

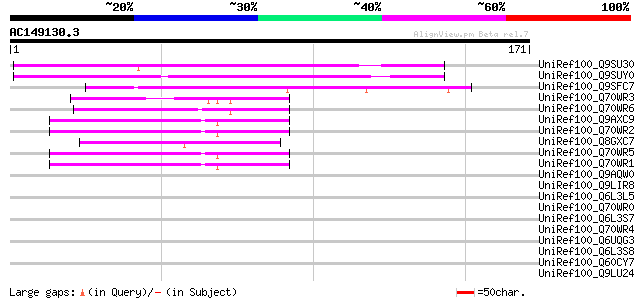
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149130.3 - phase: 0 /pseudo
(171 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SU30 Hypothetical protein AT4g12560 [Arabidopsis tha... 106 3e-22
UniRef100_Q9SUY0 Hypothetical protein AT4g22390 [Arabidopsis tha... 72 8e-12
UniRef100_Q9SFC7 F17A17.21 protein [Arabidopsis thaliana] 53 3e-06
UniRef100_Q70WR3 S locus F-box (SLF)-S1E protein [Antirrhinum hi... 48 9e-05
UniRef100_Q70WR6 S locus F-box (SLF)-S4D protein [Antirrhinum hi... 48 1e-04
UniRef100_Q9AXC9 S locus F-box (SLF)-S2-like protein [Antirrhinu... 48 1e-04
UniRef100_Q70WR2 S locus F-box (SLF)-S1 protein [Antirrhinum his... 48 1e-04
UniRef100_Q8GXC7 Hypothetical protein At3g06240/F28L1_18 [Arabid... 46 4e-04
UniRef100_Q70WR5 S locus F-box (SLF)-S4 protein [Antirrhinum his... 46 4e-04
UniRef100_Q70WR1 S locus F-box (SLF)-S5 protein [Antirrhinum his... 46 4e-04
UniRef100_Q9AQW0 SLF-S2 protein (S locus F-box (SLF)-S2 protein)... 45 0.001
UniRef100_Q9LIR8 Gb|AAF30317.1 [Arabidopsis thaliana] 43 0.003
UniRef100_Q6L3L5 Putative F-box domain containing protein [Solan... 43 0.003
UniRef100_Q70WR0 S locus F-box (SLF)-S5A protein [Antirrhinum hi... 43 0.003
UniRef100_Q6L3S7 Putative F-Box protein [Solanum demissum] 42 0.005
UniRef100_Q70WR4 S locus F-box (SLF)-S4A protein [Antirrhinum hi... 42 0.005
UniRef100_Q6UQG3 S3 self-incompatibility locus-linked putative F... 39 0.070
UniRef100_Q6L3S8 Putative F-Box protein [Solanum demissum] 39 0.070
UniRef100_Q60CY7 Putative F-box protein [Solanum demissum] 39 0.070
UniRef100_Q9LU24 Gb|AAF25964.1 [Arabidopsis thaliana] 38 0.092
>UniRef100_Q9SU30 Hypothetical protein AT4g12560 [Arabidopsis thaliana]
Length = 408
Score = 106 bits (264), Expect = 3e-22
Identities = 56/143 (39%), Positives = 83/143 (57%), Gaps = 8/143 (5%)
Query: 2 GVFVENSIRCIMAKKLDGLHPSLIVAFNLTLEIFNEVPLP-AELQGETSLLIDVAVLGGC 60
GV NS+ ++ ++ + +LIV F+L LE F V P A G + +D+ VL GC
Sbjct: 212 GVLAGNSLHWVLPRRPGLIAFNLIVRFDLALEEFEIVRFPEAVANGNVDIQMDIGVLDGC 271
Query: 61 LCITVNYETTKIDVWVMKEYGYRDSWCKLFTLVKSCFTSHLRYSNPIGYSSDGSKVLFEG 120
LC+ NY+ + +DVW+MKEY RDSW K+FT+ K Y P+ YS D K
Sbjct: 272 LCLMCNYDQSYVDVWMMKEYNVRDSWTKVFTVQKPKSVKSFSYMRPLVYSKDKKK----- 326
Query: 121 IEVLLDVHYKKLFWYDLKSERVS 143
VLL+++ KL W+DL+S+++S
Sbjct: 327 --VLLELNNTKLVWFDLESKKMS 347
>UniRef100_Q9SUY0 Hypothetical protein AT4g22390 [Arabidopsis thaliana]
Length = 394
Score = 71.6 bits (174), Expect = 8e-12
Identities = 42/142 (29%), Positives = 73/142 (50%), Gaps = 8/142 (5%)
Query: 2 GVFVENSIRCIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCL 61
GV V N + I+ ++ + + I+ ++L + + P EL E ++ D+ VL GC+
Sbjct: 211 GVVVNNHLHWILPRRQGVIAFNAIIKYDLASDDIGVLSFPQELYIEDNM--DIGVLDGCV 268
Query: 62 CITVNYETTKIDVWVMKEYGYRDSWCKLFTLVKSCFTSHLRYSNPIGYSSDGSKVLFEGI 121
C+ E + +DVWV+KEY SW KL+ + K + + P+ S D SK+L E
Sbjct: 269 CLMCYDEYSHVDVWVLKEYEDYKSWTKLYRVPKPESVESVEFIRPLICSKDRSKILLE-- 326
Query: 122 EVLLDVHYKKLFWYDLKSERVS 143
+ L W+DL+S+ ++
Sbjct: 327 ----INNAANLMWFDLESQSLT 344
>UniRef100_Q9SFC7 F17A17.21 protein [Arabidopsis thaliana]
Length = 417
Score = 53.1 bits (126), Expect = 3e-06
Identities = 43/137 (31%), Positives = 63/137 (45%), Gaps = 11/137 (8%)
Query: 26 VAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTKIDVWVMKEYGYRDS 85
V+F+L E F E+P P + G + L GCLC V K+D+WVMK YG ++S
Sbjct: 263 VSFDLEDEEFKEIPKP-DCGGLNRTNHRLVNLKGCLCAVVYGNYGKLDIWVMKTYGVKES 321
Query: 86 WCKLF---TLVKSCFTSHLRYSNPIGYSSDGSKV-----LFEGIEVLLDVHYKKLFWYDL 137
W K + T + +L I +++ KV L E E+LL+ + L YD
Sbjct: 322 WGKEYSIGTYLPKGLKQNLDRPMWIWKNAENGKVVRVLCLLENGEILLEYKSRVLVAYDP 381
Query: 138 KSERVS--YVEGIPKLF 152
K + G+P F
Sbjct: 382 KLGKFKDLLFHGLPNWF 398
>UniRef100_Q70WR3 S locus F-box (SLF)-S1E protein [Antirrhinum hispanicum]
Length = 384
Score = 48.1 bits (113), Expect = 9e-05
Identities = 30/85 (35%), Positives = 44/85 (51%), Gaps = 22/85 (25%)
Query: 21 HPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCIT--------VNY-ETTK 71
+P I+ F+++ EIF+E P + + GGCLC+T + Y ++TK
Sbjct: 241 YPGSILTFDISTEIFSEFEYPDGFR---------ELYGGCLCLTALSECLSVIRYNDSTK 291
Query: 72 ----IDVWVMKEYGYRDSWCKLFTL 92
I++WVMK YG DSW K F L
Sbjct: 292 DPQFIEIWVMKVYGNSDSWTKDFVL 316
>UniRef100_Q70WR6 S locus F-box (SLF)-S4D protein [Antirrhinum hispanicum]
Length = 374
Score = 47.8 bits (112), Expect = 1e-04
Identities = 25/73 (34%), Positives = 41/73 (55%), Gaps = 3/73 (4%)
Query: 22 PSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTK--IDVWVMKE 79
P +I+ F+++ E+F + P+ + T L + +L C +V E + I+VWVMKE
Sbjct: 238 PDVILTFDVSTEVFGQFEHPSGFKLCTGLQHNFMILNECFA-SVRSEVVRCLIEVWVMKE 296
Query: 80 YGYRDSWCKLFTL 92
YG + SW K F +
Sbjct: 297 YGIKQSWTKKFVI 309
>UniRef100_Q9AXC9 S locus F-box (SLF)-S2-like protein [Antirrhinum hispanicum]
Length = 376
Score = 47.8 bits (112), Expect = 1e-04
Identities = 26/84 (30%), Positives = 42/84 (49%), Gaps = 6/84 (7%)
Query: 14 AKKLDGLHPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNY-----E 68
A D + I+ F++ E+F E+ P L ++ + + L CL + V Y E
Sbjct: 227 ANSTDIFYADFILTFDIITEVFKEMAYPHCLAQFSNSFLSLMSLNECLAM-VRYKEWMEE 285
Query: 69 TTKIDVWVMKEYGYRDSWCKLFTL 92
D+WVMK+YG R+SW K + +
Sbjct: 286 PELFDIWVMKQYGVRESWTKQYVI 309
>UniRef100_Q70WR2 S locus F-box (SLF)-S1 protein [Antirrhinum hispanicum]
Length = 376
Score = 47.8 bits (112), Expect = 1e-04
Identities = 26/84 (30%), Positives = 42/84 (49%), Gaps = 6/84 (7%)
Query: 14 AKKLDGLHPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNY-----E 68
A D + I+ F++ E+F E+ P L ++ + + L CL + V Y E
Sbjct: 227 ANSTDIFYADFILTFDIITEVFKEMAYPHCLAQFSNSFLSLMSLNECLAM-VRYKEWMEE 285
Query: 69 TTKIDVWVMKEYGYRDSWCKLFTL 92
D+WVMK+YG R+SW K + +
Sbjct: 286 PELFDIWVMKQYGVRESWTKQYVI 309
>UniRef100_Q8GXC7 Hypothetical protein At3g06240/F28L1_18 [Arabidopsis thaliana]
Length = 427
Score = 45.8 bits (107), Expect = 4e-04
Identities = 23/68 (33%), Positives = 37/68 (53%), Gaps = 2/68 (2%)
Query: 24 LIVAFNLTLEIFNEVPLPAELQGETSLLIDVAV--LGGCLCITVNYETTKIDVWVMKEYG 81
++VAF++ E F E+P+P E + + + V L G LC+ + D+WVM EYG
Sbjct: 288 VVVAFDIQTEEFREMPVPDEAEDCSHRFSNFVVGSLNGRLCVVNSCYDVHDDIWVMSEYG 347
Query: 82 YRDSWCKL 89
SW ++
Sbjct: 348 EAKSWSRI 355
>UniRef100_Q70WR5 S locus F-box (SLF)-S4 protein [Antirrhinum hispanicum]
Length = 376
Score = 45.8 bits (107), Expect = 4e-04
Identities = 25/84 (29%), Positives = 41/84 (48%), Gaps = 6/84 (7%)
Query: 14 AKKLDGLHPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNY-----E 68
A D + I+ F++ E+F E+ P L ++ + + L CL + V Y E
Sbjct: 227 ANSTDIFYADFILTFDIITEVFKEMAYPHCLAQFSNSFLSLMSLNECLAM-VRYKEWMEE 285
Query: 69 TTKIDVWVMKEYGYRDSWCKLFTL 92
D+WVM +YG R+SW K + +
Sbjct: 286 PELFDIWVMNQYGVRESWTKQYVI 309
>UniRef100_Q70WR1 S locus F-box (SLF)-S5 protein [Antirrhinum hispanicum]
Length = 376
Score = 45.8 bits (107), Expect = 4e-04
Identities = 25/84 (29%), Positives = 41/84 (48%), Gaps = 6/84 (7%)
Query: 14 AKKLDGLHPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNY-----E 68
A D + I+ F++ E+F E+ P L ++ + + L CL + V Y E
Sbjct: 227 ANSTDIFYADFILTFDIITEVFKEMAYPHCLAQFSNSFLSLMSLNECLAM-VRYKEWMEE 285
Query: 69 TTKIDVWVMKEYGYRDSWCKLFTL 92
D+WVM +YG R+SW K + +
Sbjct: 286 PELFDIWVMNQYGVRESWTKQYVI 309
>UniRef100_Q9AQW0 SLF-S2 protein (S locus F-box (SLF)-S2 protein) [Antirrhinum
hispanicum]
Length = 376
Score = 44.7 bits (104), Expect = 0.001
Identities = 23/83 (27%), Positives = 40/83 (47%), Gaps = 4/83 (4%)
Query: 14 AKKLDGLHPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTK-- 71
A D + I+ F++ E+F E+ P L ++ + + L CL + E +
Sbjct: 227 ANSTDIFYADFILTFDIITEVFKEMAYPHCLAQFSNSFLSLMSLNECLAMVRYKEWMEDP 286
Query: 72 --IDVWVMKEYGYRDSWCKLFTL 92
D+WVM +YG R+SW K + +
Sbjct: 287 ELFDIWVMNQYGVRESWTKQYVI 309
>UniRef100_Q9LIR8 Gb|AAF30317.1 [Arabidopsis thaliana]
Length = 364
Score = 43.1 bits (100), Expect = 0.003
Identities = 31/95 (32%), Positives = 49/95 (50%), Gaps = 7/95 (7%)
Query: 25 IVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTKIDVWVMKEYGYRD 84
I++++++ + F E+P P G + + L GCL + + DVWVMKE+G
Sbjct: 235 IISYDMSRDEFKELPGPV-CCGRGCFTMTLGDLRGCLSMVCYCKGANADVWVMKEFGEVY 293
Query: 85 SWCKLFTLVKSCFTSHLRYSNPIGYSSDGSKVLFE 119
SW KL ++ T +R P+ + SDG VL E
Sbjct: 294 SWSKLLSI--PGLTDFVR---PL-WISDGLVVLLE 322
>UniRef100_Q6L3L5 Putative F-box domain containing protein [Solanum demissum]
Length = 307
Score = 43.1 bits (100), Expect = 0.003
Identities = 27/93 (29%), Positives = 47/93 (50%), Gaps = 3/93 (3%)
Query: 2 GVFVENSIRCIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCL 61
G FV I ++ +D + I++ +L E + + LP + G+ S + + V+G L
Sbjct: 144 GKFVNGKIYWALSADVDTFNMCNIISLDLADETWRRLELP-DSYGKGSYPLALGVVGSHL 202
Query: 62 CITV--NYETTKIDVWVMKEYGYRDSWCKLFTL 92
+ + E T DVW+ K+ G SW K+FT+
Sbjct: 203 SVLCLNSIEGTNSDVWIRKDCGVEVSWTKIFTV 235
>UniRef100_Q70WR0 S locus F-box (SLF)-S5A protein [Antirrhinum hispanicum]
Length = 404
Score = 43.1 bits (100), Expect = 0.003
Identities = 24/77 (31%), Positives = 38/77 (49%), Gaps = 7/77 (9%)
Query: 22 PSLIVAFNLTLEIFNEVPLPAELQGET-SLLIDVAVLGGCLCITVNYE-----TTKIDVW 75
P I+ FN+ E+F ++ + + E + + + CL + V YE ID+W
Sbjct: 251 PDTILTFNIRTEVFGQLEFIPDWEEEVYGYCVSLIAIDNCLGM-VRYEGWLEEPQLIDIW 309
Query: 76 VMKEYGYRDSWCKLFTL 92
VM EYG +SW K F +
Sbjct: 310 VMNEYGVGESWTKSFVI 326
>UniRef100_Q6L3S7 Putative F-Box protein [Solanum demissum]
Length = 372
Score = 42.4 bits (98), Expect = 0.005
Identities = 28/94 (29%), Positives = 47/94 (49%), Gaps = 5/94 (5%)
Query: 2 GVFVENSIRCIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGG-- 59
G FV I ++ +D + I++ +L E + + LP + G+ S + + V+G
Sbjct: 209 GKFVNGKIYWALSADVDTFNMCNIISLDLADETWRRLELP-DSYGKGSYPLALGVVGSHL 267
Query: 60 -CLCITVNYETTKIDVWVMKEYGYRDSWCKLFTL 92
LC+ E T DVW+ K+ G SW K+FT+
Sbjct: 268 SVLCLNC-IEGTNSDVWIRKDCGVEVSWTKIFTV 300
>UniRef100_Q70WR4 S locus F-box (SLF)-S4A protein [Antirrhinum hispanicum]
Length = 391
Score = 42.4 bits (98), Expect = 0.005
Identities = 23/77 (29%), Positives = 38/77 (48%), Gaps = 7/77 (9%)
Query: 22 PSLIVAFNLTLEIFNEVPLPAELQGET-SLLIDVAVLGGCLCITVNYE-----TTKIDVW 75
P I+ FN+ E+F ++ + + E + + + CL + + YE ID+W
Sbjct: 238 PDTILTFNIGTEVFGQLEFIPDWEEEVYGYCVSLIAIDNCLGM-IRYEGWLEEPQLIDIW 296
Query: 76 VMKEYGYRDSWCKLFTL 92
VM EYG +SW K F +
Sbjct: 297 VMNEYGVGESWTKSFVI 313
>UniRef100_Q6UQG3 S3 self-incompatibility locus-linked putative F-box protein S3-
A113 [Petunia integrifolia subsp. inflata]
Length = 376
Score = 38.5 bits (88), Expect = 0.070
Identities = 21/74 (28%), Positives = 39/74 (52%), Gaps = 11/74 (14%)
Query: 24 LIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITV--------NYETTKI-DV 74
+I+ F++ E F+ + +P + + +L C C+T+ + T K+ D+
Sbjct: 230 VILCFDMNTEKFHNLGMPDACHFDDGKCYGLVIL--CKCMTLICYPDPMPSSPTEKLTDI 287
Query: 75 WVMKEYGYRDSWCK 88
W+MKEYG ++SW K
Sbjct: 288 WIMKEYGEKESWIK 301
>UniRef100_Q6L3S8 Putative F-Box protein [Solanum demissum]
Length = 327
Score = 38.5 bits (88), Expect = 0.070
Identities = 27/93 (29%), Positives = 43/93 (46%), Gaps = 4/93 (4%)
Query: 2 GVFVENSIRCIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCL 61
G FV + + ++ I +F+L + + LP+ G+ + I+V V+G L
Sbjct: 211 GRFVNGKLYWTSSSCINNYKVCNITSFDLADGTWERLELPS--CGKDNSYINVGVVGSDL 268
Query: 62 CI--TVNYETTKIDVWVMKEYGYRDSWCKLFTL 92
+ T DVW+MK G SW KLFT+
Sbjct: 269 SLLYTCQRGAATSDVWIMKHSGVNVSWTKLFTI 301
>UniRef100_Q60CY7 Putative F-box protein [Solanum demissum]
Length = 383
Score = 38.5 bits (88), Expect = 0.070
Identities = 27/93 (29%), Positives = 43/93 (46%), Gaps = 4/93 (4%)
Query: 2 GVFVENSIRCIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCL 61
G FV + + ++ I +F+L + + LP+ G+ + I+V V+G L
Sbjct: 209 GRFVNGKLYWTSSTCINNYKVCNITSFDLADGTWGSLELPS--CGKDNSYINVGVVGSDL 266
Query: 62 CI--TVNYETTKIDVWVMKEYGYRDSWCKLFTL 92
+ T DVW+MK G SW KLFT+
Sbjct: 267 SLLYTCQLGAATSDVWIMKHSGVNVSWTKLFTI 299
>UniRef100_Q9LU24 Gb|AAF25964.1 [Arabidopsis thaliana]
Length = 360
Score = 38.1 bits (87), Expect = 0.092
Identities = 30/77 (38%), Positives = 41/77 (52%), Gaps = 8/77 (10%)
Query: 25 IVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTK--IDVWVMKEYGY 82
I+ FNL+ + ++PLP QG TS + V LCIT YE K I + VM++ G
Sbjct: 206 ILCFNLSTHEYRKLPLPVYNQGVTSSWL--GVTSQKLCIT-EYEMCKKEIRISVMEKTG- 261
Query: 83 RDSWCKLFTLVKSCFTS 99
SW K+ +L S F S
Sbjct: 262 --SWSKIISLSMSSFIS 276
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.143 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 289,331,882
Number of Sequences: 2790947
Number of extensions: 11740032
Number of successful extensions: 39726
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 35
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 39675
Number of HSP's gapped (non-prelim): 58
length of query: 171
length of database: 848,049,833
effective HSP length: 118
effective length of query: 53
effective length of database: 518,718,087
effective search space: 27492058611
effective search space used: 27492058611
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 70 (31.6 bits)
Medicago: description of AC149130.3


