

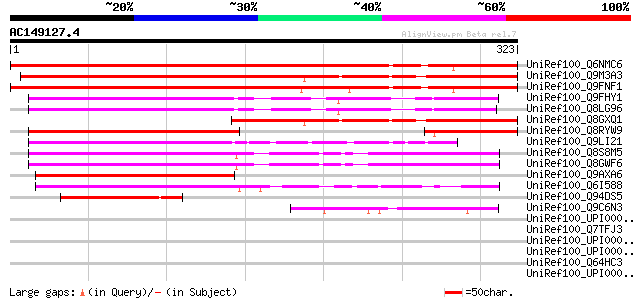
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149127.4 + phase: 0
(323 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6NMC6 At5g24310 [Arabidopsis thaliana] 386 e-106
UniRef100_Q9M3A3 Hypothetical protein F2K15.150 [Arabidopsis tha... 383 e-105
UniRef100_Q9FNF1 Emb|CAB66408.1 [Arabidopsis thaliana] 363 4e-99
UniRef100_Q9FHY1 Emb|CAB66408.1 [Arabidopsis thaliana] 190 5e-47
UniRef100_Q8LG96 Hypothetical protein [Arabidopsis thaliana] 188 2e-46
UniRef100_Q8GXQ1 Hypothetical protein At3g49290/F2K15_150 [Arabi... 181 2e-44
UniRef100_Q8RYW9 OSJNBa0066C06.8 protein [Oryza sativa] 165 2e-39
UniRef100_Q9LI21 Similar to Arabidopsis thaliana chromosome II B... 146 6e-34
UniRef100_Q8S8M5 Expressed protein [Arabidopsis thaliana] 139 8e-32
UniRef100_Q8GWF6 Hypothetical protein At2g46225/At2g46220 [Arabi... 137 5e-31
UniRef100_Q9AXA6 Hypothetical protein P0501G01.11 [Oryza sativa] 135 1e-30
UniRef100_Q6I588 Hypothetical protein OSJNBa0009C07.8 [Oryza sat... 132 1e-29
UniRef100_Q94DS5 P0460E08.23 protein [Oryza sativa] 55 3e-06
UniRef100_Q9C6N3 Transposon protein, putative [Arabidopsis thali... 48 4e-04
UniRef100_UPI000036334D UPI000036334D UniRef100 entry 46 0.002
UniRef100_Q7TFJ3 Rh148 [Rhesus cytomegalovirus] 44 0.008
UniRef100_UPI000035F990 UPI000035F990 UniRef100 entry 43 0.013
UniRef100_UPI00003C1D5C UPI00003C1D5C UniRef100 entry 43 0.013
UniRef100_Q64HC3 ASF/SF2-like pre-mRNA splicing factor SRP32 [Ze... 43 0.013
UniRef100_UPI000042F989 UPI000042F989 UniRef100 entry 42 0.017
>UniRef100_Q6NMC6 At5g24310 [Arabidopsis thaliana]
Length = 321
Score = 386 bits (992), Expect = e-106
Identities = 206/327 (62%), Positives = 243/327 (73%), Gaps = 10/327 (3%)
Query: 1 MGKIATQPLPRMASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQI 60
M AT P+PR ASNYDE+ M Q++LF DSL DLKNLRTQLYSAAEYFELSYTND+QKQI
Sbjct: 1 MSAAATMPMPREASNYDEISMQQSMLFSDSLKDLKNLRTQLYSAAEYFELSYTNDEQKQI 60
Query: 61 LVETLKDYAVKALINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMD 120
+VETLKDYA+KAL+NTVDHLGSV YKV+D +DEKV EV G +LR+SCIEQR++ CQ +MD
Sbjct: 61 VVETLKDYAIKALVNTVDHLGSVTYKVNDFVDEKVDEVAGTELRVSCIEQRLRMCQEYMD 120
Query: 121 HEGHTQQSLVISTPKHHKRYILPVGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATI 180
HEG +QQSLVI TPK HKRY LP GE G N K K + D EDDW+ FRNAVRATI
Sbjct: 121 HEGRSQQSLVIDTPKFHKRYFLPSGEIKRGGNLAKLKNVEGSFDGEDDWNQFRNAVRATI 180
Query: 181 RETPTSTSSKGNSPSPS-LQPQRVGAFSFTSPNMA-KKDLEKRTVSPHRFPLSRTGSMSS 238
RETP K SPS +PQR FSF+S A KK+ +KR VSPHRFPL R+GS++
Sbjct: 181 RETPPPPVRKPILQSPSQRKPQRSATFSFSSIATAPKKEQDKRAVSPHRFPLLRSGSVAI 240
Query: 239 RSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLS--SDVNNIRDIDQHPSK 296
R ++ R TTP+ S T +P RYPSEPR+SAS+R++ + + Q PSK
Sbjct: 241 RPSS--ISRPTTPSKSRAVTPTPK----RYPSEPRRSASVRVAFEKEAQKEPEHQQQPSK 294
Query: 297 SKRLLKSLLSRRKSKKDDTLYTYLDEY 323
SKRLLK+LLSRRK+KKDDTLYTYLDEY
Sbjct: 295 SKRLLKALLSRRKTKKDDTLYTYLDEY 321
>UniRef100_Q9M3A3 Hypothetical protein F2K15.150 [Arabidopsis thaliana]
Length = 312
Score = 383 bits (984), Expect = e-105
Identities = 201/319 (63%), Positives = 240/319 (75%), Gaps = 11/319 (3%)
Query: 8 PLPRMASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKD 67
P ASNYDEV M Q++LF D L DLKNLR QLYSAAEYFELSYT DD+KQI+VETLKD
Sbjct: 2 PASHEASNYDEVSMQQSMLFSDGLQDLKNLRAQLYSAAEYFELSYTTDDKKQIVVETLKD 61
Query: 68 YAVKALINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQ 127
YAVKAL+NTVDHLGSV YKV+D +DEKV EV +LR+SCIEQR++ CQ +MDHEG +QQ
Sbjct: 62 YAVKALVNTVDHLGSVTYKVNDFIDEKVDEVSETELRVSCIEQRLRMCQEYMDHEGRSQQ 121
Query: 128 SLVISTPKHHKRYILPVGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTS- 186
SLVI TPK HKRYILP GE + TN K KY G L+D DDW+ FRNAVRATIRETP
Sbjct: 122 SLVIDTPKFHKRYILPAGEIMTATNLEKLKYFGSSLEDADDWNQFRNAVRATIRETPPPP 181
Query: 187 -TSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKT 245
S S SP PQR FSFTS + KK+ +KR+VSPHRFPL R+GS+++R +
Sbjct: 182 VRKSTSQSSSPRQPPQRSATFSFTS-TIPKKEQDKRSVSPHRFPLLRSGSVATRKSA-SI 239
Query: 246 GRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDID-QHPSKSKRLLKSL 304
R TTP+ S T +RYPSEPR+SAS+R++ + +N ++ + Q PSKSKRLLK+L
Sbjct: 240 SRPTTPSKSRSITP------IRYPSEPRRSASVRVAFEKDNQKETEQQQPSKSKRLLKAL 293
Query: 305 LSRRKSKKDDTLYTYLDEY 323
LSRRK+KKDDTLYT+LDEY
Sbjct: 294 LSRRKTKKDDTLYTFLDEY 312
>UniRef100_Q9FNF1 Emb|CAB66408.1 [Arabidopsis thaliana]
Length = 333
Score = 363 bits (931), Expect = 4e-99
Identities = 196/339 (57%), Positives = 237/339 (69%), Gaps = 22/339 (6%)
Query: 1 MGKIATQPLPRMASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQI 60
M AT P+PR ASNYDE+ M Q++LF DSL DLKNLRTQLYSAAEYFELSYTND+QKQI
Sbjct: 1 MSAAATMPMPREASNYDEISMQQSMLFSDSLKDLKNLRTQLYSAAEYFELSYTNDEQKQI 60
Query: 61 LVETLKDYAVKALINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMD 120
+VETLKDYA+KAL+NTVDHLGSV YKV+D +DEKV EV G +LR+SCIEQR++ CQ +MD
Sbjct: 61 VVETLKDYAIKALVNTVDHLGSVTYKVNDFVDEKVDEVAGTELRVSCIEQRLRMCQEYMD 120
Query: 121 HEGHTQQSLVISTPKHHKRYILPVGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATI 180
HEG +QQSLVI TPK HKRY LP GE G N K K + D EDDW+ FRNAVRATI
Sbjct: 121 HEGRSQQSLVIDTPKFHKRYFLPSGEIKRGGNLAKLKNVEGSFDGEDDWNQFRNAVRATI 180
Query: 181 RETP------------TSTSSKGNSPSPSLQPQRVGAFSFTSPNMAK--KDLEKRTVSPH 226
RETP T TS ++ + + V + + ++KR VSPH
Sbjct: 181 RETPPPPVRYSPVLIKTDTSVSLSTETSTFSNLFVLLYRHRTKERTSVHGKIDKRAVSPH 240
Query: 227 RFPLSRTGSMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLS--SDV 284
RFPL R+GS++ R ++ R TTP+ S T +P RYPSEPR+SAS+R++ +
Sbjct: 241 RFPLLRSGSVAIRPSS--ISRPTTPSKSRAVTPTPK----RYPSEPRRSASVRVAFEKEA 294
Query: 285 NNIRDIDQHPSKSKRLLKSLLSRRKSKKDDTLYTYLDEY 323
+ Q PSKSKRLLK+LLSRRK+KKDDTLYTYLDEY
Sbjct: 295 QKEPEHQQQPSKSKRLLKALLSRRKTKKDDTLYTYLDEY 333
>UniRef100_Q9FHY1 Emb|CAB66408.1 [Arabidopsis thaliana]
Length = 279
Score = 190 bits (482), Expect = 5e-47
Identities = 115/302 (38%), Positives = 168/302 (55%), Gaps = 40/302 (13%)
Query: 13 ASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKA 72
+SN+DE+FM QTL F ++L DLKNLR QLYSAAEYFE SY + K+ ++ETLK+YA KA
Sbjct: 14 SSNHDELFMKQTLQFSETLKDLKNLRKQLYSAAEYFETSYGKAEHKETVIETLKEYAAKA 73
Query: 73 LINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVIS 132
++NTVDHLGSV+ K + L + T LRLS +EQR++ C+ +M G Q L+
Sbjct: 74 VVNTVDHLGSVSDKFNSFLSDNSTHFSTTHLRLSSLEQRMRLCRDYMGKSGTHQHLLLFQ 133
Query: 133 TPKHHKRYILPVGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSKGN 192
P+HHKRY P + GT+ + DD H F +AVR+TI E +T+ K N
Sbjct: 134 YPRHHKRYFFP--QQGRGTSFSAG----------DDSHRFTSAVRSTILENLPNTARKAN 181
Query: 193 SPSPSLQPQRVGAFSF---TSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRST 249
+ G+FSF N+ + KR+ SP RFPL R+GS+ RS++P
Sbjct: 182 ---------KTGSFSFAPIVHNNINNRTPNKRSNSPMRFPLLRSGSLLKRSSSP------ 226
Query: 250 TPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPSKSKRLLKSLLSRRK 309
+ P + P EP+++ S+ ++++ I+ K + K+L+S K
Sbjct: 227 ---------SQPKKPPLALP-EPQRAISVSRNTEIVEIKQSSSRKGKKILMFKALMSMSK 276
Query: 310 SK 311
S+
Sbjct: 277 SR 278
>UniRef100_Q8LG96 Hypothetical protein [Arabidopsis thaliana]
Length = 279
Score = 188 bits (477), Expect = 2e-46
Identities = 114/301 (37%), Positives = 167/301 (54%), Gaps = 40/301 (13%)
Query: 13 ASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKA 72
+SN+DE+FM QTL F ++L DLKNLR QLYSAAEYFE SY ++ K+ ++ETLK+YA KA
Sbjct: 14 SSNHDELFMKQTLQFSETLKDLKNLRKQLYSAAEYFETSYGKEEHKETVIETLKEYAAKA 73
Query: 73 LINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVIS 132
++NTVDHLGSV+ K + L + T LRLS +EQR++ C+ +M G Q L+
Sbjct: 74 VVNTVDHLGSVSDKFNSFLSDNSTHFSTTHLRLSSLEQRMRLCRDYMGKSGTHQHLLLFQ 133
Query: 133 TPKHHKRYILPVGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSKGN 192
P+HHKRY P + GT+ + DD H F +AVR+TI E +T+ K N
Sbjct: 134 YPRHHKRYFFP--QQGRGTSFSAG----------DDSHRFTSAVRSTILENLPNTARKAN 181
Query: 193 SPSPSLQPQRVGAFSF---TSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRST 249
+ G+FSF N+ + KR+ SP RFPL +GS+ RS++P
Sbjct: 182 ---------KTGSFSFAPIVHNNINNRTPNKRSNSPMRFPLLSSGSLLKRSSSP------ 226
Query: 250 TPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPSKSKRLLKSLLSRRK 309
+ P + P EP+++ S+ ++++ I+ K + K+L+S K
Sbjct: 227 ---------SQPKKPPLPLP-EPQRAISVSRNTEIVEIKQSSSRKGKKILMFKALMSMSK 276
Query: 310 S 310
S
Sbjct: 277 S 277
>UniRef100_Q8GXQ1 Hypothetical protein At3g49290/F2K15_150 [Arabidopsis thaliana]
Length = 180
Score = 181 bits (459), Expect = 2e-44
Identities = 101/185 (54%), Positives = 127/185 (68%), Gaps = 11/185 (5%)
Query: 142 LPVGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTS--TSSKGNSPSPSLQ 199
L GE + TN K KY G L+D DDW+ FRNAVRATIRETP S S SP
Sbjct: 4 LSAGEIMTATNLEKLKYFGSSLEDADDWNQFRNAVRATIRETPPPPVRKSTSQSSSPRQP 63
Query: 200 PQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRSTTPNSSNRATT 259
PQR FSFTS + KK+ +KR+VSPHRFPL R+GS+++R + R TTP+ S T
Sbjct: 64 PQRSATFSFTS-TIPKKEQDKRSVSPHRFPLLRSGSVATRKSA-SISRPTTPSKSRSITP 121
Query: 260 SPSNARVRYPSEPRKSASMRLSSDVNNIRDID-QHPSKSKRLLKSLLSRRKSKKDDTLYT 318
+RYPSEPR+SAS+R++ + +N ++ + Q PSKSKRLLK+LLSRRK+KKDDTLYT
Sbjct: 122 ------IRYPSEPRRSASVRVAFEKDNQKETEQQQPSKSKRLLKALLSRRKTKKDDTLYT 175
Query: 319 YLDEY 323
+LDEY
Sbjct: 176 FLDEY 180
>UniRef100_Q8RYW9 OSJNBa0066C06.8 protein [Oryza sativa]
Length = 271
Score = 165 bits (417), Expect = 2e-39
Identities = 78/134 (58%), Positives = 106/134 (78%)
Query: 13 ASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKA 72
+S +EV M +TL+F D++ DLK L++QLYSAAEYFEL+YT +D KQ ++ LK+Y+VKA
Sbjct: 20 SSTLEEVQMQETLIFSDTIKDLKMLKSQLYSAAEYFELAYTQEDDKQEVMNNLKEYSVKA 79
Query: 73 LINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVIS 132
L+NTVDHLGS+++KVS L+D++ EV +LR+SCI QR + Q MD EG +QQSLVI+
Sbjct: 80 LVNTVDHLGSISFKVSSLIDQRFDEVDDTNLRVSCIHQRAQVSQACMDKEGLSQQSLVIT 139
Query: 133 TPKHHKRYILPVGE 146
PK+HKRYILP G+
Sbjct: 140 APKYHKRYILPAGD 153
Score = 57.8 bits (138), Expect = 4e-07
Identities = 30/65 (46%), Positives = 44/65 (67%), Gaps = 6/65 (9%)
Query: 265 RVRYP------SEPRKSASMRLSSDVNNIRDIDQHPSKSKRLLKSLLSRRKSKKDDTLYT 318
R++YP SE +K AS+R +D N+ ++ +Q P K K+ LKSLLSRRKS+K++ L
Sbjct: 207 RIQYPPNEMHTSEHKKLASVRSQADRNDDKEGEQTPKKGKKFLKSLLSRRKSRKEEPLPC 266
Query: 319 YLDEY 323
Y D+Y
Sbjct: 267 YFDDY 271
>UniRef100_Q9LI21 Similar to Arabidopsis thaliana chromosome II BAC T3F17 [Oryza
sativa]
Length = 652
Score = 146 bits (369), Expect = 6e-34
Identities = 100/273 (36%), Positives = 150/273 (54%), Gaps = 15/273 (5%)
Query: 13 ASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKA 72
+++ D + + LLF DSL DL+NLR+QLYSAAEYFE+ Y N+ QK ++ +LKDY V+A
Sbjct: 345 STSEDMSSLQEGLLFSDSLKDLRNLRSQLYSAAEYFEVFYRNNSQKSTVMTSLKDYTVEA 404
Query: 73 LINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVIS 132
L++TVDHLG V+YKV +L+ E+ EV + R+S +EQR++ CQ +D EG +QQSL+I
Sbjct: 405 LVSTVDHLGFVSYKVDNLVKERSDEVNETEFRVSSVEQRVRICQQTIDQEGRSQQSLLIR 464
Query: 133 TPKHHKRYILPVGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSKGN 192
PK+H+R + V +H S +Y H+ + H ++ + T S++
Sbjct: 465 APKYHRRTDI-VESAIHPV-SEPPRYSRQHMSRK---MHKSQSISTPVGRQSTMRSAR-- 517
Query: 193 SPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRSTTPN 252
SPSPS + + S + A+ ++ SP G +SS T S PN
Sbjct: 518 SPSPSARGTHHRSRSLSPSRKAR----AKSPSPQIISTQTKGMLSSLDET--RAGSPIPN 571
Query: 253 SSNRATTSPSNARVRYPSEPRKSASMRLSSDVN 285
SN S + AR R P P+ S L ++
Sbjct: 572 -SNPLARSATVAR-RPPVHPKHFYSYALQVSIS 602
>UniRef100_Q8S8M5 Expressed protein [Arabidopsis thaliana]
Length = 298
Score = 139 bits (351), Expect = 8e-32
Identities = 94/302 (31%), Positives = 156/302 (51%), Gaps = 24/302 (7%)
Query: 13 ASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKA 72
A DEV M + F +L +LKNLR QLYSAA+Y E SY + +QKQ++++ LKDY VKA
Sbjct: 12 AMTLDEVSMERNKSFVKALQELKNLRPQLYSAADYCEKSYLHSEQKQMVLDNLKDYTVKA 71
Query: 73 LINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVIS 132
L+N VDHLG+VA K++DL D + +++ ++R SC+ Q++ TC+ ++D EG QQ L+
Sbjct: 72 LVNAVDHLGTVASKLTDLFDHQNSDISTMEMRASCVSQQLLTCRTYIDKEGLRQQQLLAV 131
Query: 133 TPKHHKRYILP--VGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSK 190
P HHK YILP V + +H + ++ + +H++ R + P S S
Sbjct: 132 IPLHHKHYILPNSVNKRVHFSPLRRT---------DTRQNHYQAISRLQPSDAPASKSLS 182
Query: 191 GNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRSTT 250
+ S + + S +P + KD + S+T + + +
Sbjct: 183 WHLGSET--KSTLKGTSTVAP--SSKDSK---------AFSKTSGVFHLLGDDENIANKK 229
Query: 251 PNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPSKSKRLLKSLLSRRKS 310
P + ++ + P+ + E K + S D N R+I Q P ++K +L + ++K+
Sbjct: 230 PLAGSQVSGVPAASTAHKDLEVPKLLTAHRSLDNNPRREIIQAPVRTKSVLSAFFVKQKT 289
Query: 311 KK 312
K
Sbjct: 290 PK 291
>UniRef100_Q8GWF6 Hypothetical protein At2g46225/At2g46220 [Arabidopsis thaliana]
Length = 298
Score = 137 bits (344), Expect = 5e-31
Identities = 93/302 (30%), Positives = 155/302 (50%), Gaps = 24/302 (7%)
Query: 13 ASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKA 72
A DEV M + F +L +LKNLR QLYSAA+Y E SY + +QKQ++++ LKDY VKA
Sbjct: 12 AMTLDEVSMERNKSFVKALQELKNLRPQLYSAADYCEKSYLHSEQKQMVLDNLKDYTVKA 71
Query: 73 LINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVIS 132
L+N VDHLG+VA K++DL D + +++ ++R SC+ Q++ TC+ ++D EG QQ L+
Sbjct: 72 LVNAVDHLGTVASKLTDLFDHQNSDISTMEMRASCVSQQLLTCRTYIDKEGLRQQQLLAV 131
Query: 133 TPKHHKRYILP--VGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSK 190
P HHK YILP V + +H + ++ + +H++ R + P S S
Sbjct: 132 IPLHHKHYILPNSVNKRVHFSPLRRT---------DTRQNHYQAISRLQPSDAPASKSLS 182
Query: 191 GNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRSTT 250
+ S + + S +P + KD + S+T + + +
Sbjct: 183 WHLGSET--KSTLKGTSTVAP--SSKDSK---------AFSKTSGVFHLLGDDENIANKK 229
Query: 251 PNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPSKSKRLLKSLLSRRKS 310
P + ++ + P+ + E K + S D N R+I P ++K +L + ++K+
Sbjct: 230 PLAGSQVSGVPAASTAHKDLEVPKLLTAHRSLDNNPRREIILAPVRTKSVLSAFFVKQKT 289
Query: 311 KK 312
K
Sbjct: 290 PK 291
>UniRef100_Q9AXA6 Hypothetical protein P0501G01.11 [Oryza sativa]
Length = 306
Score = 135 bits (340), Expect = 1e-30
Identities = 62/127 (48%), Positives = 94/127 (73%)
Query: 17 DEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKALINT 76
DE M ++ F +L +LKNLR QLYSA+EY E SY + +QKQ++++ LKDYAV+AL+N
Sbjct: 34 DEASMERSKSFVKALQELKNLRPQLYSASEYCEKSYLHSEQKQMVLDNLKDYAVRALVNA 93
Query: 77 VDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVISTPKH 136
VDHLG+VAYK++DL +++ +EV +L+++C+ Q++ TCQ + D EG QQ + + +H
Sbjct: 94 VDHLGTVAYKLTDLYEQQASEVSTLELKVACLNQQVLTCQTYTDKEGIRQQQMTGTATRH 153
Query: 137 HKRYILP 143
HK YI+P
Sbjct: 154 HKHYIVP 160
>UniRef100_Q6I588 Hypothetical protein OSJNBa0009C07.8 [Oryza sativa]
Length = 302
Score = 132 bits (333), Expect = 1e-29
Identities = 102/311 (32%), Positives = 153/311 (48%), Gaps = 55/311 (17%)
Query: 17 DEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKALINT 76
DE M ++ F +L +LKNLR QLYSA+EY E SY + +QKQ+++E LKDYAV+A++N
Sbjct: 25 DEASMERSKSFVKALQELKNLRPQLYSASEYCEKSYLHSEQKQMVLENLKDYAVRAVVNA 84
Query: 77 VDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVISTPKH 136
VDHLG+VAYK++DL +++ +EV +L+++ + Q+I TCQ F D G QQ + +T KH
Sbjct: 85 VDHLGTVAYKLTDLFEQQASEVSTVELKVARLNQQILTCQIFTDRAGLRQQKIGGTTFKH 144
Query: 137 HKRYILPVG----------ETLHGTNSTKSKY-----IGCHLDDEDDWHHFRNAVRATIR 181
HK YILP +T +G +S Y + HL E N++ T
Sbjct: 145 HKHYILPSTGHKRTQAARLQTDNGQDSKPKPYPSAKTLSWHLSSE-------NSISTTGA 197
Query: 182 ETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRST 241
+ T T S P+ S S + KD+ SP PL G+ +S
Sbjct: 198 QKYTFTLGDTISSKPA---------SNGSMYLLGKDI---PASPMHKPLQPNGN-TSFDA 244
Query: 242 TPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPSKSKRLL 301
G P + +T + + +PR R+I + P +K +L
Sbjct: 245 KKNVGSKDQPGFMHMSTFNALD-------KPRG-------------REIQKVPVSTKSML 284
Query: 302 KSLLSRRKSKK 312
+L + KS K
Sbjct: 285 ATLFIKHKSAK 295
>UniRef100_Q94DS5 P0460E08.23 protein [Oryza sativa]
Length = 142
Score = 55.1 bits (131), Expect = 3e-06
Identities = 25/78 (32%), Positives = 53/78 (67%), Gaps = 1/78 (1%)
Query: 33 DLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKALINTVDHLGSVAYKVSDLLD 92
+LK+LR+QL+ A+ E ++ + ++K++++E+ K Y A++ +DHLG+V+ K+ L
Sbjct: 60 ELKDLRSQLHQTADCCEKAFLDTEKKKLILESTKGYICDAIVAVIDHLGTVSSKLEQQLQ 119
Query: 93 EKVTEVFGEDLRLSCIEQ 110
EK+ E+ + +L+ ++Q
Sbjct: 120 EKI-EITQTEKKLNFLKQ 136
>UniRef100_Q9C6N3 Transposon protein, putative [Arabidopsis thaliana]
Length = 1148
Score = 47.8 bits (112), Expect = 4e-04
Identities = 41/143 (28%), Positives = 72/143 (49%), Gaps = 16/143 (11%)
Query: 180 IRETPTSTSSKGNSPSPSLQ--PQRVGAFSFTSPNMAKKDLEKRTVSPHR--FPLSRTG- 234
+R T +SS+GNSPSP ++ + FS +P + L R S R P SR G
Sbjct: 225 VRGTSPVSSSRGNSPSPKIKVWQSNIPGFSLDAPPNLRTSLGDRPASYVRGSSPASRNGR 284
Query: 235 -SMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDI--- 290
++S+RS +S +P++S ++S S+ R R+ S+ + S + D+++++ I
Sbjct: 285 DAVSTRSR-----KSVSPSASRSVSSSHSHERDRFSSQSKGSVASSGDDDLHSLQSIPVG 339
Query: 291 --DQHPSKSKRLLKSLLSRRKSK 311
++ SK L + + R SK
Sbjct: 340 GSERAVSKRASLSPNSRTSRSSK 362
>UniRef100_UPI000036334D UPI000036334D UniRef100 entry
Length = 665
Score = 45.8 bits (107), Expect = 0.002
Identities = 30/113 (26%), Positives = 52/113 (45%), Gaps = 6/113 (5%)
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
+P++TS+ +SP+ S + + + TSP+ T S + T SS +T+
Sbjct: 247 SPSTTSATTSSPTTSATTSQTSSSTSTSPSTTSATTSSPTTS------ATTSQTSSSTTS 300
Query: 243 PKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPS 295
P T +TT + + ATTS +++ PS + S +S N+ H S
Sbjct: 301 PSTTSATTSSPTTSATTSQTSSSTTSPSTTSATTSSPTTSATNSQTSSSTHKS 353
Score = 43.5 bits (101), Expect = 0.008
Identities = 31/113 (27%), Positives = 51/113 (44%), Gaps = 5/113 (4%)
Query: 175 AVRATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSR-- 232
+V + + TSTS NS + S + + + TSP+ T S PL+
Sbjct: 522 SVTTSQTSSSTSTSPSTNSATTSATTSQTSSSTSTSPSTTSATTSSPTTSASSVPLTTEP 581
Query: 233 ---TGSMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSS 282
T + SS +T+P T +TT + + ATTS +++ PS + S +S
Sbjct: 582 ATTTTTSSSTTTSPSTTSATTSSPTTSATTSQTSSSTTSPSTTSATTSSPTTS 634
Score = 41.2 bits (95), Expect = 0.038
Identities = 29/110 (26%), Positives = 50/110 (45%), Gaps = 4/110 (3%)
Query: 178 ATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMS 237
AT +T +ST+S S + + P S TS + T S ++ + + S
Sbjct: 471 ATTSQTSSSTTSPSTSSATTSSPTTSATTSQTSSSTTSPSTTSATTSSPTTSVTTSQTSS 530
Query: 238 SRSTTPKTG----RSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSD 283
S ST+P T +TT +S+ +TSPS S ++S+ L+++
Sbjct: 531 STSTSPSTNSATTSATTSQTSSSTSTSPSTTSATTSSPTTSASSVPLTTE 580
Score = 38.9 bits (89), Expect = 0.19
Identities = 23/82 (28%), Positives = 42/82 (51%), Gaps = 6/82 (7%)
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
+P++TS+ +SP+ S + + + TSP+ T S + T SS +T+
Sbjct: 194 SPSTTSATTSSPTTSATTSQTSSSTSTSPSTTSATTSSPTTS------ATTSQTSSSTTS 247
Query: 243 PKTGRSTTPNSSNRATTSPSNA 264
P T +TT + + ATTS +++
Sbjct: 248 PSTTSATTSSPTTSATTSQTSS 269
Score = 38.9 bits (89), Expect = 0.19
Identities = 28/100 (28%), Positives = 49/100 (49%), Gaps = 6/100 (6%)
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
+P++TS+ +SP+ S + + S TSP+ T S + + + SS ST+
Sbjct: 221 SPSTTSATTSSPTTSATTSQTSS-STTSPSTTSATTSSPTTSA-----TTSQTSSSTSTS 274
Query: 243 PKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSS 282
P T +TT + + ATTS +++ PS + S +S
Sbjct: 275 PSTTSATTSSPTTSATTSQTSSSTTSPSTTSATTSSPTTS 314
Score = 37.4 bits (85), Expect = 0.55
Identities = 27/100 (27%), Positives = 46/100 (46%), Gaps = 3/100 (3%)
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
+P++TS+ +SP+ S S TS + T S + + + SS ST+
Sbjct: 165 SPSTTSATTSSPTTSAT---AATSSQTSSSTTSPSTTSATTSSPTTSATTSQTSSSTSTS 221
Query: 243 PKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSS 282
P T +TT + + ATTS +++ PS + S +S
Sbjct: 222 PSTTSATTSSPTTSATTSQTSSSTTSPSTTSATTSSPTTS 261
Score = 37.0 bits (84), Expect = 0.72
Identities = 29/107 (27%), Positives = 51/107 (47%), Gaps = 6/107 (5%)
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
+P++TS+ +SP+ S + + S TSP+ + + + S T S+S+ S T
Sbjct: 405 SPSTTSATTSSPTTSATTSQTSS-SSTSPSTTSATTSSPSATSSQTSSSTTTSLSTTSAT 463
Query: 243 ---PKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNN 286
P T +T+ SS +TTSPS + S + + + SS +
Sbjct: 464 TSLPTTSATTSQTSS--STTSPSTSSATTSSPTTSATTSQTSSSTTS 508
Score = 37.0 bits (84), Expect = 0.72
Identities = 28/114 (24%), Positives = 49/114 (42%), Gaps = 14/114 (12%)
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRF-------------- 228
+P++TS+ +SP+ S + + TSP+ T S
Sbjct: 17 SPSTTSATTSSPTTSPTTSHTTSSTATSPSTTSATTSSPTTSATTSQTSSSTATIPLTTE 76
Query: 229 PLSRTGSMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSS 282
P + T + SS +T+P T +TT + + ATTS +++ PS + S +S
Sbjct: 77 PATTTTTSSSTTTSPSTTSATTSSPTTSATTSQTSSSTTSPSTTSATTSSPTTS 130
Score = 36.6 bits (83), Expect = 0.94
Identities = 31/108 (28%), Positives = 52/108 (47%), Gaps = 11/108 (10%)
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
+P++TS+ +SP+ S + + S TSP+ T S ++ + + SS ST+
Sbjct: 90 SPSTTSATTSSPTTSATTSQTSS-STTSPSTTSATTSSPTTS-----VTTSQTSSSTSTS 143
Query: 243 PKT----GRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNN 286
P T +TT +S+ +TSPS S P SA+ SS ++
Sbjct: 144 PSTTSATTSATTSQTSSSTSTSPSTTSAT-TSSPTTSATAATSSQTSS 190
Score = 35.8 bits (81), Expect = 1.6
Identities = 26/113 (23%), Positives = 50/113 (44%), Gaps = 9/113 (7%)
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPN--------MAKKDLEKRTVSPHRFPLSRTG 234
+P++TS+ +SP+ S+ + + + TSP+ + + SP + +
Sbjct: 508 SPSTTSATTSSPTTSVTTSQTSSSTSTSPSTNSATTSATTSQTSSSTSTSPSTTSATTSS 567
Query: 235 SMSSRSTTPKTGR-STTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNN 286
+S S+ P T +TT +S+ TTSPS S + + + SS +
Sbjct: 568 PTTSASSVPLTTEPATTTTTSSSTTTSPSTTSATTSSPTTSATTSQTSSSTTS 620
Score = 35.4 bits (80), Expect = 2.1
Identities = 22/104 (21%), Positives = 48/104 (46%), Gaps = 1/104 (0%)
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
+P++TS+ +SP+ S+ + + + TSP+ T + + S+ +++
Sbjct: 116 SPSTTSATTSSPTTSVTTSQTSSSTSTSPSTTSATTSATTSQTSSSTSTSPSTTSATTSS 175
Query: 243 PKT-GRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVN 285
P T + T + ++ +TTSPS S + + + SS +
Sbjct: 176 PTTSATAATSSQTSSSTTSPSTTSATTSSPTTSATTSQTSSSTS 219
Score = 35.0 bits (79), Expect = 2.7
Identities = 25/90 (27%), Positives = 42/90 (45%), Gaps = 3/90 (3%)
Query: 175 AVRATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTG 234
+V + + TSTS S + S + + + TSP+ T S S+T
Sbjct: 130 SVTTSQTSSSTSTSPSTTSATTSATTSQTSSSTSTSPSTTSATTSSPTTSATAATSSQT- 188
Query: 235 SMSSRSTTPKTGRSTTPNSSNRATTSPSNA 264
SS +T+P T +TT + + ATTS +++
Sbjct: 189 --SSSTTSPSTTSATTSSPTTSATTSQTSS 216
Score = 34.7 bits (78), Expect = 3.6
Identities = 25/100 (25%), Positives = 44/100 (44%), Gaps = 8/100 (8%)
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
+P++TS+ +SPS + + S A L + + T SS +T+
Sbjct: 431 SPSTTSATTSSPSATSSQTSSSTTTSLSTTSATTSLPTTSAT--------TSQTSSSTTS 482
Query: 243 PKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSS 282
P T +TT + + ATTS +++ PS + S +S
Sbjct: 483 PSTSSATTSSPTTSATTSQTSSSTTSPSTTSATTSSPTTS 522
Score = 34.3 bits (77), Expect = 4.7
Identities = 24/108 (22%), Positives = 47/108 (43%), Gaps = 3/108 (2%)
Query: 178 ATIRETPTSTSSKGNSPSPSLQPQRVGAFSF---TSPNMAKKDLEKRTVSPHRFPLSRTG 234
AT +T +STS+ ++ S + A S T P T SP + +
Sbjct: 545 ATTSQTSSSTSTSPSTTSATTSSPTTSASSVPLTTEPATTTTTSSSTTTSPSTTSATTSS 604
Query: 235 SMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSS 282
+S +T+ + +T+P++++ T+SP+ + + S S +S
Sbjct: 605 PTTSATTSQTSSSTTSPSTTSATTSSPTTSATTSQTSSSTSTSPSTTS 652
Score = 33.9 bits (76), Expect = 6.1
Identities = 26/95 (27%), Positives = 44/95 (45%), Gaps = 8/95 (8%)
Query: 170 HHFRNAVRATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFP 229
H + ++ T +TS+ +SP+ S + + + TSP+ T S
Sbjct: 365 HSATTSQTSSSTSTSPTTSATTSSPTTSATTSQTSSSTSTSPSTTSATTSSPTTS----- 419
Query: 230 LSRTGSMSSRSTTPKTGRSTTPNSSNRATTSPSNA 264
+ T SS ST+P T +TT SS AT+S +++
Sbjct: 420 -ATTSQTSSSSTSPSTTSATT--SSPSATSSQTSS 451
Score = 33.9 bits (76), Expect = 6.1
Identities = 30/113 (26%), Positives = 49/113 (42%), Gaps = 8/113 (7%)
Query: 178 ATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKK--DLEKRTVSPHRFPLSRTGS 235
AT +T +ST+S + + + P S TS + K + V+ H S+T S
Sbjct: 315 ATTSQTSSSTTSPSTTSATTSSPTTSATNSQTSSSTHKSIHNFCNYIVTNHSATTSQTSS 374
Query: 236 MSSRS------TTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSS 282
+S S T+ T +TT +S+ +TSPS S + + + SS
Sbjct: 375 STSTSPTTSATTSSPTTSATTSQTSSSTSTSPSTTSATTSSPTTSATTSQTSS 427
Score = 33.5 bits (75), Expect = 8.0
Identities = 27/112 (24%), Positives = 50/112 (44%), Gaps = 12/112 (10%)
Query: 170 HHFRNAV----RATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSP 225
H+F N + AT +T +STS+ + + + P S TS + + SP
Sbjct: 355 HNFCNYIVTNHSATTSQTSSSTSTSPTTSATTSSPTTSATTSQTSSSTS--------TSP 406
Query: 226 HRFPLSRTGSMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSAS 277
+ + +S +T+ + ST+P++++ T+SPS + S S S
Sbjct: 407 STTSATTSSPTTSATTSQTSSSSTSPSTTSATTSSPSATSSQTSSSTTTSLS 458
>UniRef100_Q7TFJ3 Rh148 [Rhesus cytomegalovirus]
Length = 356
Score = 43.5 bits (101), Expect = 0.008
Identities = 32/112 (28%), Positives = 52/112 (45%), Gaps = 4/112 (3%)
Query: 178 ATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTS---PNMAKKDLEKRTVSPHRFPLSRTG 234
AT + ++T+ ++P+ S V S T+ P+ A T + P S T
Sbjct: 14 ATSAKASSTTAGNTSTPTASTITSTVTTASTTNMSTPSPASSPATTNTTTSSPSPKSSTS 73
Query: 235 SMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNN 286
S ++ TT T +TTP SS T+SP+N ++P +S +SS +N
Sbjct: 74 STNTTITTTGTASATTPKSSTSVTSSPANT-TSTSTKPTNVSSPTVSSPASN 124
Score = 34.7 bits (78), Expect = 3.6
Identities = 30/93 (32%), Positives = 45/93 (48%), Gaps = 18/93 (19%)
Query: 178 ATIRETPTS-TSSKGNSPSPSLQPQRVGAFSFTSP--NMAKKDLEKRTVSPHRFPLSRTG 234
AT ++ TS TSS N+ S S +P V + + +SP N + TV+
Sbjct: 87 ATTPKSSTSVTSSPANTTSTSTKPTNVSSPTVSSPASNTSTTTSTTSTVN---------- 136
Query: 235 SMSSRSTTPKTGRSTTPNSS---NRATTSPSNA 264
+ STTPKT ++ PN++ N TT P N+
Sbjct: 137 --GTNSTTPKTNTTSLPNTTADMNVTTTEPYNS 167
>UniRef100_UPI000035F990 UPI000035F990 UniRef100 entry
Length = 520
Score = 42.7 bits (99), Expect = 0.013
Identities = 34/133 (25%), Positives = 59/133 (43%), Gaps = 4/133 (3%)
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
+P S SS +SPSPS + + P+ + + SP + S T S S S+T
Sbjct: 30 SPPSPSSSSSSPSPSSSSDSSSSTPPSPPSSSSPSSSSLSSSPAQSSSSSTNSSKSSSST 89
Query: 243 PKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPSKSKRLLK 302
++ S++ +SS+ +++S S + S +S+S + SS+ + S S
Sbjct: 90 SRSSSSSSKSSSSSSSSSSSKS----SSNSSRSSSSKSSSNSSCSSSSSSSKSSSSTSRS 145
Query: 303 SLLSRRKSKKDDT 315
S S KS +
Sbjct: 146 SSSSSSKSSSSSS 158
Score = 37.7 bits (86), Expect = 0.42
Identities = 34/127 (26%), Positives = 53/127 (40%), Gaps = 5/127 (3%)
Query: 186 STSSKGNSPS--PSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTP 243
STSS +S S P P + S SP+ + SP P S + S SS S++P
Sbjct: 16 STSSSSSSSSSTPPSPPSPSSSSSSPSPSSSSDSSSSTPPSP---PSSSSPSSSSLSSSP 72
Query: 244 KTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPSKSKRLLKS 303
S++ NSS ++++ ++ S S+S S N+ R S + S
Sbjct: 73 AQSSSSSTNSSKSSSSTSRSSSSSSKSSSSSSSSSSSKSSSNSSRSSSSKSSSNSSCSSS 132
Query: 304 LLSRRKS 310
S + S
Sbjct: 133 SSSSKSS 139
>UniRef100_UPI00003C1D5C UPI00003C1D5C UniRef100 entry
Length = 633
Score = 42.7 bits (99), Expect = 0.013
Identities = 34/137 (24%), Positives = 56/137 (40%), Gaps = 11/137 (8%)
Query: 178 ATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMS 237
A I++ P STS+K S + + P R + P+ + + P + T S
Sbjct: 69 AAIKQAPGSTSTKAESSATTSTPARRASTRDRKPSAKLRS---------QSPAADTKSSP 119
Query: 238 SRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPSKS 297
+TT T + +SSN A T S +V + + SA D + R P +
Sbjct: 120 QAATT--TAAAAAASSSNTAETPRSKVKVNLSRKSKPSADGATIGDSGSSRGATPQPGSA 177
Query: 298 KRLLKSLLSRRKSKKDD 314
++ + +RR K DD
Sbjct: 178 TKVTLKIGTRRNKKADD 194
>UniRef100_Q64HC3 ASF/SF2-like pre-mRNA splicing factor SRP32 [Zea mays]
Length = 285
Score = 42.7 bits (99), Expect = 0.013
Identities = 43/137 (31%), Positives = 54/137 (39%), Gaps = 16/137 (11%)
Query: 151 TNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTS 210
TN KY LDD + FRNA +G S S S P R S++
Sbjct: 157 TNYDDMKYAIKKLDDTE----FRNAFGRAYIRVKEYNGKRGRSYSRSRSPSR----SYSK 208
Query: 211 PNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGR--STTPNSSNRATTSPSNARVRY 268
K R S SR+ S+SSRS +P GR S +P S SP+N
Sbjct: 209 SRSPSKSPRTRRSSSR----SRSRSVSSRSRSPSKGRSPSRSPARSKSPNVSPANGEAAS 264
Query: 269 PSE--PRKSASMRLSSD 283
P + P +S S S D
Sbjct: 265 PKKQSPNRSPSGSRSPD 281
>UniRef100_UPI000042F989 UPI000042F989 UniRef100 entry
Length = 607
Score = 42.4 bits (98), Expect = 0.017
Identities = 44/148 (29%), Positives = 64/148 (42%), Gaps = 18/148 (12%)
Query: 186 STSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLE-------KRTVSPHRFP-----LSRT 233
ST SK P P QPQ + + P A + +R S H+ P S
Sbjct: 262 STYSKSILPQPQPQPQPSQSITNYPPASAPVQPQPFQHAQPQRLPSFHQLPQIAFKTSMF 321
Query: 234 GSMSSRSTTPKTGRSTTPNSSNRAT-TSPSNARVRYPSEPRKSASMRLSS-DVNN--IRD 289
G + +ST P +S P+S + +T P N + S P KS S R +S D+N+ +
Sbjct: 322 GKDNEKSTLPSLSKSLHPSSLSSSTPPPPENHQAAAGSSPLKSISHRSASFDINSNILPP 381
Query: 290 IDQHPSKSKRLLKSLLSRRKSKKDDTLY 317
I Q S +RL+ RR S T++
Sbjct: 382 ISQ--SNKRRLIDDQFGRRHSSVSSTIF 407
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.128 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 522,199,876
Number of Sequences: 2790947
Number of extensions: 21417199
Number of successful extensions: 81990
Number of sequences better than 10.0: 731
Number of HSP's better than 10.0 without gapping: 77
Number of HSP's successfully gapped in prelim test: 699
Number of HSP's that attempted gapping in prelim test: 78440
Number of HSP's gapped (non-prelim): 2725
length of query: 323
length of database: 848,049,833
effective HSP length: 127
effective length of query: 196
effective length of database: 493,599,564
effective search space: 96745514544
effective search space used: 96745514544
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 75 (33.5 bits)
Medicago: description of AC149127.4


