
+L
Sbjct: 752 IL 753
>UniRef100_Q7X670 OSJNBa0093P23.9 protein [Oryza sativa]
Length = 944
Score = 433 bits (1113), Expect = e-120
Identities = 220/422 (52%), Positives = 292/422 (69%), Gaps = 27/422 (6%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN-- 259
+LDT +HMCP RD F T+E V G VLMG+D C+VA IGIV K DG + TL++
Sbjct: 133 ILDTACTYHMCPNRDWFATYEAVQGGTVLMGDDTPCEVAGIGIVQIKMFDGCISTLSDVR 192
Query: 260 ------ESLVC---------KYSAEGGVLMVSKGYLVLLKAS-RIDSLYVLQGIVVTGS- 302
SL+ KYS G+L V+KG LV++KA + +LY L+G + G+
Sbjct: 193 HIPNLKRSLISLCTLDRKGYKYSGGDGILKVTKGSLVVMKADIKSANLYHLRGTTILGNV 252
Query: 303 AAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVS 362
AAVS S+ D T LWHM LGHM+E G+ LSK+G L Q I KL+FC+H +F K K+V
Sbjct: 253 AAVSDSLSNSDATNLWHMRLGHMSEIGLAELSKRGLLDGQSIGKLKFCEHCIFGKHKRVK 312
Query: 363 FSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTF 422
F+T+TH T+GILDY+HSDLWGP++ TS+GG RY+MTI+DD+ RKVW YFL++K + F F
Sbjct: 313 FNTSTHTTEGILDYVHSDLWGPARKTSFGGARYIMTIVDDYSRKVWPYFLKHKYQAFDVF 372
Query: 423 KKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMI 482
K+W+ +VE QT + VK L TDN +EFCS F +C + GI RH T PQQNGVAERM
Sbjct: 373 KEWKTMVERQTERKVKILRTDNGMEFCSKIFKSYCKSEGIVRHYT----PQQNGVAERMN 428
Query: 483 RTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSN 542
RT++ +ARCMLSNAGL + W A ST C+L+NRSP A+D K P +WSG +YS+
Sbjct: 429 RTIISKARCMLSNAGL--PKQFWAAAVSTTCYLINRSPSYAIDKKTPIKVWSGFPANYSD 486
Query: 543 LRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDA 602
LR+FGC AYA V++GKL PRA++CIFL Y+S KGY+LWC P+++K+++SR+V F+E
Sbjct: 487 LRVFGCTAYAHVDNGKLEPRAIKCIFLGYSSGVKGYKLWC--PETKKVVISRNVVFHESV 544
Query: 603 LL 604
+L
Sbjct: 545 ML 546
>UniRef100_Q7XFG2 Putative retroelement [Oryza sativa]
Length = 1225
Score = 423 bits (1087), Expect = e-117
Identities = 210/405 (51%), Positives = 279/405 (68%), Gaps = 28/405 (6%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTNES 261
+LDT +HMC RD F T+E V G VLMG+D C+VA
Sbjct: 301 ILDTACTYHMCSNRDWFATYEAVQGGTVLMGDDTPCEVAGY------------------- 341
Query: 262 LVCKYSAEGGVLMVSKGYLVLLKAS-RIDSLYVLQGIVVTGS-AAVSSSMPKKDVTKLWH 319
KYS G+L V+KG LV++KA + +LY L+G + G+ AAVS S+ D T LWH
Sbjct: 342 ---KYSGGDGILKVTKGSLVVMKADIKSANLYHLRGTTILGNVAAVSDSLSNSDATNLWH 398
Query: 320 MHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFSTATHRTKGILDYIHS 379
M LGHM+E G+ LSK+G L Q I KL+FC+H +F K K+V F+T+TH T+GILDY+HS
Sbjct: 399 MRLGHMSEIGLAELSKRGLLDGQSIKKLKFCEHCIFGKHKRVKFNTSTHTTEGILDYVHS 458
Query: 380 DLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKK 439
DLWGP++ TS+GG RYMMTI+DD+ RKVW YFL++K + F FK+W+ +VE QT + VK
Sbjct: 459 DLWGPARKTSFGGARYMMTIVDDYSRKVWPYFLKHKYQAFDVFKEWKTMVERQTERKVKI 518
Query: 440 LITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL* 499
L TDN ++FCS F +C + GI RH T+P PQQNGVAERM RT++ +ARCMLSNAGL
Sbjct: 519 LRTDNGMDFCSKIFKSYCKSEGIVRHYTVPHTPQQNGVAERMNRTIISKARCMLSNAGL- 577
Query: 500 N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYALVNDGKL 559
+ W EA STAC+L+NRSP A+D K P +WSG+ +YS+LR+FGC AYA V++ KL
Sbjct: 578 -PKQFWAEAVSTACYLINRSPSYAIDKKTPIKVWSGSPANYSDLRVFGCIAYAHVDNSKL 636
Query: 560 APRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDALL 604
PRA++CIFL Y S KGY+LWC P+++K+++SR+V F+E +L
Sbjct: 637 EPRAIKCIFLGYPSGVKGYKLWC--PETKKVVISRNVVFHESVML 679
>UniRef100_Q6L4V3 Putative polyprotein [Oryza sativa]
Length = 1243
Score = 419 bits (1076), Expect = e-115
Identities = 210/400 (52%), Positives = 281/400 (69%), Gaps = 18/400 (4%)
Query: 215 RDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN--------ESLVCKY 266
+D F T+E + G VLMG+D C+VA IG V K DG +RTL++ SL+ Y
Sbjct: 299 QDWFATYEALQGGTVLMGDDTPCEVAGIGTVQIKMFDGCIRTLSDVQHIPNLKRSLISLY 358
Query: 267 SAEGGVLMVSKGYLVLLKAS-RIDSLYVLQGIVVTGS-AAVSSSMPKKDVTKLWHMHLGH 324
G+L V+KG LV++K + +LY L+G + G+ AAV S+ D T LWHM LGH
Sbjct: 359 ----GILKVTKGSLVVMKVDIKSANLYHLRGTTILGNVAAVFDSLSNSDATNLWHMRLGH 414
Query: 325 MTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFSTATHRTKGILDYIHSDLWGP 384
M+E G+ LSK+G L Q I KL+FC+H +F K K+V F+T+TH T+GILDY+HSDLWGP
Sbjct: 415 MSEIGLAELSKRGLLDGQSIRKLKFCEHCIFGKHKRVKFNTSTHTTEGILDYVHSDLWGP 474
Query: 385 SKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN 444
+ TS+GG RYMMTI+DD+ RKVW YFL++K + F FK+W+ +VE QT + VK L TDN
Sbjct: 475 AHKTSFGGARYMMTIVDDYSRKVWPYFLKHKYQAFDGFKEWKTMVERQTERKVKILRTDN 534
Query: 445 *LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDL 504
+EFCS F +C + GI H T P PQQN VAERM RT++ +ARCMLSNAGL +
Sbjct: 535 GMEFCSKIFKSYCKSEGIVCHYTAPHTPQQNDVAERMNRTIISKARCMLSNAGL--PKQF 592
Query: 505 WVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYALVNDGKLAPRAV 564
W EA STAC+L+NRSP A+D K P ++WSG+ +YS+LR+FGC AYA V++GKL PRA+
Sbjct: 593 WAEAVSTACYLINRSPGYAIDKKTPIEVWSGSPTNYSDLRVFGCTAYAHVDNGKLEPRAI 652
Query: 565 ECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDALL 604
+CIFL YAS KGY+LWC P+++K+++SR+V F+E +L
Sbjct: 653 KCIFLGYASGVKGYKLWC--PETKKVVISRNVVFHESVIL 690
>UniRef100_Q94LG0 Putative retroelement pol polyprotein [Oryza sativa]
Length = 1326
Score = 395 bits (1016), Expect = e-108
Identities = 211/426 (49%), Positives = 281/426 (65%), Gaps = 28/426 (6%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCG-VVLMGNDVQCKVA*IGIVHFKTNDGVVRTL--- 257
+LDT +FH+C RD F++++ V G VV MG+D ++ IG V KT+DG+ RTL
Sbjct: 293 ILDTACSFHICINRDWFSSYKSVQNGDVVRMGDDNPREIVGIGSVQIKTHDGMTRTLKDV 352
Query: 258 --------------TNESLVCKYSAEGGVLMVSKGYLV-LLKASRIDSLYVLQGIVVTGS 302
T ++ KYS+ GGV+ VSKG LV ++ +LYVL+G + GS
Sbjct: 353 RHIPGMARNLISLSTLDAEGYKYSSSGGVVKVSKGSLVYMIGDMNSANLYVLRGSTLHGS 412
Query: 303 ---AAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQK 359
AAVS P K T LWHM LGHM+E GM L K+ L K++FC+H VF K K
Sbjct: 413 VTAAAVSKDEPIK--TNLWHMRLGHMSELGMAELMKRNLLDGCTQGKMKFCEHCVFGKHK 470
Query: 360 KVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETF 419
+V F+T+ HRTKGILDY+H+DLWGPS+ GG RYM+TIIDD+ RKVW YFL++K++TF
Sbjct: 471 RVKFNTSVHRTKGILDYVHTDLWGPSRKAYLGGARYMLTIIDDYSRKVWPYFLKHKDDTF 530
Query: 420 PTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAE 479
FK+W++ +E QT K VK L TDN EFCS F+++C GI RH TIP PQQNGVAE
Sbjct: 531 AAFKEWKVRIERQTEKEVKVLRTDNGGEFCSDAFDDYCRKEGIVRHHTIPYTPQQNGVAE 590
Query: 480 RMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVD 539
RM RT++ +ARCMLSNA + + W EAA+TAC+L+NRSP L+ K P ++WSG D
Sbjct: 591 RMNRTIISKARCMLSNARM--NKRFWAEAANTACYLINRSPSIPLNKKTPIEVWSGMPAD 648
Query: 540 YSNLRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFN 599
YS LR+FGC AYA V++GKL PRA++C+FL Y S K Y+LW +P++ K + R V FN
Sbjct: 649 YSQLRVFGCTAYAHVDNGKLEPRAIKCLFLGYGSGVKRYKLW--NPETNKTFMRRSVVFN 706
Query: 600 EDALLS 605
+ + +
Sbjct: 707 KSVMFN 712
>UniRef100_Q75IK3 Putative polyprotein [Oryza sativa]
Length = 1175
Score = 395 bits (1014), Expect = e-108
Identities = 208/426 (48%), Positives = 280/426 (64%), Gaps = 28/426 (6%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCG-VVLMGNDVQCKVA*IGIVHFKTNDGVVRTL--- 257
+LDT +FH+C RD F++++ V G VV MG+D ++ IG V KT+DG+ RTL
Sbjct: 188 ILDTTCSFHICINRDWFSSYKSVQNGDVVRMGDDNPREIVGIGSVQIKTHDGMTRTLKDV 247
Query: 258 --------------TNESLVCKYSAEGGVLMVSKGYLV-LLKASRIDSLYVLQGIVVTG- 301
T ++ KYS GGV+ VSKG LV ++ +LYVL+G + G
Sbjct: 248 RHIPRMARNLISLSTLDAEGYKYSGSGGVVKVSKGSLVYMIGDMNSANLYVLRGSTLHGY 307
Query: 302 --SAAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQK 359
+A VS P K T +WHM LGHM+E GM L K+ L ++FC+H VF K K
Sbjct: 308 VTAAVVSKDEPSK--TNMWHMRLGHMSELGMAELMKRNLLDGCTQGNMKFCEHCVFGKHK 365
Query: 360 KVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETF 419
+V F+T+ HRTKGILDY+H+DLWGPS+ S GG RYM+TIIDD+ RKVW YFL++K++TF
Sbjct: 366 RVKFNTSVHRTKGILDYVHADLWGPSRKPSLGGARYMLTIIDDYSRKVWPYFLKHKDDTF 425
Query: 420 PTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAE 479
FK+W+++++ QT K VK L TDN FCS F+++C GI H TIP PQQNGVAE
Sbjct: 426 AAFKEWKVMIKRQTEKEVKVLRTDNGGGFCSDAFDDYCRKEGIVMHHTIPYTPQQNGVAE 485
Query: 480 RMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVD 539
RM RT++ +ARCMLSNA + + W EAA TAC+L+NRSP +L+ K P ++WSG +
Sbjct: 486 RMNRTIISKARCMLSNARM--NKRFWAEAAKTACYLINRSPSISLNKKTPIEVWSGMPAN 543
Query: 540 YSNLRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFN 599
YS LR+FGC AYA VN+GKL PRA++C+FL Y S KGY+LW +P++ K +SR V FN
Sbjct: 544 YSQLRVFGCTAYAHVNNGKLEPRAIKCLFLGYGSGVKGYKLW--NPETNKTFMSRSVVFN 601
Query: 600 EDALLS 605
E + +
Sbjct: 602 ESVMFN 607
>UniRef100_Q75HA9 Putative polyprotein [Oryza sativa]
Length = 1322
Score = 393 bits (1009), Expect = e-108
Identities = 211/421 (50%), Positives = 279/421 (66%), Gaps = 28/421 (6%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDC-GVVLMGNDVQCKVA*IGIVHFKTNDGVVRTL--- 257
+LDT +FH+C RD F++++ V VV MG+D ++ IG V KT+DG+ RTL
Sbjct: 293 ILDTACSFHICINRDWFSSYKSVQNEDVVRMGDDNPREIVGIGSVQIKTHDGMTRTLKDV 352
Query: 258 --------------TNESLVCKYSAEGGVLMVSKGYLV-LLKASRIDSLYVLQGIVVTGS 302
T ++ KYS GGV+ VSKG LV ++ +LYVL+G + GS
Sbjct: 353 RHIPGMARNLISLSTLDAEGYKYSGSGGVVKVSKGSLVYMIGDMNSANLYVLRGSTLHGS 412
Query: 303 ---AAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQK 359
AAV+ P K T LWHM LGHM+E GM L K+ L ++FC+H VF K K
Sbjct: 413 VTAAAVTKDEPSK--TNLWHMRLGHMSELGMAELMKRNLLDGCTQGNMKFCEHCVFGKHK 470
Query: 360 KVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETF 419
+V F+T+ HRTKGILDY+H+DLWGPS+ S GG RYM+TIIDD+ RK W YFL++K++TF
Sbjct: 471 RVKFNTSVHRTKGILDYVHADLWGPSRKPSLGGARYMLTIIDDYSRKEWPYFLKHKDDTF 530
Query: 420 PTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAE 479
FK+ ++++E QT K VK L TDN EFCS F+++C GI RH TIP PQQNGVAE
Sbjct: 531 AAFKERKVMIERQTEKEVKVLCTDNGGEFCSDAFDDYCRKEGIVRHHTIPYTPQQNGVAE 590
Query: 480 RMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVD 539
RM RT++ +ARCMLSNA + + W EAA+TAC+L+NRSP L+ K P +IWSG D
Sbjct: 591 RMNRTIISKARCMLSNARM--NKRFWAEAANTACYLINRSPSIPLNKKTPIEIWSGMPAD 648
Query: 540 YSNLRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFN 599
YS LR+FGC AYA V++GKL PRA++C+FL Y S KGY+LW +P++ K +SR+V FN
Sbjct: 649 YSQLRVFGCTAYAHVDNGKLEPRAIKCLFLGYGSGVKGYKLW--NPETNKTFMSRNVIFN 706
Query: 600 E 600
E
Sbjct: 707 E 707
>UniRef100_Q9AYJ5 Putative pol polyprotein [Oryza sativa]
Length = 1005
Score = 370 bits (951), Expect = e-101
Identities = 201/412 (48%), Positives = 265/412 (63%), Gaps = 28/412 (6%)
Query: 216 DLFTTFEPVDCG-VVLMGNDVQCKVA*IGIVHFKTNDGVVRTL----------------- 257
D F++++ V G VV MG+D ++ IG V KT+DG+ TL
Sbjct: 44 DWFSSYKSVQNGDVVRMGDDNPREIVGIGSVQIKTHDGMTCTLKDVRHILGMARNLISLS 103
Query: 258 TNESLVCKYSAEGGVLMVSKGYLV-LLKASRIDSLYVLQGIVVTGS---AAVSSSMPKKD 313
T ++ KYS GGV+ VSKG LV ++ +LYVL+G + GS AAV+ P K
Sbjct: 104 TLDAEGYKYSGSGGVVKVSKGSLVYIIGDMNSANLYVLRGSTLHGSVTAAAVTKDEPSK- 162
Query: 314 VTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFSTATHRTKGI 373
T LWHM LGHM+E GM L K+ L ++FC+H VF K K V F+ + HR KGI
Sbjct: 163 -TNLWHMRLGHMSELGMAELMKRNLLDGCTQGNMKFCEHCVFGKHKWVKFNISVHRIKGI 221
Query: 374 LDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQT 433
LDY+H+DLWGPS S GG RYM+TIIDD RKVW YFL++K++TF FK+W++++E QT
Sbjct: 222 LDYVHADLWGPSHKPSLGGARYMLTIIDDHSRKVWPYFLKHKDDTFAAFKEWKVMIERQT 281
Query: 434 GKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCML 493
K VK L TDN EFCS F+++ GI RH TIP PQQNGVAERM RT++ +A CML
Sbjct: 282 EKEVKVLRTDNGGEFCSDAFDDYYRKEGIVRHYTIPYTPQQNGVAERMNRTIISKAHCML 341
Query: 494 SNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYAL 553
SNA + + W EAA+TAC+L+NRSP L+ K P ++WSG DYS LR+FGC AYA
Sbjct: 342 SNARM--NKRFWAEAANTACYLINRSPSIPLNKKTPIEVWSGMSADYSQLRVFGCTAYAH 399
Query: 554 VNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDALLS 605
V++GKL PRA++C+FL Y S KGY+LW +P++ K +SR V FNE + +
Sbjct: 400 VDNGKLEPRAIKCLFLGYGSGVKGYKLW--NPETNKTFMSRSVIFNESVMFN 449
>UniRef100_Q9ZPU5 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1335
Score = 350 bits (897), Expect = 9e-95
Identities = 187/444 (42%), Positives = 267/444 (60%), Gaps = 24/444 (5%)
Query: 183 LILAMTLLYLLSPLIRGVKLLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*I 242
++LA +++ I +LDT +FHM P +D F F+ + G V MGND V I
Sbjct: 267 VLLATDETLVVTDSIANEWVLDTGCSFHMTPRKDWFKDFKELSSGYVKMGNDTYSPVKGI 326
Query: 243 GIVHFKTNDGVVRTLTN-----------------ESLVCKYSAEGGVLMVSKGYLVLLKA 285
G + + +DG LT+ E C + ++ G+L + KG +LK
Sbjct: 327 GSIKIRNSDGSQVILTDVRYMPNMTRNLISLGTLEDRGCWFKSQDGILKIVKGCSTILKG 386
Query: 286 SRIDSLYVLQGIVVTGSAAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGID 345
+ D+LY+L G+ G + SS KD T LWH LGHM++KGM +L K+G L ++ I
Sbjct: 387 QKRDTLYILDGVTEEGES--HSSAEVKDETALWHSRLGHMSQKGMEILVKKGCLRREVIK 444
Query: 346 KLEFCKHYVFWKQKKVSFSTATHRTKGILDYIHSDLWG-PSKVTSYGGRRYMMTIIDDFP 404
+LEFC+ V+ KQ +VSF+ A H TK L Y+HSDLWG P S G +Y ++ +DD+
Sbjct: 445 ELEFCEDCVYGKQHRVSFAPAQHVTKEKLAYVHSDLWGSPHNPASLGNSQYFISFVDDYS 504
Query: 405 RKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIAR 464
RKVW+YFLR K+E F F +W+ +VE Q+ + VKKL TDN LE+C+ F +FC GI R
Sbjct: 505 RKVWIYFLRKKDEAFEKFVEWKKMVENQSDRKVKKLRTDNGLEYCNHYFEKFCKEEGIVR 564
Query: 465 HKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSAL 524
HKT PQQNG+AER+ RT++++ R MLS +G+ + W EAASTA +L+NRSP +A+
Sbjct: 565 HKTCAYTPQQNGIAERLNRTIMDKVRSMLSRSGM--EKKFWAEAASTAVYLINRSPSTAI 622
Query: 525 DFKVPEDIWSGNLVDYSNLRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSD 584
+F +PE+ W+G L D S+LR FGC AY + GKL PR+ + IF SY KGY++W +
Sbjct: 623 NFDLPEEKWTGALPDLSSLRKFGCLAYIHADQGKLNPRSKKGIFTSYPEGVKGYKVWVLE 682
Query: 585 PKSQKLILSRDVTFNEDALLSSGK 608
K K ++SR+V F E + K
Sbjct: 683 DK--KCVISRNVIFREQVMFKDLK 704
>UniRef100_Q9SH77 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1356
Score = 314 bits (804), Expect = 6e-84
Identities = 176/420 (41%), Positives = 247/420 (57%), Gaps = 24/420 (5%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN-- 259
+LDT ++HM R+ F F G V MGN +V +G + K +DG+ LTN
Sbjct: 310 ILDTGCSYHMTYKREWFHEFNEDAGGSVRMGNKTVSRVRGVGTIRVKNSDGLTIVLTNVR 369
Query: 260 ---------------ESLVCKYSAEGGVLMVSKGYLVLLKASRIDSLYVLQGIVVTGSAA 304
E K+ +E G+L + G VLL R D+LY+L V +
Sbjct: 370 YIPDMDRNLLSLGTFEKAGYKFESEDGILRIKAGNQVLLTGRRYDTLYLLNWKPVASESL 429
Query: 305 VSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFS 364
+ + + D T LWH L HM++K M +L ++G L K+ + L+ C+ ++ K K+ SFS
Sbjct: 430 --AVVKRADDTVLWHQRLCHMSQKNMEILVRKGFLDKKKVSSLDVCEDCIYGKAKRKSFS 487
Query: 365 TATHRTKGILDYIHSDLWG-PSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFK 423
A H TK L+YIHSDLWG P S G +Y M+IIDDF RKVWVYF++ K+E F F
Sbjct: 488 LAHHDTKEKLEYIHSDLWGAPFVPLSLGKCQYFMSIIDDFTRKVWVYFMKTKDEAFEKFV 547
Query: 424 KWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIR 483
+W LVE QT + VK L TDN LEFC+ F+ FC + GI RH+T PQQNGVAERM R
Sbjct: 548 EWVNLVENQTDRRVKTLRTDNGLEFCNKLFDGFCESIGIHRHRTCAYTPQQNGVAERMNR 607
Query: 484 TLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNL 543
T++E+ R MLS++GL + W EA T L+N++P SAL+F++P+ WSGN YS L
Sbjct: 608 TIMEKVRSMLSDSGL--PKRFWAEATHTTVLLINKTPSSALNFEIPDKKWSGNPPVYSYL 665
Query: 544 RIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDAL 603
R +GC A+ +DGKL PRA + + + Y KGY++W D +K ++SR++ F E+A+
Sbjct: 666 RRYGCVAFVHTDDGKLEPRAKKGVLIGYPVGVKGYKVWILD--ERKCVVSRNIIFQENAV 723
>UniRef100_Q8W5D4 Putative retrotransposon-related protein [Oryza sativa]
Length = 1229
Score = 312 bits (799), Expect = 2e-83
Identities = 182/421 (43%), Positives = 244/421 (57%), Gaps = 67/421 (15%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCG-VVLMGNDVQCKVA*IGIVHFKTNDGVVRTL--- 257
+LDT F +C RD F++ + V G VV MG++ ++ IG V KT+DG+ RTL
Sbjct: 290 ILDTACLFLICINRDWFSSHKSVQNGDVVRMGDNNPREIMGIGSVQIKTHDGMTRTLKDV 349
Query: 258 --------------TNESLVCKYSAEGGVLMVSKGYLV-LLKASRIDSLYVLQGIVVTGS 302
T ++ KYS GGV+ VSKG LV ++ +LYVL+G + GS
Sbjct: 350 RHIPGMARNLISLSTLDAEGYKYSGSGGVVKVSKGSLVYMIGDMNSANLYVLRGSTLHGS 409
Query: 303 ---AAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQK 359
AAVS P K T LWHM LGHM+E GM L K+ L ++FC+H VF K K
Sbjct: 410 LTAAAVSKDEPSK--TNLWHMRLGHMSELGMAELMKRNLLDGCTQGNMKFCEHCVFGKHK 467
Query: 360 KVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETF 419
+V F+T+ HRTKGILDY+H+DLWGPS+ S GG YM+TIIDD+ RKVW YFL++K++TF
Sbjct: 468 RVKFNTSVHRTKGILDYVHADLWGPSRKPSLGGACYMLTIIDDYSRKVWPYFLKHKDDTF 527
Query: 420 PTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAE 479
FK+W++++E Q K VK L TDN EFCS F+++C GI RH TIP PQQN VAE
Sbjct: 528 AAFKEWKVMIERQAEKEVKVLRTDNGGEFCSDAFDDYCRKEGIGRHHTIPYTPQQNSVAE 587
Query: 480 RMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVD 539
RM RT++ +ARCMLSNA + + W EAA+T C+L+NRSP L+ + P ++WSG
Sbjct: 588 RMNRTIISKARCMLSNARM--NKHYWAEAANTTCYLINRSPSILLNKETPIEVWSG---- 641
Query: 540 YSNLRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFN 599
KGY+LW +P++ K + R V FN
Sbjct: 642 -----------------------------------VKGYKLW--NPETNKTFMRRSVVFN 664
Query: 600 E 600
+
Sbjct: 665 K 665
>UniRef100_Q9SJT2 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1333
Score = 308 bits (790), Expect = 2e-82
Identities = 180/427 (42%), Positives = 245/427 (57%), Gaps = 24/427 (5%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN-- 259
++DT ++HM R+ F G V MGN KV IG + K G+V LTN
Sbjct: 288 VMDTGCSYHMTYKREWFEDLNEDAGGSVRMGNKTVSKVRGIGTIRVKNEAGMVVRLTNVR 347
Query: 260 ---------------ESLVCKYSAEGGVLMVSKGYLVLLKASRIDSLYVLQGIVVTGSAA 304
E + E G L + G VLL R +LY+LQ VT +
Sbjct: 348 YIPEMDRNLLSLGTFEKSGYSFKLENGTLSIIAGDSVLLTVRRCYTLYLLQWRPVTEESL 407
Query: 305 VSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFS 364
S + ++D T LWH LGHM++K M LL K+G L K+ + KLE C+ ++ K K++ F+
Sbjct: 408 --SVVKRQDDTILWHRRLGHMSQKNMDLLLKKGLLDKKKVSKLETCEDCIYGKAKRIGFN 465
Query: 365 TATHRTKGILDYIHSDLWG-PSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFK 423
A H T+ L+Y+HSDLWG PS S G +Y ++ IDD+ RKV +YFL+ K+E F F
Sbjct: 466 LAQHDTREKLEYVHSDLWGAPSVPFSLGKCQYFISFIDDYTRKVRIYFLKTKDEAFDKFV 525
Query: 424 KWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIR 483
+W LVE QT K +K L TDN LEFC+ F+EFC+ GI H+T PQQNGVAERM R
Sbjct: 526 EWANLVENQTDKRIKTLRTDNGLEFCNRSFDEFCSQKGILWHRTCAYTPQQNGVAERMNR 585
Query: 484 TLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNL 543
TL+E+ R MLS++GL + W EA T L+N++P SAL+++VP+ WSG YS L
Sbjct: 586 TLMEKVRSMLSDSGL--PKKFWAEATHTTAILINKTPSSALNYEVPDKRWSGKSPIYSYL 643
Query: 544 RIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDAL 603
R FGC A+ +DGKL PRA + I + Y KGY++W + K K ++SR+V F E+A
Sbjct: 644 RRFGCIAFVHTDDGKLNPRAKKGILVGYPIGVKGYKIWLLEEK--KCVVSRNVIFQENAS 701
Query: 604 LSSGKQS 610
QS
Sbjct: 702 YKDMMQS 708
>UniRef100_Q9SHR5 F28L22.3 protein [Arabidopsis thaliana]
Length = 1356
Score = 307 bits (787), Expect = 5e-82
Identities = 169/426 (39%), Positives = 247/426 (57%), Gaps = 24/426 (5%)
Query: 196 LIRGVKLLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVR 255
+++ + +LD+ HM RD F +F+ +L+G+D + G + T+ G ++
Sbjct: 303 MVKDLWILDSGCTSHMTSRRDWFISFQEKGNTTILLGDDHSVESQGQGTIRIDTHGGTIK 362
Query: 256 TLTNESLV---------------CKYSAEGGVLMVS--KGYLVLLKASRIDSLYVLQGIV 298
L N V Y EGG V K L+ S + LYVL G
Sbjct: 363 ILENVKYVPHLRRNLISTGTLDKLGYRHEGGEGKVRYFKNNKTALRGSLSNGLYVLDGST 422
Query: 299 VTGSAAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQ 358
V + ++ K T LWH LGHM+ + +L+ +G + ++ I++LEFC+H V K
Sbjct: 423 VMSE--LCNAETDKVKTALWHSRLGHMSMNNLKVLAGKGLIDRKEINELEFCEHCVMGKS 480
Query: 359 KKVSFSTATHRTKGILDYIHSDLWGPSKVT-SYGGRRYMMTIIDDFPRKVWVYFLRYKNE 417
KKVSF+ H ++ L Y+H+DLWG VT S G++Y ++IIDD RKVW+YFL+ K+E
Sbjct: 481 KKVSFNVGKHTSEDALSYVHADLWGSPNVTPSISGKQYFLSIIDDKTRKVWLYFLKSKDE 540
Query: 418 TFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGV 477
TF F +W+ LVE Q K VK L TDN LEFC+S F+ +C HGI RH+T PQQNGV
Sbjct: 541 TFDKFCEWKSLVENQVNKKVKCLRTDNGLEFCNSRFDSYCKEHGIERHRTCTYTPQQNGV 600
Query: 478 AERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNL 537
AERM RT++E+ RC+L+ +G+ W EAA+TA +L+NRSP SA++ VPE++W
Sbjct: 601 AERMNRTIMEKVRCLLNKSGV--EEVFWAEAAATAAYLINRSPASAINHNVPEEMWLNRK 658
Query: 538 VDYSNLRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVT 597
Y +LR FG AY + GKL PRA++ FL Y + +KGY++W + +K ++SR+V
Sbjct: 659 PGYKHLRKFGSIAYVHQDQGKLKPRALKGFFLGYPAGTKGYKVWLLE--EEKCVISRNVV 716
Query: 598 FNEDAL 603
F E +
Sbjct: 717 FQESVV 722
>UniRef100_Q9SZY0 Putative retrotransposon [Arabidopsis thaliana]
Length = 1230
Score = 304 bits (779), Expect = 4e-81
Identities = 168/406 (41%), Positives = 242/406 (59%), Gaps = 15/406 (3%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTNES 261
++DT N+HM ++ F G V MGN K I N + TL
Sbjct: 313 VMDTGCNYHMTHKKEWFEELSEDAGGTVRMGNKSTSKFRVKYIPDMDRNLLSMGTLEEHG 372
Query: 262 LVCKYSAEGGVLMVSKGYLVLLKASRIDSLYVLQGIVVTGSAAVSSSMP---KKDVTKLW 318
+ ++ GVL+V +G LL SR + LY+LQG VS SM + D T LW
Sbjct: 373 Y--SFESKNGVLVVKEGTRTLLIGSRHEKLYLLQG-----KPEVSHSMTVERRNDDTVLW 425
Query: 319 HMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFSTATHRTKGILDYIH 378
H LGH+++K M +L K+G L + + KLE C+ ++ K +++SF ATH T+ L+Y+H
Sbjct: 426 HRRLGHISQKNMDILVKKGYLDGKKVSKLELCEDCIYGKARRLSFVVATHNTEDKLNYVH 485
Query: 379 SDLWG-PSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQTGKNV 437
SDLWG PS S G +Y ++ ID + RK WVYFL++K+E F TF +W ++VE QTG+ +
Sbjct: 486 SDLWGAPSVPLSLGKCQYFISFIDVYSRKTWVYFLKHKDEAFGTFAEWSVMVENQTGRKI 545
Query: 438 KKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAG 497
K L DN LEFC+ FN+FC GI RH+T PQQNGVAERM T++E+ R MLS +G
Sbjct: 546 KILRIDNGLEFCNQQFNDFCKEKGIVRHQTCAYTPQQNGVAERMNHTIMEKVRRMLSYSG 605
Query: 498 L*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYALVNDG 557
L + W EA +T L+N++P SA++F++ + WSG Y+ L+ FGC A+ ++G
Sbjct: 606 L--PKTFWAEATNTVVTLINKTPSSAVNFEISDKRWSGKSPVYNYLKRFGCVAFTYADEG 663
Query: 558 KLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDAL 603
KL PRA + +FL Y S KGY++W + +K +SR+VTF E+A+
Sbjct: 664 KLVPRAKKGVFLGYLSGEKGYKVWLLE--ERKCSVSRNVTFQENAV 707
>UniRef100_O81903 Putative transposable element [Arabidopsis thaliana]
Length = 1308
Score = 304 bits (778), Expect = 6e-81
Identities = 171/427 (40%), Positives = 247/427 (57%), Gaps = 34/427 (7%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN-- 259
++D+ +HM D F+ F + ++L+G+D + G V T+ G +R L N
Sbjct: 310 VIDSGCTYHMTSRMDWFSEFNENETTMILLGDDHTVESKGSGTVKVNTHGGSIRVLKNVR 369
Query: 260 ---------------ESLVCKYSAEGGVLMVSKGYLVLLKASRIDSLYVLQG-IVVTGSA 303
+ L K+ G + K L + ++ LYVL G VV +
Sbjct: 370 FVPNLRRNLISTGTLDKLGYKHEGGDGKVRFYKENKTALCGNLVNGLYVLDGHTVVNENC 429
Query: 304 AVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSF 363
V S K T+LWH LGHM+ M +L+++G L K+ I +L FC++ V K KK+SF
Sbjct: 430 NVEGSNEK---TELWHCRLGHMSLNNMKILAEKGLLEKKDIKELSFCENCVMGKSKKLSF 486
Query: 364 STATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFK 423
+ H T +L YIH+DLWG ++Y ++IIDD RKVW+ FL+ K+ETF F
Sbjct: 487 NVGKHITDEVLGYIHADLWG---------KQYFLSIIDDKSRKVWLMFLKTKDETFERFC 537
Query: 424 KWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIR 483
+W+ LVE Q K VK L TDN LEFC+ F+EFC +GI RH+T PQQNGVA+RM R
Sbjct: 538 EWKELVENQVNKKVKILRTDNGLEFCNLKFDEFCKQNGIERHRTCTYTPQQNGVAKRMNR 597
Query: 484 TLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNL 543
TL+E+ RC+L+ +GL W EAA+TA +LVNRSP SA+D VPE++W Y +L
Sbjct: 598 TLMEKVRCLLNESGL--EEVFWAEAAATAAYLVNRSPASAVDHNVPEELWLDKKPGYKHL 655
Query: 544 RIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDAL 603
R FGC AY ++ GKL PRA++ +FL Y +KGY++W D +K ++SR++ FNE+ +
Sbjct: 656 RRFGCIAYVHLDQGKLKPRALKGVFLGYPQGTKGYKVWLLD--EEKCVISRNIVFNENQV 713
Query: 604 LSSGKQS 610
++S
Sbjct: 714 YKDIRES 720
>UniRef100_Q9FFM0 Copia-like retrotransposable element [Arabidopsis thaliana]
Length = 1342
Score = 302 bits (773), Expect = 2e-80
Identities = 168/420 (40%), Positives = 234/420 (55%), Gaps = 40/420 (9%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN-- 259
+LDT +FHM +D F+ G V MGND +V IG V K DG LT+
Sbjct: 313 ILDTGCSFHMTCRKDWIIDFKETASGKVRMGNDTYSEVKGIGDVRIKNEDGSTILLTDVR 372
Query: 260 ---------------ESLVCKYSAEGGVLMVSKGYLVLLKASRIDSLYVLQGIVVTGSAA 304
E C + ++ G+L + K L +L + +LY LQG + G A
Sbjct: 373 YIPEMSKNLISLGTLEDKGCWFESKKGILTIFKNDLTVLTGKKESTLYFLQGTTLAGEAN 432
Query: 305 VSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFS 364
V +KD T LWH LGH+ KG+ +L +G L K + +SF
Sbjct: 433 VIDK--EKDETSLWHSRLGHIGAKGLQVLVSKGHLDKNIM----------------ISFG 474
Query: 365 TATHRTKGILDYIHSDLWGPSKVT-SYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFK 423
A H TK LDY+HSDLWG + V S G +Y +T IDDF R+ W+YF+R K+E F F
Sbjct: 475 AAKHVTKDKLDYVHSDLWGSTNVPFSIGKCQYFITFIDDFTRRTWIYFIRTKDEAFSKFV 534
Query: 424 KWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIR 483
+W+ +E Q K +K LITDN LEFC+ +F+ FC G+ RH+T PQQNGVAERM R
Sbjct: 535 EWKTQIENQQDKKLKILITDNGLEFCNQEFDSFCRKEGVIRHRTCAYTPQQNGVAERMNR 594
Query: 484 TLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNL 543
T++ + RCMLS +GL + W EAASTA L+N+SP S+++F +PE+ W+G+ DY L
Sbjct: 595 TIMNKVRCMLSESGL--GKQFWAEAASTAVFLINKSPSSSIEFDIPEEKWTGHPPDYKIL 652
Query: 544 RIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDAL 603
+ FG AY + GKL PRA + IFL Y K +++W + +K ++SRD+ F E+ +
Sbjct: 653 KKFGSVAYIHSDQGKLNPRAKKGIFLGYPDGVKRFKVWLLE--DRKCVVSRDIVFQENQM 710
>UniRef100_Q9AUZ1 Polyprotein, putative [Arabidopsis thaliana]
Length = 855
Score = 301 bits (772), Expect = 3e-80
Identities = 168/411 (40%), Positives = 233/411 (55%), Gaps = 24/411 (5%)
Query: 211 MCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTNESLVCK----- 265
M RD F +F+ D +L+G+D + G + T+ G + L N V
Sbjct: 1 MTSRRDWFCSFQEKDTTKILLGDDHSVESQGQGSIRLDTHGGTITILENVKYVPNLRRNL 60
Query: 266 ----------YSAEGGVLMVS--KGYLVLLKASRIDSLYVLQGIVVTGSAAVSSSMPKKD 313
Y EGG V K L+ S LYVL G V + ++ K+
Sbjct: 61 ISTGTLDRLGYKHEGGDGQVRYYKNNKTALRGSLSGGLYVLDGNTVIAESCIAER--SKE 118
Query: 314 VTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFSTATHRTKGI 373
+T LWH LGHM M +L+ +G + LEF +H V K KKVSF+ H ++ I
Sbjct: 119 LTTLWHSRLGHMGGNNMKILAGKGLIKPSEATSLEFYEHCVMGKAKKVSFNIGKHNSEEI 178
Query: 374 LDYIHSDLWGPSKVT-SYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQ 432
L Y+H+DLWG VT S G +Y ++IIDD RKVW+YFLR K+ETF F +W+ LVE Q
Sbjct: 179 LSYVHADLWGSQNVTPSMSGNKYFLSIIDDKSRKVWLYFLRSKDETFDKFCEWKELVENQ 238
Query: 433 TGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCM 492
T K VK L TDN LEFC+ F+ +C +GI RHKT PQQNGVAERM RT++E+ RC+
Sbjct: 239 TDKRVKCLRTDNGLEFCNIKFDSYCKKYGIERHKTCTYTPQQNGVAERMNRTVMEKVRCL 298
Query: 493 LSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYA 552
L+ +GL + W E A+TA +++NRSP +A+D VPE++W Y +LR AY
Sbjct: 299 LNESGL--EEEFWAEVATTAVYIINRSPSAAIDHNVPEELWLNRKPGYKHLRRLRAVAYV 356
Query: 553 LVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDAL 603
V+ GKL PRA++ IF+ Y S +KGY++W + QK ++SR+V F E+ +
Sbjct: 357 HVDQGKLKPRAIKGIFIGYPSGTKGYKVWLLE--EQKCVISRNVIFQEEVV 405
>UniRef100_Q6BCY1 Gag-Pol [Ipomoea batatas]
Length = 1298
Score = 298 bits (763), Expect = 3e-79
Identities = 168/424 (39%), Positives = 251/424 (58%), Gaps = 29/424 (6%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN-- 259
L+D+ +HM ++ F +EP+ G V +D ++ IG + K DG V+T+ +
Sbjct: 283 LIDSGATYHMTSRKEWFHHYEPISGGSVYSCDDHALEIIGIGTIKLKMYDGTVQTVQDVR 342
Query: 260 ---------------ESLVCKYSAEGGVLMVSKGYLVLLKASRIDS-LYVLQGIVVT-GS 302
++ + + GV+ + +G LV++K +I + LY+L+G +
Sbjct: 343 HVKGLKKNLLSYGILDNSATQIETQKGVMKIFQGALVVMKGEKIAANLYMLKGETLQEAE 402
Query: 303 AAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVS 362
A+V++ P D T LWH LGHM+++GM +L +Q + L C+H + KQ ++
Sbjct: 403 ASVAACSP--DSTLLWHQKLGHMSDQGMKILVEQKLIPGLTKVSLPLCEHCITSKQHRLK 460
Query: 363 FSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTF 422
FST+ R K +L+ +HSD+W + V S GG +Y ++ IDD+ R+ WVY ++ K++ F TF
Sbjct: 461 FSTSNSRGKVVLELVHSDVW-QAPVPSLGGAKYFVSFIDDYSRRCWVYPIKKKSDVFATF 519
Query: 423 KKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMI 482
K ++ VE +GK +K TDN E+ S +F++FC GI R T+ PQQNGVAERM
Sbjct: 520 KAFKARVELDSGKKIKCFRTDNGGEYTSEEFDDFCKKEGIKRQFTVAYTPQQNGVAERMN 579
Query: 483 RTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSN 542
RTLLER R ML AGL + W EA +TAC+LVNR+P +A++ K P ++W+G VDYSN
Sbjct: 580 RTLLERTRAMLRAAGL--EKSFWAEAVNTACYLVNRAPSTAIELKTPMEMWTGKPVDYSN 637
Query: 543 LRIFGCPAYALVND---GKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFN 599
L IFG YA+ N KL P++ +C FL YA KGYRLW DP + K+++SRDV F
Sbjct: 638 LHIFGSIVYAMYNAQEITKLDPKSRKCRFLGYADGVKGYRLW--DPTAHKVVISRDVIFV 695
Query: 600 EDAL 603
ED L
Sbjct: 696 EDRL 699
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.331 0.144 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 953,721,491
Number of Sequences: 2790947
Number of extensions: 38477714
Number of successful extensions: 98633
Number of sequences better than 10.0: 1375
Number of HSP's better than 10.0 without gapping: 982
Number of HSP's successfully gapped in prelim test: 393
Number of HSP's that attempted gapping in prelim test: 95788
Number of HSP's gapped (non-prelim): 1560
length of query: 610
length of database: 848,049,833
effective HSP length: 133
effective length of query: 477
effective length of database: 476,853,882
effective search space: 227459301714
effective search space used: 227459301714
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 78 (34.7 bits)
Medicago: description of AC149197.1


