

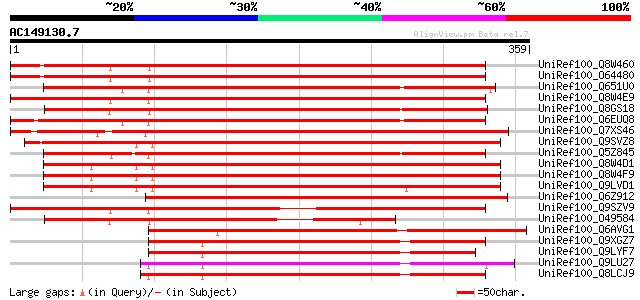
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149130.7 - phase: 0 /pseudo
(359 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8W460 Hypothetical protein At2g19160; T20K24.18 [Arab... 380 e-104
UniRef100_O64480 Hypothetical protein At2g19160 [Arabidopsis tha... 380 e-104
UniRef100_Q651U0 Hypothetical protein OSJNBa0051O02.41 [Oryza sa... 370 e-101
UniRef100_Q8W4E9 Hypothetical protein At4g30060; F6G3.90 [Arabid... 366 e-100
UniRef100_Q8GS18 Hypothetical protein [Arabidopsis thaliana] 364 2e-99
UniRef100_Q6EUQ8 Hypothetical protein OJ1077_E05.12 [Oryza sativa] 363 3e-99
UniRef100_Q7XS46 OSJNBa0035M09.18 protein [Oryza sativa] 360 3e-98
UniRef100_Q9SVZ8 Hypothetical protein AT4g25870 [Arabidopsis tha... 358 1e-97
UniRef100_Q5Z845 Hypothetical protein P0468G03.24 [Oryza sativa] 357 3e-97
UniRef100_Q8W4D1 Hypothetical protein At5g57270; MJB24.8 [Arabid... 356 6e-97
UniRef100_Q8W4F9 Hypothetical protein At5g57270; MJB24.8 [Arabid... 353 5e-96
UniRef100_Q9LVD1 Arabidopsis thaliana genomic DNA, chromosome 5,... 352 9e-96
UniRef100_Q6Z912 Hypothetical protein P0494D11.22 [Oryza sativa] 331 2e-89
UniRef100_Q9SZV9 Hypothetical protein F6G3.90 [Arabidopsis thali... 323 6e-87
UniRef100_O49584 Predicted protein [Arabidopsis thaliana] 240 5e-62
UniRef100_Q6AVG1 Expressed protein [Oryza sativa] 227 3e-58
UniRef100_Q9XGZ7 T1N24.19 protein [Arabidopsis thaliana] 215 1e-54
UniRef100_Q9LYF7 Hypothetical protein T22P22_120 [Arabidopsis th... 210 6e-53
UniRef100_Q9LU27 Gb|AAD40142.1 [Arabidopsis thaliana] 209 1e-52
UniRef100_Q8LCJ9 Hypothetical protein [Arabidopsis thaliana] 207 4e-52
>UniRef100_Q8W460 Hypothetical protein At2g19160; T20K24.18 [Arabidopsis thaliana]
Length = 394
Score = 380 bits (976), Expect = e-104
Identities = 187/339 (55%), Positives = 244/339 (71%), Gaps = 12/339 (3%)
Query: 1 MKTAKFWRLS-MGDMEILLGSPVLHRPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTSC 59
MK K WRL + M+ L G+ HR +KP+WI+ ++S I +F+ AY++ + +C
Sbjct: 1 MKAIKGWRLGKINYMQSLPGAR--HRAPTRKPIWIIAVLSLIAMFVIGAYMFPHHSKAAC 58
Query: 60 NMFYSKPCI-----IDISVLAVSHIPVSVYIERGITLDMPQ----NPKIAFMFLTPGSLP 110
MF SK C + S+ S ++ + L P+ + KIAFMFLTPG+LP
Sbjct: 59 YMFSSKGCKGLTDWLPPSLREYSDDEIAARVVISEILSSPRVIKKSSKIAFMFLTPGTLP 118
Query: 111 FEKLWDNFFQGHEGKFSVYVHASKAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLA 170
FEKLWD FFQGHEGKFSVY+HASK PVH SRYF+NR+IRSD++VWG++S+++AERRLL
Sbjct: 119 FEKLWDLFFQGHEGKFSVYIHASKDTPVHTSRYFLNREIRSDEVVWGRISMIDAERRLLT 178
Query: 171 NALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPE 230
NAL+DP NQ FVLLSDSCVPL +F Y+++Y+M+++ S+VD F DPGP G GR+ +HMLPE
Sbjct: 179 NALRDPENQQFVLLSDSCVPLRSFEYMYNYMMHSNVSYVDCFDDPGPHGTGRHMDHMLPE 238
Query: 231 VEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTI 290
+ +DFR GAQWFS+KRQHAV +AD+LYYSKF+ C KNCI DEHYLPTFF +
Sbjct: 239 IPREDFRKGAQWFSMKRQHAVVTVADNLYYSKFRDYCGPGVEGNKNCIADEHYLPTFFYM 298
Query: 291 VDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNI 329
+DP GIA W+VTYVD SE+K HP+ Y +DIT EL+KNI
Sbjct: 299 LDPTGIANWTVTYVDWSERKWHPRKYMPEDITLELIKNI 337
>UniRef100_O64480 Hypothetical protein At2g19160 [Arabidopsis thaliana]
Length = 378
Score = 380 bits (976), Expect = e-104
Identities = 187/339 (55%), Positives = 244/339 (71%), Gaps = 12/339 (3%)
Query: 1 MKTAKFWRLS-MGDMEILLGSPVLHRPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTSC 59
MK K WRL + M+ L G+ HR +KP+WI+ ++S I +F+ AY++ + +C
Sbjct: 1 MKAIKGWRLGKINYMQSLPGAR--HRAPTRKPIWIIAVLSLIAMFVIGAYMFPHHSKAAC 58
Query: 60 NMFYSKPCI-----IDISVLAVSHIPVSVYIERGITLDMPQ----NPKIAFMFLTPGSLP 110
MF SK C + S+ S ++ + L P+ + KIAFMFLTPG+LP
Sbjct: 59 YMFSSKGCKGLTDWLPPSLREYSDDEIAARVVISEILSSPRVIKKSSKIAFMFLTPGTLP 118
Query: 111 FEKLWDNFFQGHEGKFSVYVHASKAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLA 170
FEKLWD FFQGHEGKFSVY+HASK PVH SRYF+NR+IRSD++VWG++S+++AERRLL
Sbjct: 119 FEKLWDLFFQGHEGKFSVYIHASKDTPVHTSRYFLNREIRSDEVVWGRISMIDAERRLLT 178
Query: 171 NALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPE 230
NAL+DP NQ FVLLSDSCVPL +F Y+++Y+M+++ S+VD F DPGP G GR+ +HMLPE
Sbjct: 179 NALRDPENQQFVLLSDSCVPLRSFEYMYNYMMHSNVSYVDCFDDPGPHGTGRHMDHMLPE 238
Query: 231 VEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTI 290
+ +DFR GAQWFS+KRQHAV +AD+LYYSKF+ C KNCI DEHYLPTFF +
Sbjct: 239 IPREDFRKGAQWFSMKRQHAVVTVADNLYYSKFRDYCGPGVEGNKNCIADEHYLPTFFYM 298
Query: 291 VDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNI 329
+DP GIA W+VTYVD SE+K HP+ Y +DIT EL+KNI
Sbjct: 299 LDPTGIANWTVTYVDWSERKWHPRKYMPEDITLELIKNI 337
>UniRef100_Q651U0 Hypothetical protein OSJNBa0051O02.41 [Oryza sativa]
Length = 378
Score = 370 bits (950), Expect = e-101
Identities = 183/325 (56%), Positives = 231/325 (70%), Gaps = 14/325 (4%)
Query: 24 HRPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTSCNMFYSKPCIIDISVLAV------- 76
HR KKP WI++LVS + L AY++ + + C +F S C L
Sbjct: 9 HRAVAKKPKWIIILVSLVCFVLIGAYVFPPRRYSQCYLFGSGACATFKDWLPSVTRRERT 68
Query: 77 -SHIPVSVYIERGITLDMP--QNPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHAS 133
I SV + + + MP +NPKIA MFLTPG+LPFEKLW+ F QG EG++S+YVHAS
Sbjct: 69 DEEIISSVVLRDILAMPMPVSKNPKIALMFLTPGTLPFEKLWEKFLQGQEGRYSIYVHAS 128
Query: 134 KAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYN 193
+ KPVH S FV RDI SD +VWGK+S+V+AE+RLLANAL D +NQ FVLLSDSCVPL+
Sbjct: 129 REKPVHTSSLFVGRDIHSDAVVWGKISMVDAEKRLLANALADVDNQFFVLLSDSCVPLHT 188
Query: 194 FNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKV 253
F+Y+++YLM T+ SF+D FRDPGP GNGRYS MLPE+E KDFR GAQWF++ R+HA+ +
Sbjct: 189 FDYVYNYLMGTNISFIDCFRDPGPHGNGRYSPEMLPEIEEKDFRKGAQWFAITRRHALLI 248
Query: 254 MADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHP 313
+AD LYY KF+ C+ + DG+NCI DEHYLPT F +VDP GIA WSVT+VD SE K HP
Sbjct: 249 LADSLYYKKFKLYCKPA--DGRNCIADEHYLPTLFNMVDPGGIANWSVTHVDWSEGKWHP 306
Query: 314 KSYRTQDITYELLKNIKL--HNFHL 336
+SYR D+TY+LLKNI NFH+
Sbjct: 307 RSYRAADVTYDLLKNITAVDENFHV 331
>UniRef100_Q8W4E9 Hypothetical protein At4g30060; F6G3.90 [Arabidopsis thaliana]
Length = 401
Score = 366 bits (940), Expect = e-100
Identities = 182/340 (53%), Positives = 241/340 (70%), Gaps = 11/340 (3%)
Query: 1 MKTAKFWRL-SMGDMEILL-GSPVLHRPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTS 58
MK K W + ++ D+ + L G+ P ++ +WI++++S I +F AY+Y + +
Sbjct: 1 MKAVKRWSIGNLADIPVSLPGARYRAPPPGRRRVWIIMVLSLITMFFIMAYMYPHHSKRA 60
Query: 59 CNMFYSKPCI-----IDISVLAVSHIPVSVYIERGITLDMP----QNPKIAFMFLTPGSL 109
C M S+ C + S+ S ++ + L P +N KIAFMFLTPG+L
Sbjct: 61 CYMISSRGCKALADWLPPSLREYSDDEIAARVVIREILSSPPVIRKNSKIAFMFLTPGTL 120
Query: 110 PFEKLWDNFFQGHEGKFSVYVHASKAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLL 169
PFE+LWD FF GHEGKFSVY+HASK +PVH SRYF+NR+IRSD++VWG++S+V+AERRLL
Sbjct: 121 PFERLWDRFFLGHEGKFSVYIHASKERPVHYSRYFLNREIRSDEVVWGRISMVDAERRLL 180
Query: 170 ANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLP 229
ANAL+D +NQ FVLLSDSCVPL +F YI++YLM+++ S+VD F DPG G GR+ HMLP
Sbjct: 181 ANALRDTSNQQFVLLSDSCVPLRSFEYIYNYLMHSNLSYVDCFDDPGQHGAGRHMNHMLP 240
Query: 230 EVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFT 289
E+ KDFR GAQWF++KRQHAV MAD LYYSKF+ C + KNCI DEHYLPTFF
Sbjct: 241 EIPKKDFRKGAQWFTMKRQHAVATMADSLYYSKFRDYCGPGIENNKNCIADEHYLPTFFH 300
Query: 290 IVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNI 329
++DP GIA W+VT VD SE+K HPK+Y +DIT+ELL N+
Sbjct: 301 MLDPGGIANWTVTQVDWSERKWHPKTYMPEDITHELLNNL 340
>UniRef100_Q8GS18 Hypothetical protein [Arabidopsis thaliana]
Length = 376
Score = 364 bits (935), Expect = 2e-99
Identities = 170/315 (53%), Positives = 229/315 (71%), Gaps = 10/315 (3%)
Query: 25 RPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTSCNMFYSKPC-----IIDISVLAVSHI 79
RP K P WI+ LV + + + A++Y +N+ +C MF C + + ++
Sbjct: 7 RPPFKGPRWIITLVVLVTVVVITAFIYPPRNSVACYMFSGPGCPLYQQFLFVPTRELTDS 66
Query: 80 PVSVYIERGITLDMPQ----NPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKA 135
+ + +++PQ NPK+AFMFLTPG+LPFE LW+ FF+GHE KFSVYVHASK
Sbjct: 67 EAAAQVVMNEIMNLPQSKTANPKLAFMFLTPGTLPFEPLWEMFFRGHENKFSVYVHASKK 126
Query: 136 KPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFN 195
PVH S YFV RDI S ++ WG++S+V+AERRLLA+AL DP+NQHF+LLSDSCVPL++FN
Sbjct: 127 SPVHTSSYFVGRDIHSHKVAWGQISMVDAERRLLAHALVDPDNQHFILLSDSCVPLFDFN 186
Query: 196 YIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMA 255
YI+++L++ + SF+D F DPGP G+GRYS+HMLPEVE KDFR G+QWFS+KR+HA+ VMA
Sbjct: 187 YIYNHLIFANLSFIDCFEDPGPHGSGRYSQHMLPEVEKKDFRKGSQWFSMKRRHAIVVMA 246
Query: 256 DHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKS 315
D LYY+KF+ C + ++G+NC DEHY PT F ++DP+GIA WSVT+VD SE K HPK
Sbjct: 247 DSLYYTKFKLYCRPN-MEGRNCYADEHYFPTLFNMIDPDGIANWSVTHVDWSEGKWHPKL 305
Query: 316 YRTQDITYELLKNIK 330
Y +DIT L++ IK
Sbjct: 306 YNARDITPYLIRKIK 320
>UniRef100_Q6EUQ8 Hypothetical protein OJ1077_E05.12 [Oryza sativa]
Length = 390
Score = 363 bits (933), Expect = 3e-99
Identities = 179/339 (52%), Positives = 239/339 (69%), Gaps = 14/339 (4%)
Query: 1 MKTAKFWRLSMGDMEILLGSPVLHRPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTSCN 60
MK ++ W+ DM + P R KKPMWI+VL+S + + L AY Y + ++C
Sbjct: 1 MKASQVWQRGSKDMTAM--PPPRQRGAAKKPMWIIVLLSLVCVALIGAYAYPPRRYSACY 58
Query: 61 MFYSKPCIIDISVLAV--------SHIPVSVYIERGITLDMP--QNPKIAFMFLTPGSLP 110
F S C L I SV + + + MP +NPKIA MFLTPGSLP
Sbjct: 59 FFASSVCTPFKDWLPTVTRRERTDEEIVSSVVMRDLLAMPMPVSKNPKIALMFLTPGSLP 118
Query: 111 FEKLWDNFFQGHEGKFSVYVHASKAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLA 170
FEKLW+ F QGHE ++S+Y+HAS+ +PVH S FV R+I S+++VWG++S+V+AE+RLLA
Sbjct: 119 FEKLWEKFLQGHEDRYSIYIHASRERPVHSSSLFVGREIHSEKVVWGRISMVDAEKRLLA 178
Query: 171 NALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPE 230
NAL+D +NQ FVLLSDSCVPL+ F+YI+++LM T+ SF+D F DPGP G+GRYS MLPE
Sbjct: 179 NALEDVDNQFFVLLSDSCVPLHTFDYIYNFLMGTNVSFIDCFLDPGPHGSGRYSVEMLPE 238
Query: 231 VEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTI 290
+E +DFR GAQWF++ R+HA+ ++ADHLYY+KF+ C+ + +G+NCI DEHYLPT F +
Sbjct: 239 IEQRDFRKGAQWFAVTRRHALLILADHLYYNKFELYCKPA--EGRNCIADEHYLPTLFNM 296
Query: 291 VDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNI 329
VDP GI+ WSVT+VD SE K HP+SYR D+TY LLKNI
Sbjct: 297 VDPGGISNWSVTHVDWSEGKWHPRSYRAIDVTYALLKNI 335
>UniRef100_Q7XS46 OSJNBa0035M09.18 protein [Oryza sativa]
Length = 382
Score = 360 bits (925), Expect = 3e-98
Identities = 178/349 (51%), Positives = 246/349 (70%), Gaps = 11/349 (3%)
Query: 1 MKTAKFWRLSMGDMEILLGSPVLHRPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTSC- 59
MK + W+L + DM+ +P+ K +++V+F+ + AYLY Q+ TS
Sbjct: 1 MKAHQLWQLPIKDMK---SAPLPRGRTSPKKHLCILVVAFVSIVTLWAYLYPPQHYTSPM 57
Query: 60 -NMFYSKPCIIDISVLAVSHIPVSVYIERGITLD--MPQNPKIAFMFLTPGSLPFEKLWD 116
+ ++P + L V ++ ++ + K+AFMFLTPG+LPFE+LW+
Sbjct: 58 RDWLPAEP----VRELTDQERASQVVFKQILSTPPVKSKRSKVAFMFLTPGTLPFERLWE 113
Query: 117 NFFQGHEGKFSVYVHASKAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLANALQDP 176
FF+GHEG++++YVHAS+ KP H S F++RDIRS+++VWGK+S+V+AERRLLANAL+D
Sbjct: 114 KFFEGHEGRYTIYVHASREKPEHASPLFIDRDIRSEKVVWGKISMVDAERRLLANALEDV 173
Query: 177 NNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDF 236
+NQHFVLLSDSCVPL+NF+Y+++YL+ T+ SF+DSF DPGP GN RYS+HMLPEV DF
Sbjct: 174 DNQHFVLLSDSCVPLHNFDYVYNYLIGTNISFIDSFYDPGPHGNFRYSKHMLPEVRESDF 233
Query: 237 RTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGI 296
R G+QWFS+KRQHA+ ++AD LYY+KF+ C+ DG+NC DEHYLPT F ++DPNGI
Sbjct: 234 RKGSQWFSVKRQHALMIIADSLYYTKFKLHCKPGMEDGRNCYADEHYLPTLFHMIDPNGI 293
Query: 297 AKWSVTYVDRSEQKRHPKSYRTQDITYELLKNIKLHNFHLFLTERSTKM 345
A WSVT+VD SE K HPK+YR D+TYELLKNI + +T S K+
Sbjct: 294 ANWSVTHVDWSEGKWHPKAYRANDVTYELLKNITSIDMSYHITSDSKKV 342
>UniRef100_Q9SVZ8 Hypothetical protein AT4g25870 [Arabidopsis thaliana]
Length = 389
Score = 358 bits (919), Expect = 1e-97
Identities = 170/341 (49%), Positives = 242/341 (70%), Gaps = 13/341 (3%)
Query: 11 MGDMEILLGSPVLHRPQMKKPMWIVVLVSFIILFLTCAYLY-RMQNTTSCNMFYS-KPCI 68
MG+ + +L P H +KKP+W+V+ VS + L C ++Y + ++SC+ YS + C
Sbjct: 1 MGETQKILQGP-RHHTSLKKPLWVVLTVSVTSMLLICTHMYPKHGKSSSCHGLYSTRGCE 59
Query: 69 IDISVLAVSHIPVSVYIE---RGITLDMPQNP-------KIAFMFLTPGSLPFEKLWDNF 118
+S H+ E R + D+ + P KIAF+FLTPG+LPFEKLWD F
Sbjct: 60 DALSKWLPVHVRKFTDEEIAARAVVRDILRTPPFITNNSKIAFLFLTPGTLPFEKLWDEF 119
Query: 119 FQGHEGKFSVYVHASKAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLANALQDPNN 178
F+GHEGKFS+Y+H SK +PVH+SR+F +R+I SD++ WG++S+V+AE+RLL +AL+DP+N
Sbjct: 120 FKGHEGKFSIYIHPSKERPVHISRHFSDREIHSDEVTWGRISMVDAEKRLLVSALEDPDN 179
Query: 179 QHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRT 238
QHFVL+S+SC+PL+ F+Y + YL+Y++ SF++SF DPGP G GR+ EHMLPE+ +DFR
Sbjct: 180 QHFVLVSESCIPLHTFDYTYRYLLYSNVSFIESFVDPGPHGTGRHMEHMLPEIAKEDFRK 239
Query: 239 GAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAK 298
GAQWF++KRQHA+ VMAD LYYSKF+ C KNCI DEHYLPTFF ++DP GI+
Sbjct: 240 GAQWFTMKRQHAIIVMADGLYYSKFREYCGPGIEADKNCIADEHYLPTFFNMIDPMGISN 299
Query: 299 WSVTYVDRSEQKRHPKSYRTQDITYELLKNIKLHNFHLFLT 339
WSVT+VD SE++ HPK+Y +I+ E +KN+ + + +T
Sbjct: 300 WSVTFVDWSERRWHPKTYGGNEISLEFMKNVTSEDMSVHVT 340
>UniRef100_Q5Z845 Hypothetical protein P0468G03.24 [Oryza sativa]
Length = 372
Score = 357 bits (916), Expect = 3e-97
Identities = 175/314 (55%), Positives = 226/314 (71%), Gaps = 10/314 (3%)
Query: 24 HRPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTSCNMFYSKPCII-----DISVLAVSH 78
+R ++P+WIV+L++F+ AYLY Q+ T C + S C V
Sbjct: 5 NRSSARRPLWIVILIAFVCAVGIGAYLYTPQHYTPCYLVSSNSCSSRPPPEPARVYTDDE 64
Query: 79 IPVSVYIERGITLDMP---QNPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKA 135
I + I R I L P +NPKIAFMFLTP SLPFEKLW+ FF GHE ++++YVHAS+
Sbjct: 65 IAARIVI-RDIILAQPVQSKNPKIAFMFLTPSSLPFEKLWEKFFMGHEDRYTIYVHASRE 123
Query: 136 KPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFN 195
+PVH S F RDIRS+++VWG +S+++AERRLLANALQDP+NQHFVLLS+SCVPL+NF+
Sbjct: 124 RPVHASPIFNGRDIRSEKVVWGTISMIDAERRLLANALQDPDNQHFVLLSESCVPLHNFD 183
Query: 196 YIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMA 255
Y++ YLM T+ SFVD F DPGP G GRYS+HMLPE+ +D+R GAQWF++KRQHAV +++
Sbjct: 184 YVYSYLMETNISFVDCFDDPGPHGAGRYSDHMLPEIVKRDWRKGAQWFTVKRQHAVLILS 243
Query: 256 DHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKS 315
D LYY+KF+ C+ + NC DEHYLPT F +VDP GIA WSVT+VD SE K HPK+
Sbjct: 244 DFLYYAKFKRYCKPG-NEWHNCYSDEHYLPTLFNMVDPTGIANWSVTHVDWSEGKWHPKA 302
Query: 316 YRTQDITYELLKNI 329
YR D ++ELLKNI
Sbjct: 303 YRAVDTSFELLKNI 316
>UniRef100_Q8W4D1 Hypothetical protein At5g57270; MJB24.8 [Arabidopsis thaliana]
Length = 388
Score = 356 bits (913), Expect = 6e-97
Identities = 173/329 (52%), Positives = 232/329 (69%), Gaps = 13/329 (3%)
Query: 24 HRPQMKKPMWIVVLVSFIILFLTCAYLYRMQN---TTSCNMFYSKPCIIDISVLAVSHIP 80
HR +KKP+ IV+LV + L Y+Y N +++C S+ C +S H+
Sbjct: 11 HRSSLKKPLLIVLLVCITSVLLVITYMYPQHNNSKSSACAGLSSRGCQAALSGWLPVHVR 70
Query: 81 VSVYIE---RGITLDMPQNP-------KIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYV 130
E R + D+ + P KIAFMFLTPG+LPFEKLWD FFQG EG+FS+Y+
Sbjct: 71 KFTDEEVAARVVIKDILRLPPALTAKSKIAFMFLTPGTLPFEKLWDKFFQGQEGRFSIYI 130
Query: 131 HASKAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVP 190
H S+ +PVH+SR+F +R+I SD + WG++S+V+AERRLLANAL+DP+NQHFVLLS+SC+P
Sbjct: 131 HPSRLRPVHISRHFSDREIHSDHVTWGRISMVDAERRLLANALEDPDNQHFVLLSESCIP 190
Query: 191 LYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHA 250
L+ F+Y + YLM+ + SF+DSF D GP G GR+ +HMLPE+ +DFR GAQWF++KRQHA
Sbjct: 191 LHTFDYTYRYLMHANVSFIDSFEDLGPHGTGRHMDHMLPEIPRQDFRKGAQWFTMKRQHA 250
Query: 251 VKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQK 310
V VMAD LYYSKF+ C KNCI DEHYLPTFF ++DP GI+ WSVTYVD SE++
Sbjct: 251 VIVMADGLYYSKFREYCRPGVEANKNCIADEHYLPTFFHMLDPGGISNWSVTYVDWSERR 310
Query: 311 RHPKSYRTQDITYELLKNIKLHNFHLFLT 339
HPK+YR +D++ +LLKNI + + +T
Sbjct: 311 WHPKTYRARDVSLKLLKNITSDDMSVHVT 339
>UniRef100_Q8W4F9 Hypothetical protein At5g57270; MJB24.8 [Arabidopsis thaliana]
Length = 388
Score = 353 bits (905), Expect = 5e-96
Identities = 172/329 (52%), Positives = 231/329 (69%), Gaps = 13/329 (3%)
Query: 24 HRPQMKKPMWIVVLVSFIILFLTCAYLYRMQN---TTSCNMFYSKPCIIDISVLAVSHIP 80
HR +KKP+ IV+LV + L Y+Y N +++C S+ C +S H+
Sbjct: 11 HRSSLKKPLLIVLLVCITSVLLVITYMYPQHNNSKSSACAGLSSRGCQAALSGWLPVHVR 70
Query: 81 VSVYIE---RGITLDMPQNP-------KIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYV 130
E R + D+ + P KIAFMFLTPG+LPFEKLWD FFQG EG+FS+Y+
Sbjct: 71 KFTDEEVAARVVIKDILRLPPALTAKSKIAFMFLTPGTLPFEKLWDKFFQGQEGRFSIYI 130
Query: 131 HASKAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVP 190
H S+ + VH+SR+F +R+I SD + WG++S+V+AERRLLANAL+DP+NQHFVLLS+SC+P
Sbjct: 131 HPSRLRTVHISRHFSDREIHSDHVTWGRISMVDAERRLLANALEDPDNQHFVLLSESCIP 190
Query: 191 LYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHA 250
L+ F+Y + YLM+ + SF+DSF D GP G GR+ +HMLPE+ +DFR GAQWF++KRQHA
Sbjct: 191 LHTFDYTYRYLMHANVSFIDSFEDLGPHGTGRHMDHMLPEIPRQDFRKGAQWFTMKRQHA 250
Query: 251 VKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQK 310
V VMAD LYYSKF+ C KNCI DEHYLPTFF ++DP GI+ WSVTYVD SE++
Sbjct: 251 VIVMADGLYYSKFREYCRPGVEANKNCIADEHYLPTFFHMLDPGGISNWSVTYVDWSERR 310
Query: 311 RHPKSYRTQDITYELLKNIKLHNFHLFLT 339
HPK+YR +D++ +LLKNI + + +T
Sbjct: 311 WHPKTYRARDVSLKLLKNITSDDMSVHVT 339
>UniRef100_Q9LVD1 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MJB24
[Arabidopsis thaliana]
Length = 426
Score = 352 bits (903), Expect = 9e-96
Identities = 174/332 (52%), Positives = 233/332 (69%), Gaps = 16/332 (4%)
Query: 24 HRPQMKKPMWIVVLVSFIILFLTCAYLYRMQN---TTSCNMFYSKPCIIDISVLAVSHIP 80
HR +KKP+ IV+LV + L Y+Y N +++C S+ C +S H+
Sbjct: 11 HRSSLKKPLLIVLLVCITSVLLVITYMYPQHNNSKSSACAGLSSRGCQAALSGWLPVHVR 70
Query: 81 VSVYIE---RGITLDMPQNP-------KIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYV 130
E R + D+ + P KIAFMFLTPG+LPFEKLWD FFQG EG+FS+Y+
Sbjct: 71 KFTDEEVAARVVIKDILRLPPALTAKSKIAFMFLTPGTLPFEKLWDKFFQGQEGRFSIYI 130
Query: 131 HASKAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVP 190
H S+ +PVH+SR+F +R+I SD + WG++S+V+AERRLLANAL+DP+NQHFVLLS+SC+P
Sbjct: 131 HPSRLRPVHISRHFSDREIHSDHVTWGRISMVDAERRLLANALEDPDNQHFVLLSESCIP 190
Query: 191 LYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHA 250
L+ F+Y + YLM+ + SF+DSF D GP G GR+ +HMLPE+ +DFR GAQWF++KRQHA
Sbjct: 191 LHTFDYTYRYLMHANVSFIDSFEDLGPHGTGRHMDHMLPEIPRQDFRKGAQWFTMKRQHA 250
Query: 251 VKVMADHLYYSKFQAQCEQSCVD---GKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRS 307
V VMAD LYYSKF+ C S KNCI DEHYLPTFF ++DP GI+ WSVTYVD S
Sbjct: 251 VIVMADGLYYSKFREYCRVSSPGVEANKNCIADEHYLPTFFHMLDPGGISNWSVTYVDWS 310
Query: 308 EQKRHPKSYRTQDITYELLKNIKLHNFHLFLT 339
E++ HPK+YR +D++ +LLKNI + + +T
Sbjct: 311 ERRWHPKTYRARDVSLKLLKNITSDDMSVHVT 342
>UniRef100_Q6Z912 Hypothetical protein P0494D11.22 [Oryza sativa]
Length = 376
Score = 331 bits (848), Expect = 2e-89
Identities = 154/250 (61%), Positives = 196/250 (77%)
Query: 95 QNPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKAKPVHVSRYFVNRDIRSDQL 154
+NPKIAFMFLTPG LPFEKLW+ FF+GHEG++++YVHAS+ KP HVS FV RDI SD++
Sbjct: 86 RNPKIAFMFLTPGKLPFEKLWELFFKGHEGRYTIYVHASREKPEHVSPVFVGRDIHSDKV 145
Query: 155 VWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRD 214
WG +S+V+AERRLLA AL+D +NQ FVLLSDSCVPL+NF+Y++D+LM + SF+D F D
Sbjct: 146 GWGMISMVDAERRLLAKALEDTDNQLFVLLSDSCVPLHNFDYVYDFLMGSRHSFLDCFDD 205
Query: 215 PGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDG 274
PGP G RYS+HMLPEV DFR G+QWF++KRQHA+ V+AD LYY+KF+ C+ +G
Sbjct: 206 PGPHGVFRYSKHMLPEVREIDFRKGSQWFAIKRQHAMVVVADSLYYTKFRRFCKPGMEEG 265
Query: 275 KNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNIKLHNF 334
+NC DEHYLPT F ++DP GIA WSVTYVD SE K HP+S+R +D+TYELLKN+ +
Sbjct: 266 RNCYADEHYLPTLFLMMDPAGIANWSVTYVDWSEGKWHPRSFRAKDVTYELLKNMTSVDI 325
Query: 335 HLFLTERSTK 344
+T K
Sbjct: 326 SYHITSDEKK 335
>UniRef100_Q9SZV9 Hypothetical protein F6G3.90 [Arabidopsis thaliana]
Length = 318
Score = 323 bits (827), Expect = 6e-87
Identities = 169/340 (49%), Positives = 221/340 (64%), Gaps = 35/340 (10%)
Query: 1 MKTAKFWRL-SMGDMEILL-GSPVLHRPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTS 58
MK K W + ++ D+ + L G+ P ++ +WI++++S I +F AY+Y + +
Sbjct: 1 MKAVKRWSIGNLADIPVSLPGARYRAPPPGRRRVWIIMVLSLITMFFIMAYMYPHHSKRA 60
Query: 59 CNMFYSKPCI-----IDISVLAVSHIPVSVYIERGITLDMP----QNPKIAFMFLTPGSL 109
C M S+ C + S+ S ++ + L P +N KIAFMFLTPG+L
Sbjct: 61 CYMISSRGCKALADWLPPSLREYSDDEIAARVVIREILSSPPVIRKNSKIAFMFLTPGTL 120
Query: 110 PFEKLWDNFFQGHEGKFSVYVHASKAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLL 169
PFE+LWD FF GHEGKFSVY+HASK +PVH SRYF+NR+IRSD++VWG++S+V+AERRLL
Sbjct: 121 PFERLWDRFFLGHEGKFSVYIHASKERPVHYSRYFLNREIRSDEVVWGRISMVDAERRLL 180
Query: 170 ANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLP 229
ANAL+D +NQ FVLLSDS F DPG G GR+ HMLP
Sbjct: 181 ANALRDTSNQQFVLLSDS------------------------FDDPGQHGAGRHMNHMLP 216
Query: 230 EVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFT 289
E+ KDFR GAQWF++KRQHAV MAD LYYSKF+ C + KNCI DEHYLPTFF
Sbjct: 217 EIPKKDFRKGAQWFTMKRQHAVATMADSLYYSKFRDYCGPGIENNKNCIADEHYLPTFFH 276
Query: 290 IVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNI 329
++DP GIA W+VT VD SE+K HPK+Y +DIT+ELL N+
Sbjct: 277 MLDPGGIANWTVTQVDWSERKWHPKTYMPEDITHELLNNL 316
>UniRef100_O49584 Predicted protein [Arabidopsis thaliana]
Length = 711
Score = 240 bits (612), Expect = 5e-62
Identities = 124/264 (46%), Positives = 163/264 (60%), Gaps = 45/264 (17%)
Query: 25 RPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTSCNMFYSKPC-----IIDISVLAVSHI 79
RP K P WI+ LV + + + A++Y +N+ +C MF C + + ++
Sbjct: 373 RPPFKGPRWIITLVVLVTVVVITAFIYPPRNSVACYMFSGPGCPLYQQFLFVPTRELTDS 432
Query: 80 PVSVYIERGITLDMPQ----NPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKA 135
+ + +++PQ NPK+AFMFLTPG+LPFE LW+ FF+GHE KFSVYVHASK
Sbjct: 433 EAAAQVVMNEIMNLPQSKTANPKLAFMFLTPGTLPFEPLWEMFFRGHENKFSVYVHASKK 492
Query: 136 KPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFN 195
PVH S YFV RDI S ++ WG++S+V+AERRLLA+AL DP+NQHF+LLS
Sbjct: 493 SPVHTSSYFVGRDIHSHKVAWGQISMVDAERRLLAHALVDPDNQHFILLS---------- 542
Query: 196 YIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQ------------WF 243
DSF DPGP G+GRYS+HMLPEVE KDFR G+Q WF
Sbjct: 543 --------------DSFEDPGPHGSGRYSQHMLPEVEKKDFRKGSQEGFTVQKKTMVNWF 588
Query: 244 SLKRQHAVKVMADHLYYSKFQAQC 267
S+KR+HA+ VMAD LYY+KF+ C
Sbjct: 589 SMKRRHAIVVMADSLYYTKFKLYC 612
>UniRef100_Q6AVG1 Expressed protein [Oryza sativa]
Length = 398
Score = 227 bits (579), Expect = 3e-58
Identities = 115/263 (43%), Positives = 164/263 (61%), Gaps = 8/263 (3%)
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKAKPVHVSR--YFVNRDIRSDQL 154
PK+AFMFLT G LP LW+ FF+GH+G +SVYVHA + + + F R I S
Sbjct: 128 PKVAFMFLTRGPLPLAPLWERFFRGHDGLYSVYVHALPSYRANFTTDSVFYRRQIPSKVA 187
Query: 155 VWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRD 214
WG+M++ +AERRLLANAL D +N+ FVL+S+SC+P++NFN + YL + +SFV +F D
Sbjct: 188 EWGEMTMCDAERRLLANALLDISNEWFVLVSESCIPIFNFNTTYRYLQNSSQSFVMAFDD 247
Query: 215 PGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDG 274
PGP G GRY+ +M PEVE+ +R G+QWF + R+ A++++ D LYY KF+ C
Sbjct: 248 PGPYGRGRYNWNMTPEVELTQWRKGSQWFEVNRELAIEIVRDTLYYPKFKEFCR------ 301
Query: 275 KNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNIKLHNF 334
+C +DEHY PT TI P +A S+T+VD S HP ++ DIT E L+ ++
Sbjct: 302 PHCYVDEHYFPTMLTIEAPQSLANRSITWVDWSRGGAHPATFGRGDITEEFLRRVQEGRT 361
Query: 335 HLFLTERSTKMDLFLEWISEAVL 357
L+ + ST LF + + L
Sbjct: 362 CLYNGQNSTMCFLFARKFAPSAL 384
>UniRef100_Q9XGZ7 T1N24.19 protein [Arabidopsis thaliana]
Length = 436
Score = 215 bits (548), Expect = 1e-54
Identities = 110/235 (46%), Positives = 145/235 (60%), Gaps = 8/235 (3%)
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHA--SKAKPVHVSRYFVNRDIRSDQL 154
PKIAFMFLT G LP LW+ +GHE +SVY+H+ S + S F R I S
Sbjct: 167 PKIAFMFLTMGPLPLAPLWERLLKGHEKLYSVYIHSPVSSSAKFPASSVFYRRHIPSQVA 226
Query: 155 VWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRD 214
WG+M++ +AERRLLANAL D +N+ FVLLS+SC+PL+NF I+ Y+ ++ SF+ SF D
Sbjct: 227 EWGRMTMCDAERRLLANALLDISNEWFVLLSESCIPLFNFTTIYTYMTKSEHSFMGSFDD 286
Query: 215 PGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDG 274
PG G GRY +M PEV I +R G+QWF + R+ AV ++ D LYY KF+ C+ +
Sbjct: 287 PGAYGRGRYHGNMAPEVFIDQWRKGSQWFEINRELAVSIVKDTLYYPKFKEFCQPA---- 342
Query: 275 KNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNI 329
C +DEHY PT TI P +A SVT+VD S HP ++ QDI E I
Sbjct: 343 --CYVDEHYFPTMLTIEKPAALANRSVTWVDWSRGGAHPATFGAQDINEEFFARI 395
>UniRef100_Q9LYF7 Hypothetical protein T22P22_120 [Arabidopsis thaliana]
Length = 386
Score = 210 bits (534), Expect = 6e-53
Identities = 106/228 (46%), Positives = 141/228 (61%), Gaps = 8/228 (3%)
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHA--SKAKPVHVSRYFVNRDIRSDQL 154
PK+AFMFLT G LP LW+ F +GH+G +SVY+H S S F R I S
Sbjct: 117 PKVAFMFLTKGPLPLASLWERFLKGHKGLYSVYLHPHPSFTAKFPASSVFHRRQIPSQVA 176
Query: 155 VWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRD 214
WG+MS+ +AE+RLLANAL D +N+ FVL+S+SC+PLYNF I+ YL + SF+ +F D
Sbjct: 177 EWGRMSMCDAEKRLLANALLDVSNEWFVLVSESCIPLYNFTTIYSYLSRSKHSFMGAFDD 236
Query: 215 PGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDG 274
PGP G GRY+ +M PEV + +R G+QWF + R A ++ D LYY KF+ C +
Sbjct: 237 PGPFGRGRYNGNMEPEVPLTKWRKGSQWFEVNRDLAATIVKDTLYYPKFKEFCRPA---- 292
Query: 275 KNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDIT 322
C +DEHY PT TI P +A S+T+VD S HP ++ DIT
Sbjct: 293 --CYVDEHYFPTMLTIEKPTVLANRSLTWVDWSRGGPHPATFGRSDIT 338
>UniRef100_Q9LU27 Gb|AAD40142.1 [Arabidopsis thaliana]
Length = 384
Score = 209 bits (531), Expect = 1e-52
Identities = 113/275 (41%), Positives = 162/275 (58%), Gaps = 22/275 (8%)
Query: 91 LDMP--QNPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHA--SKAKPVHVSRYFVN 146
LD P + PK+AFMFLT G LPF LW+ FF+GHEG +S+YVH + S F
Sbjct: 106 LDYPFKRVPKMAFMFLTKGPLPFAPLWERFFKGHEGFYSIYVHTLPNYRSDFPSSSVFYR 165
Query: 147 RDIRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDK 206
R I S + WG+MS+ +AERRLLANAL D +N+ FVLLS++C+PL FN+++ Y+ +
Sbjct: 166 RQIPSQHVAWGEMSMCDAERRLLANALLDISNEWFVLLSEACIPLRGFNFVYRYVSRSRY 225
Query: 207 SFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQ 266
SF+ S + GP G GRYS M PEV + ++R G+QWF + R AV ++ D +YY+KF+
Sbjct: 226 SFMGSVDEDGPYGRGRYSYAMGPEVSLNEWRKGSQWFEINRALAVDIVEDMVYYNKFKEF 285
Query: 267 CEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELL 326
C C +DEHY PT +I P+ +A ++T+ D S HP ++ DIT + +
Sbjct: 286 CRPP------CYVDEHYFPTMLSIGYPDFLANRTLTWTDWSRGGAHPATFGKADITEKFI 339
Query: 327 KN------------IKLHNFHLFLTERSTKMDLFL 349
K I + +FH+ + S +LFL
Sbjct: 340 KKLSRASYSMDTTLILIVSFHIDNIDTSMAKELFL 374
>UniRef100_Q8LCJ9 Hypothetical protein [Arabidopsis thaliana]
Length = 381
Score = 207 bits (527), Expect = 4e-52
Identities = 106/243 (43%), Positives = 151/243 (61%), Gaps = 10/243 (4%)
Query: 91 LDMP--QNPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHA--SKAKPVHVSRYFVN 146
LD P + PK+AFMFLT G LPF LW+ FF+GHEG +S+YVH + S F
Sbjct: 104 LDYPFKRVPKMAFMFLTKGPLPFAPLWERFFKGHEGFYSIYVHTLPNYRSDFPSSSVFYR 163
Query: 147 RDIRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDK 206
R I S + WG+MS+ +AERRLLANAL D +N+ FVLLS++C+PL FN+++ Y+ +
Sbjct: 164 RQIPSQHVAWGEMSMCDAERRLLANALLDISNEWFVLLSEACIPLRGFNFVYRYVSRSRY 223
Query: 207 SFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQ 266
SF+ S + GP G GRYS M PEV + ++R G+QWF + R AV ++ D +YY+KF+
Sbjct: 224 SFMGSVDEDGPYGRGRYSYAMGPEVSLNEWRKGSQWFEINRALAVDIVEDMVYYNKFKEF 283
Query: 267 CEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELL 326
C C +DEHY PT +I P+ +A ++T+ D S HP ++ DIT + +
Sbjct: 284 CRPP------CYVDEHYFPTMLSIGYPDFLANRTLTWTDWSRGGAHPATFGKADITEKFI 337
Query: 327 KNI 329
K +
Sbjct: 338 KKL 340
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.139 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 631,105,820
Number of Sequences: 2790947
Number of extensions: 26715028
Number of successful extensions: 71331
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 55
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 71153
Number of HSP's gapped (non-prelim): 68
length of query: 359
length of database: 848,049,833
effective HSP length: 128
effective length of query: 231
effective length of database: 490,808,617
effective search space: 113376790527
effective search space used: 113376790527
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 75 (33.5 bits)
Medicago: description of AC149130.7


