

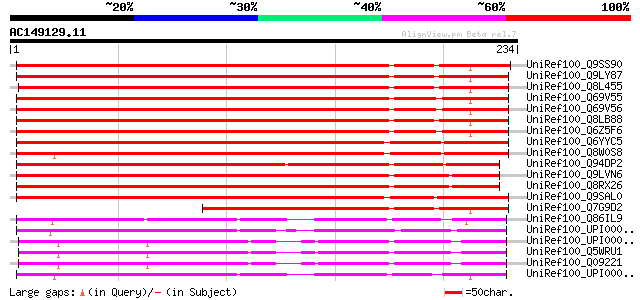
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149129.11 - phase: 0 /pseudo
(234 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SS90 F4P13.19 protein [Arabidopsis thaliana] 267 2e-70
UniRef100_Q9LY87 Hypothetical protein F18O22_210 [Arabidopsis th... 266 3e-70
UniRef100_Q8L455 P0497A05.19 protein [Oryza sativa] 265 9e-70
UniRef100_Q69V55 Hypothetical protein P0556B08.19-2 [Oryza sativa] 263 3e-69
UniRef100_Q69V56 Putative copine I [Oryza sativa] 263 3e-69
UniRef100_Q8LB88 Hypothetical protein [Arabidopsis thaliana] 254 2e-66
UniRef100_Q6Z5F6 Copine I-like [Oryza sativa] 245 6e-64
UniRef100_Q6YYC5 Copine I-like protein [Oryza sativa] 243 3e-63
UniRef100_Q8W0S8 Hypothetical protein [Sorghum bicolor] 240 2e-62
UniRef100_Q94DP2 P0483G10.1 protein [Oryza sativa] 232 7e-60
UniRef100_Q9LVN6 Gb|AAF01562.1 [Arabidopsis thaliana] 223 4e-57
UniRef100_Q8RX26 Hypothetical protein At5g63970 [Arabidopsis tha... 223 4e-57
UniRef100_Q9SAL0 T8K14.20 protein [Arabidopsis thaliana] 220 3e-56
UniRef100_Q7G9D2 F12A21.7 [Arabidopsis thaliana] 162 7e-39
UniRef100_Q86IL9 Similar to B0228.2.p [Dictyostelium discoideum] 159 4e-38
UniRef100_UPI000049A1C7 UPI000049A1C7 UniRef100 entry 159 6e-38
UniRef100_UPI00000851C9 UPI00000851C9 UniRef100 entry 153 4e-36
UniRef100_Q5WRU1 Hypothetical protein B0228.4 [Caenorhabditis el... 153 4e-36
UniRef100_Q09221 Hypothetical protein B0228.4 in chromosome II [... 153 4e-36
UniRef100_UPI0000498847 UPI0000498847 UniRef100 entry 152 5e-36
>UniRef100_Q9SS90 F4P13.19 protein [Arabidopsis thaliana]
Length = 489
Score = 267 bits (682), Expect = 2e-70
Identities = 142/229 (62%), Positives = 170/229 (74%), Gaps = 5/229 (2%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+G S R+SLH + N PN E+AI+IIG+ + F +DNLIPC+GFGDAST D DVFSFY
Sbjct: 170 TGAKSFNRKSLHHLSNTPNPYEQAITIIGRTLAAFDEDNLIPCYGFGDASTHDQDVFSFY 229
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
+ RFCNG EE+L+RYREI P ++ AG TSFAPIIEMA +V+QS G++HVLVII+DGQV
Sbjct: 230 PEGRFCNGFEEVLARYREIVPQLKLAGPTSFAPIIEMAMTVVEQSSGQYHVLVIIADGQV 289
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D +HG LS Q+ TVDAIV+A PLSI+LVGVGDGPWDM++EF DNI R F
Sbjct: 290 TRSVDTEHGRLSPQEQKTVDAIVKASTLPLSIVLVGVGDGPWDMMQEFDDNI--PARAFD 347
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILL-AIREILFQYKAAKELGLLGL 231
NF+FVNFTEIMSK N S KE E L A+ EI QYKA EL LLG+
Sbjct: 348 NFQFVNFTEIMSK--NKDQSRKETEFALSALMEIPPQYKATIELNLLGV 394
>UniRef100_Q9LY87 Hypothetical protein F18O22_210 [Arabidopsis thaliana]
Length = 468
Score = 266 bits (680), Expect = 3e-70
Identities = 142/228 (62%), Positives = 173/228 (75%), Gaps = 5/228 (2%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+G S R+SLH I + PN E+AI+IIG+ + F +DNLIPC+GFGDAST D DVFSF
Sbjct: 136 TGARSFNRKSLHFIGSSPNPYEQAITIIGRTLAAFDEDNLIPCYGFGDASTHDQDVFSFN 195
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
++RFCNG EE+LSRY+EI P ++ AG TSFAPII+MA IV+QSGG++HVLVII+DGQV
Sbjct: 196 SEDRFCNGFEEVLSRYKEIVPQLKLAGPTSFAPIIDMAMTIVEQSGGQYHVLVIIADGQV 255
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D ++G LS Q+ TVDAIV+A K PLSI+LVGVGDGPWDM++EF DNI R F
Sbjct: 256 TRSVDTENGQLSPQEQKTVDAIVQASKLPLSIVLVGVGDGPWDMMREFDDNI--PARAFD 313
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILL-AIREILFQYKAAKELGLLG 230
NF+FVNFTEIM+K N + SLKE E L A+ EI QYKA EL LLG
Sbjct: 314 NFQFVNFTEIMAK--NKAQSLKETEFALSALMEIPQQYKATIELNLLG 359
>UniRef100_Q8L455 P0497A05.19 protein [Oryza sativa]
Length = 495
Score = 265 bits (676), Expect = 9e-70
Identities = 143/227 (62%), Positives = 169/227 (73%), Gaps = 5/227 (2%)
Query: 5 GKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFYR 64
GK+S +SLH I N PN E+AISIIG+ S F +DNLIPCFGFGDA+T D DVF FY
Sbjct: 142 GKFSFHGRSLHHISNAPNPYEQAISIIGQTLSKFDEDNLIPCFGFGDATTHDQDVFCFYP 201
Query: 65 DERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQVI 124
D R CNG E LSRYRE+ P+++ AG TSFAPIIEMA IV+QSGG++HVL+II+DGQV
Sbjct: 202 DLRPCNGFSEALSRYRELVPHLRLAGPTSFAPIIEMAMTIVEQSGGQYHVLLIIADGQVT 261
Query: 125 RRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFHN 184
R D G LSSQ+ TVDAIV A + PLSI+LVGVGDGPWDM+KEF DNI +R F N
Sbjct: 262 RSVDTASGQLSSQEQKTVDAIVRASELPLSIVLVGVGDGPWDMMKEFDDNI--PSRAFDN 319
Query: 185 FKFVNFTEIMSKSNNISPSLKEAEILL-AIREILFQYKAAKELGLLG 230
F+FVNF+EIM+K N+ S KEA L A+ EI QYKA ELG+LG
Sbjct: 320 FQFVNFSEIMNK--NMPQSRKEAAFALSALMEIPQQYKATVELGILG 364
>UniRef100_Q69V55 Hypothetical protein P0556B08.19-2 [Oryza sativa]
Length = 415
Score = 263 bits (672), Expect = 3e-69
Identities = 141/228 (61%), Positives = 169/228 (73%), Gaps = 5/228 (2%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+GK S R+ LHDI N PN E+AISIIG+ S F +DNLIPCFGFGDAST D +VFSFY
Sbjct: 104 TGKLSFNRRCLHDIGNTPNPYEQAISIIGRTLSAFDEDNLIPCFGFGDASTHDQEVFSFY 163
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
+ R CNG EE L RYREI P ++ AG TSFAP+IE A IVD +GG++HVL+II+DGQV
Sbjct: 164 PENRPCNGFEEALERYREIVPTLRLAGPTSFAPMIETAIGIVDSTGGQYHVLLIIADGQV 223
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D Q G LS Q+ T+DAIV+A +FPLSI+LVGVGDGPWDM+ +F DNI R+F
Sbjct: 224 TRSVDTQSGQLSPQERDTIDAIVKASQFPLSIVLVGVGDGPWDMMHQFDDNI--PARSFD 281
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILL-AIREILFQYKAAKELGLLG 230
NF+FVNFT+IMSKS I+ KEAE L A+ EI QYKA +L LLG
Sbjct: 282 NFQFVNFTDIMSKS--IAADRKEAEFALSALMEIPTQYKATLDLQLLG 327
>UniRef100_Q69V56 Putative copine I [Oryza sativa]
Length = 447
Score = 263 bits (672), Expect = 3e-69
Identities = 141/228 (61%), Positives = 169/228 (73%), Gaps = 5/228 (2%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+GK S R+ LHDI N PN E+AISIIG+ S F +DNLIPCFGFGDAST D +VFSFY
Sbjct: 104 TGKLSFNRRCLHDIGNTPNPYEQAISIIGRTLSAFDEDNLIPCFGFGDASTHDQEVFSFY 163
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
+ R CNG EE L RYREI P ++ AG TSFAP+IE A IVD +GG++HVL+II+DGQV
Sbjct: 164 PENRPCNGFEEALERYREIVPTLRLAGPTSFAPMIETAIGIVDSTGGQYHVLLIIADGQV 223
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D Q G LS Q+ T+DAIV+A +FPLSI+LVGVGDGPWDM+ +F DNI R+F
Sbjct: 224 TRSVDTQSGQLSPQERDTIDAIVKASQFPLSIVLVGVGDGPWDMMHQFDDNI--PARSFD 281
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILL-AIREILFQYKAAKELGLLG 230
NF+FVNFT+IMSKS I+ KEAE L A+ EI QYKA +L LLG
Sbjct: 282 NFQFVNFTDIMSKS--IAADRKEAEFALSALMEIPTQYKATLDLQLLG 327
>UniRef100_Q8LB88 Hypothetical protein [Arabidopsis thaliana]
Length = 433
Score = 254 bits (648), Expect = 2e-66
Identities = 134/228 (58%), Positives = 167/228 (72%), Gaps = 5/228 (2%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+G S R+SLH I PN ++AISIIGK S F +DNLIPC+GFGDA+T D DVFSF
Sbjct: 107 TGARSFGRKSLHFIGTTPNPYQQAISIIGKTLSVFDEDNLIPCYGFGDATTHDQDVFSFN 166
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
++ +CNG EE+L YREI P ++ +G TSFAPIIE A IV++SGG++HVL+II+DGQV
Sbjct: 167 PNDTYCNGFEEVLMCYREIVPQLRLSGPTSFAPIIERAMTIVEESGGQYHVLLIIADGQV 226
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D +G S Q+ T+DAIV A ++PLSI+LVGVGDGPWD +++F DNI R F
Sbjct: 227 TRSVDTDNGGFSPQEQQTIDAIVRASEYPLSIVLVGVGDGPWDTMRQFDDNI--PARAFD 284
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILL-AIREILFQYKAAKELGLLG 230
NF+FVNFT+IMSK NI P+ KEAE L A+ EI QYKA ELGLLG
Sbjct: 285 NFQFVNFTDIMSK--NIDPARKEAEFALSALMEIPSQYKATLELGLLG 330
>UniRef100_Q6Z5F6 Copine I-like [Oryza sativa]
Length = 396
Score = 245 bits (626), Expect = 6e-64
Identities = 131/228 (57%), Positives = 166/228 (72%), Gaps = 5/228 (2%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+G+ S QSLH + ++ N E+AISIIG+ S F +DNLIPC+GFGDA+T D VFSFY
Sbjct: 74 TGRVSFNNQSLHSLGHISNPYEQAISIIGQTLSRFDEDNLIPCYGFGDATTHDQKVFSFY 133
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
D + C+G E+ L RYREI P ++ AG TSFAP+IE A IVD SGG++HVL+II+DGQV
Sbjct: 134 PDNKPCDGFEQALDRYREIVPQLRLAGPTSFAPMIETAIGIVDSSGGQYHVLLIIADGQV 193
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D +G LS Q+ T+DAIV+A +PLSI+LVGVGDGPWDM+++F DNI +R F
Sbjct: 194 TRSVDTGNGQLSPQERETIDAIVKASDYPLSIVLVGVGDGPWDMMRQFDDNI--PSRAFD 251
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILL-AIREILFQYKAAKELGLLG 230
NF+FVNFTEIMS+ + S KEAE L A+ EI Q+KAA L LLG
Sbjct: 252 NFQFVNFTEIMSRP--VPASKKEAEFALSALMEIPEQFKAAINLQLLG 297
>UniRef100_Q6YYC5 Copine I-like protein [Oryza sativa]
Length = 401
Score = 243 bits (620), Expect = 3e-63
Identities = 125/227 (55%), Positives = 163/227 (71%), Gaps = 3/227 (1%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+GK S QSLH I + PN E+AISIIGK +PF DDNLIPCFGFGDA+T D++VFSF+
Sbjct: 84 TGKRSFSGQSLHKIGSTPNPYEQAISIIGKTLAPFDDDNLIPCFGFGDATTHDYNVFSFH 143
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
D C+G EE+LS Y++I P+++ +G TSFAPI+E A IVD+SGG++HVLVI++DGQV
Sbjct: 144 PDNSPCHGFEEVLSCYKKIVPHLRLSGPTSFAPIVEAAVDIVDRSGGQYHVLVIVADGQV 203
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D LS Q+ TVD++V A +PLSI+LVGVGDGPW+ +++F D K+ R F
Sbjct: 204 TRSVDTSDNDLSPQERRTVDSVVMASSYPLSIVLVGVGDGPWEDMQKFDD--KIPARQFD 261
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILLAIREILFQYKAAKELGLLG 230
NF+FVNFT IMS+S + A L A+ E+ QYKA ELG+LG
Sbjct: 262 NFQFVNFTSIMSRSTT-QQQKESAFALAALMEVPIQYKATMELGILG 307
>UniRef100_Q8W0S8 Hypothetical protein [Sorghum bicolor]
Length = 449
Score = 240 bits (612), Expect = 2e-62
Identities = 126/228 (55%), Positives = 164/228 (71%), Gaps = 4/228 (1%)
Query: 4 SGKYSLIRQSLHDIRN-VPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSF 62
+GK S QSLH I + PN E+AISIIGK +PF +DNLIPCFGFGDA+T D++VFSF
Sbjct: 95 TGKQSFGGQSLHRISDDAPNPYEQAISIIGKTLAPFDEDNLIPCFGFGDATTHDNNVFSF 154
Query: 63 YRDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQ 122
+RD C+G EE+L+ YR + P+++ +G TSFAPI+E A IVD+SGG++HVLVI++DGQ
Sbjct: 155 HRDNTPCHGFEEVLACYRTVVPHLRLSGPTSFAPIVEAAVDIVDRSGGQYHVLVIVADGQ 214
Query: 123 VIRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTF 182
V R D G LS Q+ TVD+IV A +PLSI+LVGVGDGPWD ++ F D K+ R F
Sbjct: 215 VTRSVDTGDGDLSPQEKRTVDSIVMASSYPLSIVLVGVGDGPWDDMRRFDD--KLPARDF 272
Query: 183 HNFKFVNFTEIMSKSNNISPSLKEAEILLAIREILFQYKAAKELGLLG 230
NF+FVNFT IMS++ + + A L A+ E+ QYKA ELG+LG
Sbjct: 273 DNFQFVNFTSIMSRAAT-AQQKESAFALAALMEVPIQYKATMELGILG 319
>UniRef100_Q94DP2 P0483G10.1 protein [Oryza sativa]
Length = 402
Score = 232 bits (591), Expect = 7e-60
Identities = 125/223 (56%), Positives = 156/223 (69%), Gaps = 4/223 (1%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
SG+YS R+SLH I PN E+AISIIG+ SPF DDNLIPCFGFGDAST D VFSFY
Sbjct: 71 SGRYSFGRKSLHAISATPNPYEQAISIIGRTLSPFDDDNLIPCFGFGDASTHDQSVFSFY 130
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
+D R C G EE+L RYR+I P++ +G TSFAP+I A +V+ S ++HVLVII+DGQV
Sbjct: 131 QDSRSCCGFEEVLERYRQIVPHLNLSGPTSFAPLIYAAISVVENSNLQYHVLVIIADGQV 190
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
N + G LS Q+ T+ AIV+A +PLSI++VGVGDGPWD ++ F D I R F
Sbjct: 191 TTSN-TKDGKLSPQEQATIQAIVDASYYPLSIVMVGVGDGPWDAMQHFDDCI--PDRAFD 247
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILLAIREILFQYKAAKEL 226
NF+FVNFTEIMS S ++ P + A L A+ EI QYKA + L
Sbjct: 248 NFQFVNFTEIMSTSKDM-PKKEAAFALAALMEIPSQYKATQGL 289
>UniRef100_Q9LVN6 Gb|AAF01562.1 [Arabidopsis thaliana]
Length = 387
Score = 223 bits (567), Expect = 4e-57
Identities = 118/223 (52%), Positives = 156/223 (69%), Gaps = 3/223 (1%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+G+YS R+SLH I N EKAISIIG+ SPF +D+LIPCFGFGD +T D VFSFY
Sbjct: 51 TGRYSFNRKSLHAIGKRQNPYEKAISIIGRTLSPFDEDDLIPCFGFGDVTTRDQYVFSFY 110
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
+ + C+G E + RYREI P+++ +G TSFAP+I+ A IV+Q+ ++HVLVII+DGQV
Sbjct: 111 PENKSCDGLENAVKRYREIVPHLKLSGPTSFAPVIDAAINIVEQNNMQYHVLVIIADGQV 170
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D G LS Q+ T+++I+ A +PLSI+LVGVGDGPWD +K+F DNI R F
Sbjct: 171 TRNPDVPLGRLSPQEEATMNSIMAASHYPLSIVLVGVGDGPWDTMKQFDDNI--PHREFD 228
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILLAIREILFQYKAAKEL 226
NF+FVNFT+IMS+ + + + A L A+ EI FQYKA L
Sbjct: 229 NFQFVNFTKIMSEHKDAAKK-EAAFALAALMEIPFQYKATLSL 270
>UniRef100_Q8RX26 Hypothetical protein At5g63970 [Arabidopsis thaliana]
Length = 367
Score = 223 bits (567), Expect = 4e-57
Identities = 118/223 (52%), Positives = 156/223 (69%), Gaps = 3/223 (1%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+G+YS R+SLH I N EKAISIIG+ SPF +D+LIPCFGFGD +T D VFSFY
Sbjct: 51 TGRYSFNRKSLHAIGKRQNPYEKAISIIGRTLSPFDEDDLIPCFGFGDVTTRDQYVFSFY 110
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
+ + C+G E + RYREI P+++ +G TSFAP+I+ A IV+Q+ ++HVLVII+DGQV
Sbjct: 111 PENKSCDGLENAVKRYREIVPHLKLSGPTSFAPVIDAAINIVEQNNMQYHVLVIIADGQV 170
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D G LS Q+ T+++I+ A +PLSI+LVGVGDGPWD +K+F DNI R F
Sbjct: 171 TRNPDVPLGRLSPQEEATMNSIMAASHYPLSIVLVGVGDGPWDTMKQFDDNI--PHREFD 228
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILLAIREILFQYKAAKEL 226
NF+FVNFT+IMS+ + + + A L A+ EI FQYKA L
Sbjct: 229 NFQFVNFTKIMSEHKDAAKK-EAAFALAALMEIPFQYKATLSL 270
>UniRef100_Q9SAL0 T8K14.20 protein [Arabidopsis thaliana]
Length = 401
Score = 220 bits (560), Expect = 3e-56
Identities = 113/227 (49%), Positives = 157/227 (68%), Gaps = 3/227 (1%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+GK S + LH + N EKAI +IG+ +PF +DNLIPCFGFGD++T D +VF F+
Sbjct: 93 TGKTSFDGKCLHALGETSNPYEKAIFVIGQTLAPFDEDNLIPCFGFGDSTTHDEEVFGFH 152
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
D C+G EE+L+ Y+ I PN++ +G TS+ P+I+ A IV+++ G+ HVLVI++DGQV
Sbjct: 153 SDNSPCHGFEEVLACYKRIAPNLRLSGPTSYGPLIDAAVDIVEKNNGQFHVLVIVADGQV 212
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D G LS Q+ T+DAIV A + LSI+LVGVGDGPW+ +++F D K+ R F
Sbjct: 213 TRGTDMAEGELSQQEKTTIDAIVNASSYALSIVLVGVGDGPWEDMRKFDD--KIPKREFD 270
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILLAIREILFQYKAAKELGLLG 230
NF+FVNFTEIM++ N+ + + A L A+ EI FQY+AA EL LLG
Sbjct: 271 NFQFVNFTEIMTR-NSPESAKETAFALAALMEIPFQYQAAIELRLLG 316
>UniRef100_Q7G9D2 F12A21.7 [Arabidopsis thaliana]
Length = 347
Score = 162 bits (410), Expect = 7e-39
Identities = 87/142 (61%), Positives = 106/142 (74%), Gaps = 5/142 (3%)
Query: 90 GTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQVIRRNDAQHGSLSSQKLHTVDAIVEAR 149
G TSFAPIIE A IV++SGG++HVL+II+DGQV R D +G S Q+ T+DAIV A
Sbjct: 128 GPTSFAPIIERAMTIVEESGGQYHVLLIIADGQVTRSVDTDNGGFSPQEQQTIDAIVRAS 187
Query: 150 KFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFHNFKFVNFTEIMSKSNNISPSLKEAEI 209
++PLSI+LVGVGDGPWD +++F DNI R F NF+FVNFT+IMSK NI P+ KEAE
Sbjct: 188 EYPLSIVLVGVGDGPWDTMRQFDDNI--PARAFDNFQFVNFTDIMSK--NIDPARKEAEF 243
Query: 210 LL-AIREILFQYKAAKELGLLG 230
L A+ EI QYKA ELGLLG
Sbjct: 244 ALSALMEIPSQYKATLELGLLG 265
>UniRef100_Q86IL9 Similar to B0228.2.p [Dictyostelium discoideum]
Length = 284
Score = 159 bits (403), Expect = 4e-38
Identities = 98/228 (42%), Positives = 139/228 (59%), Gaps = 23/228 (10%)
Query: 4 SGKYSLIRQSLHDIR-NVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSF 62
+GK + + LHDI+ + N ++AI IIGK S F DDNL+P +GFGD+ T+D VFSF
Sbjct: 78 NGKKTFGGKCLHDIQPGMLNPYQEAIEIIGKTLSVFDDDNLLPTYGFGDSRTTDKSVFSF 137
Query: 63 YRDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQ 122
ER C G E+L +Y EI P + +G TSFAPII I + +H+LVII+DG+
Sbjct: 138 -TTERPCFGFNEVLQKYNEITPLVSLSGPTSFAPIIRETINIT-AAAKAYHILVIIADGE 195
Query: 123 VIRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTF 182
V N+ T+ AI EA K+P+SII +GVGDGPWD +++F D+ + R F
Sbjct: 196 VSVVNE------------TIKAIEEASKYPISIICIGVGDGPWDTMEKFDDD--LPERQF 241
Query: 183 HNFKFVNFTEIMSKSNNISPSLKEAEI-LLAIREILFQYKAAKELGLL 229
NF+FV F M+K+ N +E + + A++EI Q++ K+LGLL
Sbjct: 242 DNFQFVPFHTTMAKAEN-----REVDFAVAALQEIPEQFQYIKQLGLL 284
>UniRef100_UPI000049A1C7 UPI000049A1C7 UniRef100 entry
Length = 273
Score = 159 bits (402), Expect = 6e-38
Identities = 93/227 (40%), Positives = 135/227 (58%), Gaps = 18/227 (7%)
Query: 4 SGKYSLIRQSLHDI-RNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSF 62
+G S SLH + NV N ++ I IIG+ PF DDNLIP +GFGD +T+D VFSF
Sbjct: 63 TGAKSFNGYSLHGLFENVMNPYQRVIDIIGRTLEPFDDDNLIPVYGFGDVTTTDKSVFSF 122
Query: 63 YRDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQ 122
++ CNG +E+L RY EI P+++ +G T+FAP+I A +IV Q ++H+L+II+DGQ
Sbjct: 123 MPNDTPCNGFQEVLKRYNEITPSVRLSGPTNFAPLINKAVQIV-QRTRQYHILLIIADGQ 181
Query: 123 VIRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTF 182
V DA+ T AI A +PLSI+ +GVGDGP+D ++ F D + F
Sbjct: 182 V----DAERA--------TKAAIENASNYPLSIVTIGVGDGPFDKMENFDDELGQS--KF 227
Query: 183 HNFKFVNFTEIMSKSNNISPSLKEAEILLAIREILFQYKAAKELGLL 229
NF FVNF +++ + +P + A A+ E+ Q+ K+LG L
Sbjct: 228 DNFNFVNFQKVVDGPHVENPEI--AFATAAMNEVPTQFLCCKKLGYL 272
>UniRef100_UPI00000851C9 UPI00000851C9 UniRef100 entry
Length = 1788
Score = 153 bits (386), Expect = 4e-36
Identities = 94/229 (41%), Positives = 133/229 (58%), Gaps = 23/229 (10%)
Query: 5 GKYSLIRQSLHDIRNVP-NSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSF- 62
G+ + ++ LH I N ++ I I+GK S F D IP +GFGD +DH +F+
Sbjct: 1573 GERTFDKRPLHTIDPAEMNPYQQVIQIVGKTLSSFDADGQIPAYGFGDEEFTDHGIFNIA 1632
Query: 63 --YRDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISD 120
Y E+ CNG EE+L Y E+ P I+ +G T+F P+I+ A +I + +H+LVI++D
Sbjct: 1633 ERYDLEKDCNGFEEVLRVYNEVTPTIEMSGPTNFVPLIDRAIEICKEKHS-YHILVIVAD 1691
Query: 121 GQVIRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTR 180
GQV +++K++ AI A +PLSII+VGVGDGPW+M+ F DNI R
Sbjct: 1692 GQV-----------TNEKINQ-KAIAAASHYPLSIIMVGVGDGPWNMMGRFDDNI--PKR 1737
Query: 181 TFHNFKFVNFTEIMSKSNNISPSLKEAEILLAIREILFQYKAAKELGLL 229
F NF FV+F ++M + N S L A+ EI QYKA KELGLL
Sbjct: 1738 LFDNFHFVDFHKVMFNAPNADASF----ALNALMEIPDQYKAIKELGLL 1782
>UniRef100_Q5WRU1 Hypothetical protein B0228.4 [Caenorhabditis elegans]
Length = 7548
Score = 153 bits (386), Expect = 4e-36
Identities = 94/229 (41%), Positives = 133/229 (58%), Gaps = 23/229 (10%)
Query: 5 GKYSLIRQSLHDIRNVP-NSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSF- 62
G+ + ++ LH I N ++ I I+GK S F D IP +GFGD +DH +F+
Sbjct: 7333 GERTFDKRPLHTIDPAEMNPYQQVIQIVGKTLSSFDADGQIPAYGFGDEEFTDHGIFNIA 7392
Query: 63 --YRDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISD 120
Y E+ CNG EE+L Y E+ P I+ +G T+F P+I+ A +I + +H+LVI++D
Sbjct: 7393 ERYDLEKDCNGFEEVLRVYNEVTPTIEMSGPTNFVPLIDRAIEICKEKHS-YHILVIVAD 7451
Query: 121 GQVIRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTR 180
GQV +++K++ AI A +PLSII+VGVGDGPW+M+ F DNI R
Sbjct: 7452 GQV-----------TNEKINQ-KAIAAASHYPLSIIMVGVGDGPWNMMGRFDDNI--PKR 7497
Query: 181 TFHNFKFVNFTEIMSKSNNISPSLKEAEILLAIREILFQYKAAKELGLL 229
F NF FV+F ++M + N S L A+ EI QYKA KELGLL
Sbjct: 7498 LFDNFHFVDFHKVMFNAPNADASF----ALNALMEIPDQYKAIKELGLL 7542
>UniRef100_Q09221 Hypothetical protein B0228.4 in chromosome II [Caenorhabditis
elegans]
Length = 1633
Score = 153 bits (386), Expect = 4e-36
Identities = 94/229 (41%), Positives = 133/229 (58%), Gaps = 23/229 (10%)
Query: 5 GKYSLIRQSLHDIRNVP-NSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSF- 62
G+ + ++ LH I N ++ I I+GK S F D IP +GFGD +DH +F+
Sbjct: 1418 GERTFDKRPLHTIDPAEMNPYQQVIQIVGKTLSSFDADGQIPAYGFGDEEFTDHGIFNIA 1477
Query: 63 --YRDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISD 120
Y E+ CNG EE+L Y E+ P I+ +G T+F P+I+ A +I + +H+LVI++D
Sbjct: 1478 ERYDLEKDCNGFEEVLRVYNEVTPTIEMSGPTNFVPLIDRAIEICKEKHS-YHILVIVAD 1536
Query: 121 GQVIRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTR 180
GQV +++K++ AI A +PLSII+VGVGDGPW+M+ F DNI R
Sbjct: 1537 GQV-----------TNEKINQ-KAIAAASHYPLSIIMVGVGDGPWNMMGRFDDNI--PKR 1582
Query: 181 TFHNFKFVNFTEIMSKSNNISPSLKEAEILLAIREILFQYKAAKELGLL 229
F NF FV+F ++M + N S L A+ EI QYKA KELGLL
Sbjct: 1583 LFDNFHFVDFHKVMFNAPNADASF----ALNALMEIPDQYKAIKELGLL 1627
>UniRef100_UPI0000498847 UPI0000498847 UniRef100 entry
Length = 273
Score = 152 bits (385), Expect = 5e-36
Identities = 93/230 (40%), Positives = 132/230 (56%), Gaps = 23/230 (10%)
Query: 4 SGKYSLIRQSLHDIRN---VPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVF 60
+GK S Q LH I + N ++ I IG+ PF DD LIP +GFGD +T++ +F
Sbjct: 61 TGKVSFGGQCLHYINPQYPIFNPYQRVIDAIGRALEPFDDDRLIPTYGFGDVTTTNKSIF 120
Query: 61 SFYRDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISD 120
SF +E C G E+L RY EI P++Q +G T+FAP+I A IV ++ +H+L+II D
Sbjct: 121 SFMPNEAPCAGVAEVLRRYNEITPSVQMSGPTNFAPLIRKAIDIV-RTTHSYHILLIICD 179
Query: 121 GQVIRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTR 180
GQV D T AI+EA +PLSI+ +GVGDGP+D ++ + DN +
Sbjct: 180 GQVDNEKD------------TTAAIIEASNYPLSIVAIGVGDGPFDKMENYDDN--LTES 225
Query: 181 TFHNFKFVNFTEIMSKSNNISPSLKEAEILL-AIREILFQYKAAKELGLL 229
F NF+FVNF E++S P L++ + A++EI QY KE G L
Sbjct: 226 KFDNFQFVNFYEVVS----ARPDLQDIVFAVKALQEIPDQYAFIKEHGYL 271
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.139 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 377,610,725
Number of Sequences: 2790947
Number of extensions: 15229756
Number of successful extensions: 35868
Number of sequences better than 10.0: 241
Number of HSP's better than 10.0 without gapping: 103
Number of HSP's successfully gapped in prelim test: 138
Number of HSP's that attempted gapping in prelim test: 35368
Number of HSP's gapped (non-prelim): 248
length of query: 234
length of database: 848,049,833
effective HSP length: 123
effective length of query: 111
effective length of database: 504,763,352
effective search space: 56028732072
effective search space used: 56028732072
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 73 (32.7 bits)
Medicago: description of AC149129.11


