

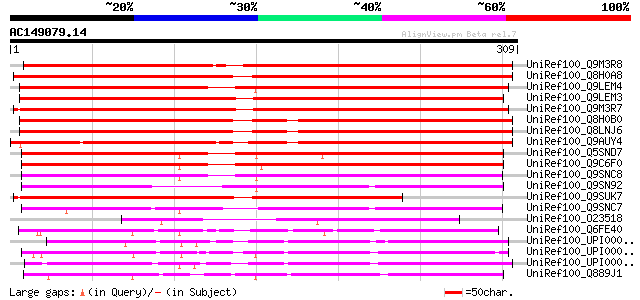
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149079.14 - phase: 0
(309 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9M3R8 Hypothetical protein [Arabidopsis thaliana] 355 7e-97
UniRef100_Q8H0A8 Putative gibberellin 20-oxidase [Oryza sativa] 340 3e-92
UniRef100_Q9LEM4 Hypothetical protein [Capsella rubella] 326 6e-88
UniRef100_Q9LEM3 Hypothetical protein [Capsella rubella] 323 4e-87
UniRef100_Q9M3R7 Hypothetical protein [Arabidopsis thaliana] 314 2e-84
UniRef100_Q8H0B0 Putative gibberellin 20-oxidase [Oryza sativa] 308 9e-83
UniRef100_Q8LNJ6 Putative gibberellin oxidase [Oryza sativa] 308 9e-83
UniRef100_Q9AUY4 Putative anthocyanidin hydroxylase [Oryza sativa] 299 7e-80
UniRef100_Q5SND7 Putative oxidoreductase [Oryza sativa] 267 3e-70
UniRef100_Q9C6F0 Hyoscyamine 6-dioxygenase hydroxylase, putative... 263 6e-69
UniRef100_Q9SNC8 Hypothetical protein F12A12.10 [Arabidopsis tha... 235 1e-60
UniRef100_Q9SN92 Hypothetical protein F18L15.200 [Arabidopsis th... 223 7e-57
UniRef100_Q9SUK7 Gibberellin oxidase-like protein [Arabidopsis t... 217 4e-55
UniRef100_Q9SNC7 Hypothetical protein F12A12.20 [Arabidopsis tha... 211 2e-53
UniRef100_O23518 APETALA2 domain TINY homolog [Arabidopsis thali... 181 3e-44
UniRef100_Q6FE40 Putative oxidoreductase [Acinetobacter sp.] 177 3e-43
UniRef100_UPI00002F02EE UPI00002F02EE UniRef100 entry 160 3e-38
UniRef100_UPI0000336F25 UPI0000336F25 UniRef100 entry 156 8e-37
UniRef100_UPI00002C3F0D UPI00002C3F0D UniRef100 entry 155 2e-36
UniRef100_Q889J1 Oxidoreductase, 2OG-Fe(II) oxygenase family [Ps... 144 4e-33
>UniRef100_Q9M3R8 Hypothetical protein [Arabidopsis thaliana]
Length = 305
Score = 355 bits (912), Expect = 7e-97
Identities = 178/301 (59%), Positives = 224/301 (74%), Gaps = 14/301 (4%)
Query: 9 LPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKIK 68
LPIIDLSSP +ISTAK I QAC+E+GFFY+ NHG+ E+ ++ F +S FF+LP+E+K+
Sbjct: 1 LPIIDLSSPGKISTAKLIRQACLEHGFFYVKNHGISEELMEGVFKESKGFFNLPLEEKMA 60
Query: 69 LNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRL-NQWPSNGVFSFFPAM 127
L R++ GYTPLYAEKLDPS S GD KES+Y G L + R NQWPS G+ +
Sbjct: 61 LLRRDLLGYTPLYAEKLDPSLSSIGDSKESFYFGSLEGVLAQRYPNQWPSEGILPSW-RQ 119
Query: 128 TLQNYSQIGDPQWNPYFGKYS-GKELLSLIALSLNLDENYFEKISALNKPEAFLRLLRYP 186
T++ Y Y S G++LL LIAL+L+LDE++FEK+ ALN P A +RLLRYP
Sbjct: 120 TMETY----------YKNVLSVGRKLLGLIALALDLDEDFFEKVGALNDPTAVVRLLRYP 169
Query: 187 GE-LGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGAIIVNI 245
GE + + E GASAHSDYGM+TLLLT+GVPGLQ+C++K KQP +WEDV ++GA IVNI
Sbjct: 170 GEVISSDVETYGASAHSDYGMVTLLLTDGVPGLQVCRDKSKQPHIWEDVPGIKGAFIVNI 229
Query: 246 GDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCSESSPPRYRKVL 305
GDMMERWTN L+RSTLHRVMP GKERYSV FF+DP DC V+C ESCCSE+ PPR+ +L
Sbjct: 230 GDMMERWTNGLFRSTLHRVMPVGKERYSVVFFLDPNPDCNVKCLESCCSETCPPRFPPIL 289
Query: 306 S 306
+
Sbjct: 290 A 290
>UniRef100_Q8H0A8 Putative gibberellin 20-oxidase [Oryza sativa]
Length = 357
Score = 340 bits (872), Expect = 3e-92
Identities = 174/308 (56%), Positives = 220/308 (70%), Gaps = 15/308 (4%)
Query: 3 GTATLSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLP 62
G L LP++DL+S D + A+SI +ACVE GFFY+VNHGV+E +K+ F +S+ FF LP
Sbjct: 48 GGNRLDLPVVDLASSDPRAAAESIRKACVESGFFYVVNHGVEEGLLKRLFAESSKFFELP 107
Query: 63 IEDKIKLNRKE-YRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVR-LNQWPSNGV 120
+E+KI L R +RGYTP YAEKLDPSS +GD KES+YIGP+ D NQWPS
Sbjct: 108 MEEKIALRRNSNHRGYTPPYAEKLDPSSKFEGDLKESFYIGPIGDEGLQNDANQWPSEER 167
Query: 121 F-SFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKPEAF 179
S + + + S + +GK +LSLIALSLNLD +FE I A + P AF
Sbjct: 168 LPSRRETIKMYHASALS-----------TGKRILSLIALSLNLDAEFFENIGAFSCPSAF 216
Query: 180 LRLLRYPGELGPNEE-ICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVE 238
LRLL YPGE+ +++ GASAHSDYGMITLL T+G PGLQIC+EK + PQ+WEDV H++
Sbjct: 217 LRLLHYPGEVDDSDDGNYGASAHSDYGMITLLATDGTPGLQICREKNRNPQLWEDVHHID 276
Query: 239 GAIIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCSESSP 298
GA+IVNIGD++ERWTNC+YRST+HRV+ GKERYS AFF+DP D VV+C ESCCSES P
Sbjct: 277 GALIVNIGDLLERWTNCIYRSTVHRVVAVGKERYSAAFFLDPNPDLVVQCLESCCSESCP 336
Query: 299 PRYRKVLS 306
PR+ + S
Sbjct: 337 PRFSPIKS 344
>UniRef100_Q9LEM4 Hypothetical protein [Capsella rubella]
Length = 315
Score = 326 bits (835), Expect = 6e-88
Identities = 161/306 (52%), Positives = 214/306 (69%), Gaps = 24/306 (7%)
Query: 7 LSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDK 66
L LPIIDLSSP+++ST+ I QAC+++GFFYL NHGV E+ ++ +S FSLP+++K
Sbjct: 5 LKLPIIDLSSPEKLSTSSLIRQACLDHGFFYLTNHGVSEELMEGVLMESKKLFSLPLDEK 64
Query: 67 IKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTS-VRLNQWPSNGVFSFFP 125
+ + R+ +RGY+PLY EK S+L+KGD KE + G + + N+WP +
Sbjct: 65 MVMARRGFRGYSPLYEEKPHSSTLAKGDSKEMFTFGSSEGVLAQIYPNEWPREELL---- 120
Query: 126 AMTLQNYSQIGDPQWNPYFGKYS------GKELLSLIALSLNLDENYFEKISALNKPEAF 179
P W P Y GK+LL L+AL+LNL+EN+FE++ N A
Sbjct: 121 ------------PLWRPTMECYYKTVMDVGKKLLGLVALALNLEENFFEQVGGFNAQAAV 168
Query: 180 LRLLRYPGELGPNEEI-CGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVE 238
+RLLRYPGEL + E+ CGASAHSDYGMITLL T+GVPGLQ+C++K K+P+VWEDV+ V+
Sbjct: 169 VRLLRYPGELNSSGEVTCGASAHSDYGMITLLATDGVPGLQVCRDKDKEPKVWEDVAGVK 228
Query: 239 GAIIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCSESSP 298
GA +VNIGD+MERWTN L+RST+HRV+ GKERYSVA F+DP +CVVEC ESCCSE+SP
Sbjct: 229 GAFVVNIGDLMERWTNGLFRSTMHRVVSVGKERYSVAVFVDPDPNCVVECLESCCSETSP 288
Query: 299 PRYRKV 304
PR+ V
Sbjct: 289 PRFPPV 294
>UniRef100_Q9LEM3 Hypothetical protein [Capsella rubella]
Length = 315
Score = 323 bits (828), Expect = 4e-87
Identities = 157/297 (52%), Positives = 213/297 (70%), Gaps = 12/297 (4%)
Query: 7 LSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDK 66
L LPIIDLSSP++IST++ I QAC+++GFFYL NHGV E+ ++ +S FSLP+++K
Sbjct: 5 LKLPIIDLSSPEKISTSRLIRQACLDHGFFYLTNHGVSEELMEGVLMESKKLFSLPLDEK 64
Query: 67 IKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTS-VRLNQWPSNGVFSFFP 125
+ + R+ +RGY+PLY EKLDPS+ SKGD KE + G + + N+WP + +
Sbjct: 65 MVMARRGFRGYSPLYDEKLDPSATSKGDSKEIFTFGSSEGVLAQIYPNEWPLEELLPLWR 124
Query: 126 AMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKPEAFLRLLRY 185
Y + D GK+LL L+AL+L+L+E +FE++ N A +RLLRY
Sbjct: 125 PTMECYYKSVMDV----------GKKLLGLVALALDLEEKFFEQVEGFNVQAAVVRLLRY 174
Query: 186 PGELGPNEEI-CGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGAIIVN 244
PGEL + E+ CGASAHSDYGM+TLL T+GVPGLQ+C++K K+P+VWEDV+ ++GA +VN
Sbjct: 175 PGELNSSGEVTCGASAHSDYGMLTLLATDGVPGLQVCRDKDKEPRVWEDVAGIKGAFLVN 234
Query: 245 IGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCSESSPPRY 301
IGD+MERWTN L+RST+HRV+ GKERYSVA F DP + VVEC ESCCSE+SPPR+
Sbjct: 235 IGDLMERWTNGLFRSTMHRVVSVGKERYSVAVFFDPDPNSVVECLESCCSETSPPRF 291
>UniRef100_Q9M3R7 Hypothetical protein [Arabidopsis thaliana]
Length = 315
Score = 314 bits (805), Expect = 2e-84
Identities = 158/304 (51%), Positives = 212/304 (68%), Gaps = 13/304 (4%)
Query: 3 GTATLSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLP 62
GTA L LPIIDLSSP+++ST++ I QAC+++GFFYL NHGV E+ ++ +S FSLP
Sbjct: 2 GTA-LKLPIIDLSSPEKLSTSRLIRQACLDHGFFYLTNHGVSEELMEGVLIESKKLFSLP 60
Query: 63 IEDKIKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTS-VRLNQWPSNGVF 121
+++K+ + R +RGY+PLY EKL+ SS S GD KE + G + N+WP +
Sbjct: 61 LDEKMVMARHGFRGYSPLYDEKLESSSTSIGDSKEMFTFGSSEGVLGQLYPNKWPLEELL 120
Query: 122 SFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKPEAFLR 181
+ Y + D GK+L L+AL+LNL+ENYFE++ A N A +R
Sbjct: 121 PLWRPTMECYYKNVMDV----------GKKLFGLVALALNLEENYFEQVGAFNDQAAVVR 170
Query: 182 LLRYPGELGPN-EEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGA 240
LLRY GE + EE CGASAHSD+GMITLL T+GV GLQ+C++K K+P+VWEDV+ ++G
Sbjct: 171 LLRYSGESNSSGEETCGASAHSDFGMITLLATDGVAGLQVCRDKDKEPKVWEDVAGIKGT 230
Query: 241 IIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCSESSPPR 300
+VNIGD+MERWTN L+RSTLHRV+ GKER+SVA F+DP +CVVEC ESCCSE+SPP+
Sbjct: 231 FVVNIGDLMERWTNGLFRSTLHRVVSVGKERFSVAVFVDPDPNCVVECLESCCSETSPPK 290
Query: 301 YRKV 304
+ V
Sbjct: 291 FPPV 294
>UniRef100_Q8H0B0 Putative gibberellin 20-oxidase [Oryza sativa]
Length = 308
Score = 308 bits (790), Expect = 9e-83
Identities = 157/304 (51%), Positives = 212/304 (69%), Gaps = 21/304 (6%)
Query: 7 LSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDK 66
L LP++DL+S D S+A+SI +ACVE GFFY+VNHG++E +++ F ++ FF P+E+K
Sbjct: 9 LDLPVVDLASSDLRSSAESIRKACVECGFFYVVNHGIEEGLLEKVFAENRRFFEQPLEEK 68
Query: 67 IKLNRKE-YRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVR-LNQWPSNGVFSFF 124
+ L R + GYTP Y EKLD SS +GD E++ IGP+ D NQWPS +
Sbjct: 69 MALLRNSSHLGYTPPYTEKLDASSKFRGDLSETFKIGPIGDEGFQNDANQWPSEERLPCW 128
Query: 125 P-AMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKPEAFLRLL 183
M L + +G +GK +LSL+ALSLNLD +F+ P AFLRLL
Sbjct: 129 KETMKLYRGTALG-----------TGKRILSLVALSLNLDAEFFDC------PLAFLRLL 171
Query: 184 RYPGELGPNEE-ICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGAII 242
YPGE+ +++ GASAHSDYG++TLL T+G+PGLQIC+EK + PQ+WEDV H++GA+I
Sbjct: 172 HYPGEINESDDGNYGASAHSDYGILTLLATDGIPGLQICREKDRHPQLWEDVHHIDGALI 231
Query: 243 VNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCSESSPPRYR 302
+NIGD++ERWTNC++RSTLHRV+ GKERYSVAFF+DP D VV+C ESCCSE+ PPR+
Sbjct: 232 INIGDLLERWTNCVFRSTLHRVVAVGKERYSVAFFLDPDPDLVVQCLESCCSEACPPRFP 291
Query: 303 KVLS 306
+ S
Sbjct: 292 PIKS 295
>UniRef100_Q8LNJ6 Putative gibberellin oxidase [Oryza sativa]
Length = 381
Score = 308 bits (790), Expect = 9e-83
Identities = 157/304 (51%), Positives = 212/304 (69%), Gaps = 21/304 (6%)
Query: 7 LSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDK 66
L LP++DL+S D S+A+SI +ACVE GFFY+VNHG++E +++ F ++ FF P+E+K
Sbjct: 82 LDLPVVDLASSDLRSSAESIRKACVECGFFYVVNHGIEEGLLEKVFAENRRFFEQPLEEK 141
Query: 67 IKLNRKE-YRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVR-LNQWPSNGVFSFF 124
+ L R + GYTP Y EKLD SS +GD E++ IGP+ D NQWPS +
Sbjct: 142 MALLRNSSHLGYTPPYTEKLDASSKFRGDLSETFKIGPIGDEGFQNDANQWPSEERLPCW 201
Query: 125 P-AMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKPEAFLRLL 183
M L + +G +GK +LSL+ALSLNLD +F+ P AFLRLL
Sbjct: 202 KETMKLYRGTALG-----------TGKRILSLVALSLNLDAEFFDC------PLAFLRLL 244
Query: 184 RYPGELGPNEE-ICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGAII 242
YPGE+ +++ GASAHSDYG++TLL T+G+PGLQIC+EK + PQ+WEDV H++GA+I
Sbjct: 245 HYPGEINESDDGNYGASAHSDYGILTLLATDGIPGLQICREKDRHPQLWEDVHHIDGALI 304
Query: 243 VNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCSESSPPRYR 302
+NIGD++ERWTNC++RSTLHRV+ GKERYSVAFF+DP D VV+C ESCCSE+ PPR+
Sbjct: 305 INIGDLLERWTNCVFRSTLHRVVAVGKERYSVAFFLDPDPDLVVQCLESCCSEACPPRFP 364
Query: 303 KVLS 306
+ S
Sbjct: 365 PIKS 368
>UniRef100_Q9AUY4 Putative anthocyanidin hydroxylase [Oryza sativa]
Length = 308
Score = 299 bits (765), Expect = 7e-80
Identities = 161/312 (51%), Positives = 212/312 (67%), Gaps = 23/312 (7%)
Query: 1 MAGTA---TLSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSAN 57
MAG +L LP++DL+S D + AKS+ +ACVEYGFFY+VNHG E ++ F +S+
Sbjct: 1 MAGAGAGESLDLPVVDLASSDLAAAAKSVRKACVEYGFFYVVNHGA-EGLAEKVFGESSK 59
Query: 58 FFSLPIEDKIKLNR-KEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLT-DTTSVRLNQW 115
FF P+ +K+ L R + Y GYTPL A+KLD SS KGD E+Y IGP+ + NQW
Sbjct: 60 FFEQPLGEKMALLRNRNYLGYTPLGADKLDASSKFKGDLNENYCIGPIRKEGYQNDANQW 119
Query: 116 PSNGVFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNK 175
PS F + T++ Y + +GK +LSLIALSLNLD +F+
Sbjct: 120 PSEENFPCWKE-TMKLYHETA---------LATGKRILSLIALSLNLDVEFFDC------ 163
Query: 176 PEAFLRLLRYPGELGPNEE-ICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDV 234
P AFLRLL YPGE +++ GASAHSDYG++TL+ T+G PGLQIC+EK + PQ+WEDV
Sbjct: 164 PVAFLRLLHYPGEANESDDGNYGASAHSDYGVLTLVATDGTPGLQICREKDRCPQLWEDV 223
Query: 235 SHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCS 294
H+EGA+IVNIGD+++RWTNC++RSTLHRV+ GKERYSVAFF+ D VV+C ESCCS
Sbjct: 224 HHIEGALIVNIGDLLQRWTNCVFRSTLHRVVAVGKERYSVAFFLHTNPDLVVQCLESCCS 283
Query: 295 ESSPPRYRKVLS 306
E+ PPR+ + S
Sbjct: 284 EACPPRFPPIRS 295
>UniRef100_Q5SND7 Putative oxidoreductase [Oryza sativa]
Length = 334
Score = 267 bits (682), Expect = 3e-70
Identities = 142/314 (45%), Positives = 192/314 (60%), Gaps = 36/314 (11%)
Query: 8 SLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKI 67
+L I L PD + + QAC++ GFFY+V+HG+ ++ + F QS FF LPI +K+
Sbjct: 9 NLNCISLGDPDTKKSVALLKQACLDSGFFYVVDHGISQELMDDVFAQSKKFFDLPINEKM 68
Query: 68 KLNRKEY-RGYTPLYAEKLDPSSLSKGDPKESYYIG-------PLTDTTSVRLNQWPSNG 119
+L R E RGYTP+ E LDP + GD KE YYIG P + NQWPS+
Sbjct: 69 ELLRDEKNRGYTPMLDEILDPENQVNGDYKEGYYIGIEIPEDDPQANRPFYGPNQWPSDE 128
Query: 120 VFSFFPAMTLQNYSQIGDPQWNPYFGKYSG------KELLSLIALSLNLDENYFEKISAL 173
V P+W +Y K + +IAL+LNL+E++F+K L
Sbjct: 129 VL----------------PRWRKVMEQYHSEALRVAKSIARIIALALNLEEDFFDKPEML 172
Query: 174 NKPEAFLRLLRYPGEL------GPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQ 227
+P A LRLL Y G P E + GA AHSDYG+ITLL T+ V GLQICK++ Q
Sbjct: 173 GEPIATLRLLHYEGGQKTGKVSNPAEGVFGAGAHSDYGLITLLATDDVVGLQICKDRNAQ 232
Query: 228 PQVWEDVSHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVE 287
PQVWE V+ V+G IVN+GDM+ERW+NC++RSTLHRV+ G+ER+S+AFF++P DCVVE
Sbjct: 233 PQVWEYVAPVKGGFIVNLGDMLERWSNCIFRSTLHRVVLDGRERHSIAFFVEPSHDCVVE 292
Query: 288 CFESCCSESSPPRY 301
C +C SES+PP++
Sbjct: 293 CLPTCKSESNPPKF 306
>UniRef100_Q9C6F0 Hyoscyamine 6-dioxygenase hydroxylase, putative [Arabidopsis
thaliana]
Length = 329
Score = 263 bits (671), Expect = 6e-69
Identities = 139/308 (45%), Positives = 187/308 (60%), Gaps = 30/308 (9%)
Query: 8 SLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKI 67
SL IDL++ D + S+ QAC++ GFFY++NHG+ E+F+ F+QS F+LP+E+K+
Sbjct: 11 SLNCIDLANDDLNHSVVSLKQACLDCGFFYVINHGISEEFMDDVFEQSKKLFALPLEEKM 70
Query: 68 KLNRKE-YRGYTPLYAEKLDPSSLSKGDPKESYYIG-------PLTDTTSVRLNQWPSNG 119
K+ R E +RGYTP+ E LDP + GD KE YYIG P D N WP
Sbjct: 71 KVLRNEKHRGYTPVLDELLDPKNQINGDHKEGYYIGIEVPKDDPHWDKPFYGPNPWPDAD 130
Query: 120 VFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKEL------LSLIALSLNLDENYFEKISAL 173
V P W KY + L L+AL+L+LD YF++ L
Sbjct: 131 VL----------------PGWRETMEKYHQEALRVSMAIARLLALALDLDVGYFDRTEML 174
Query: 174 NKPEAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWED 233
KP A +RLLRY G P++ I AHSD+GM+TLL T+GV GLQICK+K PQ WE
Sbjct: 175 GKPIATMRLLRYQGISDPSKGIYACGAHSDFGMMTLLATDGVMGLQICKDKNAMPQKWEY 234
Query: 234 VSHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCC 293
V ++GA IVN+GDM+ERW+N ++STLHRV+ G+ERYS+ FF++P DC+VEC +C
Sbjct: 235 VPPIKGAFIVNLGDMLERWSNGFFKSTLHRVLGNGQERYSIPFFVEPNHDCLVECLPTCK 294
Query: 294 SESSPPRY 301
SES P+Y
Sbjct: 295 SESELPKY 302
>UniRef100_Q9SNC8 Hypothetical protein F12A12.10 [Arabidopsis thaliana]
Length = 306
Score = 235 bits (600), Expect = 1e-60
Identities = 124/307 (40%), Positives = 180/307 (58%), Gaps = 30/307 (9%)
Query: 8 SLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKI 67
SL IDL + D +A + QAC++ GFFY++NHG+ E+ +AF+ S FF+LP+E+K+
Sbjct: 16 SLTCIDLDNSDLHQSAVLLKQACLDSGFFYVINHGISEELKDEAFEHSKKFFALPLEEKM 75
Query: 68 KLNRKE-YRGYTPLYAEKLDPSSLSKGDPKESYYIG-------PLTDTTSVRLNQWPSNG 119
K+ R E YRGY P + LDP + +GD KE + IG P D N WP+
Sbjct: 76 KVLRNEKYRGYAPFHDSLLDPENQVRGDYKEGFTIGFEGSKDGPHWDKPFHSPNIWPNPD 135
Query: 120 VFSFFPAMTLQNYSQIGDPQWNPYFGKYSG------KELLSLIALSLNLDENYFEKISAL 173
V P W KY K + ++AL+L+LD +YF L
Sbjct: 136 VL----------------PGWRETMEKYYQEALRVCKSIAKIMALALDLDVDYFNTPEML 179
Query: 174 NKPEAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWED 233
P A + L Y G+ P++ I AHSD+GM++LL T+GV GLQICK+K +PQ WE
Sbjct: 180 GNPIADMVLFHYEGKSDPSKGIYACGAHSDFGMMSLLATDGVMGLQICKDKDVKPQKWEY 239
Query: 234 VSHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCC 293
++GA IVN+GD++ERW+N ++STLHRV+ G++RYS+ FF+ P DC++EC +C
Sbjct: 240 TPSIKGAYIVNLGDLLERWSNGYFKSTLHRVLGNGQDRYSIPFFLKPSHDCIIECLPTCQ 299
Query: 294 SESSPPR 300
SE++ P+
Sbjct: 300 SENNLPK 306
>UniRef100_Q9SN92 Hypothetical protein F18L15.200 [Arabidopsis thaliana]
Length = 297
Score = 223 bits (567), Expect = 7e-57
Identities = 121/301 (40%), Positives = 176/301 (58%), Gaps = 52/301 (17%)
Query: 8 SLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKI 67
SL IDL++P+ +A S+ QAC++ GFFY+ NHG+ E+ +AF+QS FF+LP+E+K+
Sbjct: 16 SLTCIDLANPNFQQSAVSLKQACLDCGFFYVTNHGISEELKDEAFEQSKKFFALPLEEKM 75
Query: 68 KLNRKE-YRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVFSFFPA 126
K+ R E +RGY+P+ + LDP
Sbjct: 76 KVLRNEKHRGYSPVLDQILDP--------------------------------------- 96
Query: 127 MTLQNYSQIGDPQWNPYFGKYSG------KELLSLIALSLNLDENYFEKISALNKPEAFL 180
+N + P W KY K + L+AL+L+LD NYF+K L P A +
Sbjct: 97 ---ENQVDVVLPGWRATMEKYHQEALRVCKAIARLLALALDLDTNYFDKPEMLGNPIAVM 153
Query: 181 RLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGA 240
RLLRY G P + I G AHSDYGM+TLL T+ V GLQ +K +P+ WE V ++GA
Sbjct: 154 RLLRYEGMSDPLKGIFGCGAHSDYGMLTLLATDSVTGLQ---DKDVKPRKWEYVPSIKGA 210
Query: 241 IIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCSESSPPR 300
IVN+GD++ERW+N +++STLHRV+ G++RYS+ FF++P DC+VEC +C SE++ P+
Sbjct: 211 YIVNLGDLLERWSNGIFKSTLHRVLGNGQDRYSIPFFIEPSHDCLVECLPTCQSENNLPK 270
Query: 301 Y 301
Y
Sbjct: 271 Y 271
>UniRef100_Q9SUK7 Gibberellin oxidase-like protein [Arabidopsis thaliana]
Length = 243
Score = 217 bits (552), Expect = 4e-55
Identities = 114/239 (47%), Positives = 158/239 (65%), Gaps = 13/239 (5%)
Query: 3 GTATLSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLP 62
GTA L LPIIDLSSP+++ST++ I QAC+++GFFYL NHGV E+ ++ +S FSLP
Sbjct: 2 GTA-LKLPIIDLSSPEKLSTSRLIRQACLDHGFFYLTNHGVSEELMEGVLIESKKLFSLP 60
Query: 63 IEDKIKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTT-SVRLNQWPSNGVF 121
+++K+ + R +RGY+PLY EKL+ SS S GD KE + G + N+WP +
Sbjct: 61 LDEKMVMARHGFRGYSPLYDEKLESSSTSIGDSKEMFTFGSSEGVLGQLYPNKWPLEELL 120
Query: 122 SFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKPEAFLR 181
+ Y + D GK+L L+AL+LNL+ENYFE++ A N A +R
Sbjct: 121 PLWRPTMECYYKNVMD----------VGKKLFGLVALALNLEENYFEQVGAFNDQAAVVR 170
Query: 182 LLRYPGELGPN-EEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEG 239
LLRY GE + EE CGASAHSD+GMITLL T+GV GLQ+C++K K+P+VWEDV+ ++G
Sbjct: 171 LLRYSGESNSSGEETCGASAHSDFGMITLLATDGVAGLQVCRDKDKEPKVWEDVAGIKG 229
>UniRef100_Q9SNC7 Hypothetical protein F12A12.20 [Arabidopsis thaliana]
Length = 296
Score = 211 bits (538), Expect = 2e-53
Identities = 116/307 (37%), Positives = 184/307 (59%), Gaps = 40/307 (13%)
Query: 8 SLPIIDLSSPDRISTAKSIHQACVEY-----------GFFYLVNHGVDEDFIKQAFDQSA 56
SL IDL++ + +A S+ QA + + GFFY+ NHG+ E+ +AF+QS
Sbjct: 16 SLACIDLANSNLHRSAASLKQAWLFFFLFLNVNDYDCGFFYVTNHGISEELKDEAFEQSK 75
Query: 57 NFFSLPIEDKIK-LNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIG--PLTDTTSVRLN 113
FF+LP+++K+K L ++++GY+P+ ++ D + GD KES++IG DT R N
Sbjct: 76 KFFALPLDEKMKVLKNEKHQGYSPVLSQISD--NQIHGDYKESFFIGIEGSNDTPFCRAN 133
Query: 114 QWPSNGVFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISAL 173
WP+ G + ++ + ++AL+LN+D +YF+ L
Sbjct: 134 IWPNPGDHGEISSRSIA---------------------IARVLALALNVDGDYFDTPEML 172
Query: 174 NKPEAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWED 233
P F+RLL Y G P++ I G HSD+GM+TLL T+ V GLQ ++ +P+ WE
Sbjct: 173 GNPLTFMRLLHYEGMSDPSKGIYGCGPHSDFGMMTLLGTDSVMGLQ---DRDVKPRKWEY 229
Query: 234 VSHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCC 293
+ ++GA IVNIGD++ERW+N +++STLHRV+ G++RYS+AFF+ P DC+VEC +C
Sbjct: 230 ILSIKGAYIVNIGDLLERWSNGIFKSTLHRVLGNGQDRYSIAFFLQPSHDCIVECLPTCQ 289
Query: 294 SESSPPR 300
SE++PP+
Sbjct: 290 SENNPPK 296
>UniRef100_O23518 APETALA2 domain TINY homolog [Arabidopsis thaliana]
Length = 895
Score = 181 bits (458), Expect = 3e-44
Identities = 110/248 (44%), Positives = 130/248 (52%), Gaps = 86/248 (34%)
Query: 69 LNRKEYRGYTPLYAEKLDPSSLS---------KGDPKESYYIGPLTDTTSVRL-NQWPSN 118
L R++ GYTPLYAEKLDPS S GD KES+Y G L + R NQWPS
Sbjct: 3 LLRRDLLGYTPLYAEKLDPSLSSIVYKVDISLLGDSKESFYFGSLEGVLAQRYPNQWPSE 62
Query: 119 GVFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKPEA 178
DE++FEK+ ALN P A
Sbjct: 63 --------------------------------------------DEDFFEKVGALNDPTA 78
Query: 179 FLRLLRYP-------------------------------GE-LGPNEEICGASAHSDYGM 206
+RLLRYP GE + + E GASAHSDYGM
Sbjct: 79 VVRLLRYPELDRNICNQWKLKPHSGSLSQVLISALVMIIGEVISSDVETYGASAHSDYGM 138
Query: 207 ITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGAIIVNIGDMMERWTNCLYRSTLHRVMP 266
+TLLLT+GVPGLQ+C++K KQP +WEDV ++GA IVNIGDMMERWTN L+RSTLHRVMP
Sbjct: 139 VTLLLTDGVPGLQVCRDKSKQPHIWEDVPGIKGAFIVNIGDMMERWTNGLFRSTLHRVMP 198
Query: 267 TGKERYSV 274
GKERYS+
Sbjct: 199 VGKERYSL 206
>UniRef100_Q6FE40 Putative oxidoreductase [Acinetobacter sp.]
Length = 316
Score = 177 bits (449), Expect = 3e-43
Identities = 120/315 (38%), Positives = 169/315 (53%), Gaps = 43/315 (13%)
Query: 6 TLSLPIIDLS---SP---DRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFF 59
T +PII +S SP DR+ A + QAC + GFFY+ +HGV ++ ++ F+QS FF
Sbjct: 2 TQHIPIISISGLFSPLLEDRLQVAMQMKQACEDNGFFYIADHGVSKNLQQKVFEQSRQFF 61
Query: 60 SLPIEDKIKLNRKE---YRGYTPLYAEKLDPSSLSKGDPKESYYIG---------PLTDT 107
LP +K K+++K RGY PL + L+ + D KE +Y G L
Sbjct: 62 DLPTNEKEKIHKKNSIANRGYEPLKNQTLEAG--TPADLKEGFYAGREYALDSQAVLNKR 119
Query: 108 TSVRLNQWPSNGVFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYF 167
+ NQWP F F + T+Q+Y + K L+ +ALSLNLDENYF
Sbjct: 120 FNHGPNQWPEQ--FPEFES-TMQHYQAELE---------VLAKHLMRGLALSLNLDENYF 167
Query: 168 EKISALNKPEAFLRLLRYPGELG---PNEEICGASAHSDYGMITLLLTNGVPGLQICKEK 224
+ LRLL YP + PNE+ CG AH+D+G +TLLL + GLQ+
Sbjct: 168 DDF--CQDSLVTLRLLHYPPQPANPEPNEKGCG--AHTDFGALTLLLQDQQGGLQVWD-- 221
Query: 225 LKQPQVWEDVSHVEGAIIVNIGDMMERWTNCLYRSTLHRVM-PTGKERYSVAFFMDPPSD 283
K + W D +EG ++N+GD++ RWTN Y+ST+HRV+ +GK+RYSV FF D
Sbjct: 222 -KHTESWIDAPPIEGTYVINLGDLIARWTNNHYKSTVHRVINKSGKQRYSVPFFFGGNPD 280
Query: 284 CVVECFESCCSESSP 298
+V+C C SES P
Sbjct: 281 YLVKCLPDCMSESEP 295
>UniRef100_UPI00002F02EE UPI00002F02EE UniRef100 entry
Length = 292
Score = 160 bits (406), Expect = 3e-38
Identities = 106/295 (35%), Positives = 157/295 (52%), Gaps = 33/295 (11%)
Query: 23 AKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKIKL---NRKEYRGYTP 79
A + +AC E GFF L +HGV + I++ FD + F P+E+K ++ + + +RGY P
Sbjct: 5 ALQVGKACREIGFFTLTDHGVSLELIERVFDANREFHQQPLEEKEEILISHSEHHRGYHP 64
Query: 80 LYAEKLDPSSLSKGDPKESYYIGP--LTDTTSVRL-------NQWPSNGVFSFFPAMTLQ 130
AE DP++ K D KE++ + D V+ N WP N TL+
Sbjct: 65 FSAEATDPNA--KPDLKEAFDMALELSLDDPDVKAGKPFHGPNVWPRNPPQF---RKTLE 119
Query: 131 NYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKPEAFLRLLRYPG-EL 189
Y Q G+++ AL L LDE+YF + +KP A LRLL YP ++
Sbjct: 120 GYYQAL---------LLLGEQICRCFALDLGLDEDYF--VDKHSKPLAQLRLLHYPELDM 168
Query: 190 GPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGAIIVNIGDMM 249
+ + GA AH+DYG + +L + + GL+I K ++ Q W + VEGA + N+G +M
Sbjct: 169 SRHPDQQGAGAHTDYGSVAILTQDSIGGLEI---KTREGQ-WIPIEPVEGAFVCNVGHIM 224
Query: 250 ERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCSESSPPRYRKV 304
E WTN LY +T HRV G++RYS F DP DC+VE + C SE+ PP Y+ +
Sbjct: 225 EIWTNGLYPATWHRVANHGRDRYSQVLFYDPNFDCLVEPLKCCISETHPPAYQPI 279
>UniRef100_UPI0000336F25 UPI0000336F25 UniRef100 entry
Length = 316
Score = 156 bits (394), Expect = 8e-37
Identities = 115/311 (36%), Positives = 165/311 (52%), Gaps = 32/311 (10%)
Query: 8 SLPIID---LSSPD---RISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSL 61
SLP+I L+S D R TA+ + AC E GFFY V+HG+ + + AF + F L
Sbjct: 3 SLPLISVAGLNSEDPAVRAETARQLGAACREIGFFYAVDHGIPAEVMAGAFANAKRVFEL 62
Query: 62 PIEDKIKLNRKEY---RGYTPLYAEKLDPSSLSKGDPKESYYIGP--LTDTTSVRLNQWP 116
P+E K ++ K+ RGY + EKL+P + D KE++ IG D V + P
Sbjct: 63 PLETKQDMSIKKSPHNRGYVAMADEKLNPEA--GADMKEAFNIGTDFPADHPDVLAGK-P 119
Query: 117 SNGVFSFFPAMTLQNYSQIGDPQWNPYFGKYS--GKELLSLIALSLNLDENYFEKISALN 174
GV +F+P + + +G YF G+ + A+ L L E++F + L
Sbjct: 120 FRGV-NFWPPIDGWRDAMLG------YFDGCLNLGRLIHKGFAIDLGLPEDFF--VQHLT 170
Query: 175 KPEAFLRLLRYPGELGP-NEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWED 233
P A LR+LRYP G + E GA AH+DYG +TLL T+ V GLQ+ L + W D
Sbjct: 171 SPIATLRMLRYPASAGQIDREDGGAGAHTDYGNVTLLATDKVAGLQV----LTRQGQWID 226
Query: 234 VSHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCC 293
HV GA + NIGD + RW+N Y ST HRV P +ERYS+AFF++ D +V+ +
Sbjct: 227 APHVPGAFVCNIGDCLMRWSNDTYVSTPHRVRPPEQERYSIAFFLEVNPDSIVDPRDVIP 286
Query: 294 SESSPPRYRKV 304
E+ P+Y +
Sbjct: 287 DEA--PKYEPI 295
>UniRef100_UPI00002C3F0D UPI00002C3F0D UniRef100 entry
Length = 305
Score = 155 bits (391), Expect = 2e-36
Identities = 105/309 (33%), Positives = 162/309 (51%), Gaps = 36/309 (11%)
Query: 10 PIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKIKL 69
P++D S+ + AK + AC + GFFY VNHGV I AF+ S FF P+E ++++
Sbjct: 5 PLVDGSNIQEL--AKELRSACKDIGFFYAVNHGVSNTVITNAFEASRRFFDQPLEWRMQV 62
Query: 70 NRKEY-RGYTPLYAEKLDPSSLSKGDPKESYYIG---PLTDTTSVR------LNQWPSNG 119
++ + RGY PL K D K+S+ IG PLT NQWP
Sbjct: 63 HKDRFHRGYLPLGTTKYPG---KPADLKDSFDIGVDLPLTHPDVEEGLPLHGPNQWP--- 116
Query: 120 VFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKPEAF 179
V +F +S + K+ G +LL L A SL+L++++F + + P
Sbjct: 117 VAEWFKEPAELYFSAV----------KFFGFKLLLLFAKSLDLEDDFF--LRHYDNPTIL 164
Query: 180 LRLLRYPGELGPNEE-ICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVE 238
+R+L YP + E+ GA AH+DYG+IT+L + + GL++ ++ W ++
Sbjct: 165 MRMLHYPPQSEAKEQGSIGALAHTDYGVITVLAQDPLGGLEL----ERRDGSWIKAPYIP 220
Query: 239 GAIIVNIGDMMERWTNCLYRSTLHRVM-PTGKERYSVAFFMDPPSDCVVECFESCCSESS 297
G +VNIGD+M RWTN +YRS H+V+ G ER+S+ FF +P V+EC +C +
Sbjct: 221 GTFVVNIGDLMGRWTNDVYRSNKHQVINRLGVERFSIPFFFNPNHRSVIECIPTCKTLER 280
Query: 298 PPRYRKVLS 306
P RY V++
Sbjct: 281 PVRYPPVVA 289
>UniRef100_Q889J1 Oxidoreductase, 2OG-Fe(II) oxygenase family [Pseudomonas syringae]
Length = 321
Score = 144 bits (362), Expect = 4e-33
Identities = 101/310 (32%), Positives = 155/310 (49%), Gaps = 35/310 (11%)
Query: 9 LPIIDLSSPDRIST------AKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLP 62
LPIID+S T A I AC E+GFFY+ H + + I+Q + +FF+
Sbjct: 4 LPIIDISPLYGTDTQAWQAVATQIDSACREWGFFYIKGHPITAERIEQVQSAAKDFFARD 63
Query: 63 IEDKIKLNRKE---YRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNG 119
+K++++ + +RGY + E+LDP S D KE++ +G DT
Sbjct: 64 AAEKLRIDITQSTHHRGYGAIATEQLDPGLPS--DLKETFDMGLHLDTNHP--------D 113
Query: 120 VFSFFPAMTLQNYSQIGDPQWNPYFGKYS------GKELLSLIALSLNLDENYFEKISAL 173
V + P + I P W ++ + LL + L+L ++ ++F++
Sbjct: 114 VLAGKPLRGPNRHPAI--PGWKDLMEQHYLDMQALAQTLLRAMTLALGIERDFFDQ--RF 169
Query: 174 NKPEAFLRLLRYPGE-LGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWE 232
P + LR++ YP + E GA AH+DYG ITLL + GLQ+ + + W
Sbjct: 170 QDPVSVLRMIHYPPRHTATSSEQQGAGAHTDYGCITLLYQDTAGGLQVRDVRGE----WI 225
Query: 233 DVSHVEGAIIVNIGDMMERWTNCLYRSTLHRVM-PTGKERYSVAFFMDPPSDCVVECFES 291
D ++G +VN+GDMM RW+N Y ST HRV+ P G +RYS+ FF +P D +EC
Sbjct: 226 DAPPIDGTFVVNLGDMMARWSNDRYLSTPHRVVSPLGVDRYSMPFFAEPHPDTRIECLPG 285
Query: 292 CCSESSPPRY 301
C SE+ P RY
Sbjct: 286 CQSETHPARY 295
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 558,305,384
Number of Sequences: 2790947
Number of extensions: 24089493
Number of successful extensions: 54639
Number of sequences better than 10.0: 1293
Number of HSP's better than 10.0 without gapping: 1089
Number of HSP's successfully gapped in prelim test: 204
Number of HSP's that attempted gapping in prelim test: 51352
Number of HSP's gapped (non-prelim): 1709
length of query: 309
length of database: 848,049,833
effective HSP length: 127
effective length of query: 182
effective length of database: 493,599,564
effective search space: 89835120648
effective search space used: 89835120648
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC149079.14


