

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149050.11 - phase: 0
(163 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
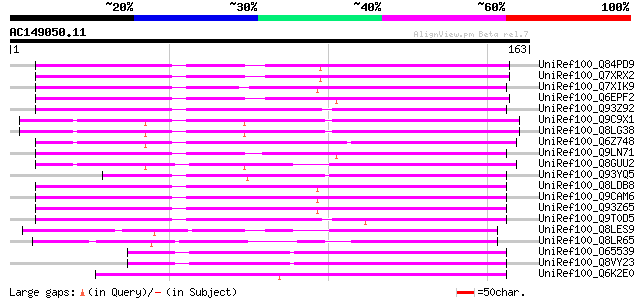
UniRef100_Q84PD9 Ring zinc finger protein-like protein [Oryza sa... 116 2e-25
UniRef100_Q7XRX2 OSJNBb0032E06.5 protein [Oryza sativa] 116 2e-25
UniRef100_Q7XIK9 Putative RES protein [Oryza sativa] 115 4e-25
UniRef100_Q6EPF2 Ring zinc finger protein-like [Oryza sativa] 115 5e-25
UniRef100_Q93Z92 AT4g11680/T5C23_110 [Arabidopsis thaliana] 113 2e-24
UniRef100_Q9C9X1 Putative RING zinc finger protein; 27623-28978 ... 113 2e-24
UniRef100_Q8LG38 Putative RING zinc finger protein [Arabidopsis ... 113 2e-24
UniRef100_Q6Z748 RING zinc finger protein-like [Oryza sativa] 107 8e-23
UniRef100_Q9LN71 T12C24.29 [Arabidopsis thaliana] 103 2e-21
UniRef100_Q8GUU2 RES protein [Arabidopsis thaliana] 102 3e-21
UniRef100_Q93YQ5 Hypothetical protein At3g61180; T20K12.80 [Arab... 100 1e-20
UniRef100_Q8LDB8 Hypothetical protein [Arabidopsis thaliana] 100 2e-20
UniRef100_Q9CAM6 Hypothetical protein F16M19.7 [Arabidopsis thal... 99 5e-20
UniRef100_Q93Z65 At1g63170/F16M19_7 [Arabidopsis thaliana] 99 5e-20
UniRef100_Q9T0D5 Hypothetical protein AT4g11680 [Arabidopsis tha... 98 7e-20
UniRef100_Q8LES9 Hypothetical protein [Arabidopsis thaliana] 86 3e-16
UniRef100_Q8LR65 P0421H07.18 protein [Oryza sativa] 85 6e-16
UniRef100_O65539 Hypothetical protein F4D11.200 [Arabidopsis tha... 83 2e-15
UniRef100_Q8VY23 Hypothetical protein At4g32600 [Arabidopsis tha... 83 2e-15
UniRef100_Q6K2E0 Putative RING zinc finger protein [Oryza sativa] 82 5e-15
>UniRef100_Q84PD9 Ring zinc finger protein-like protein [Oryza sativa]
Length = 250
Score = 116 bits (291), Expect = 2e-25
Identities = 65/156 (41%), Positives = 84/156 (53%), Gaps = 17/156 (10%)
Query: 9 LTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQIIALAYALTQPLRIREGASEDDIKSLP 68
L +V L+ + FF +I C+ IIA+ YA++ +EGASEDDI+ +P
Sbjct: 94 LCIVFLAFDVFFVVFCVALACIIGIAVCCCLPCIIAILYAVSD----QEGASEDDIRQIP 149
Query: 69 MYRFCQPNVMIMVDKNKTQLEARTGSHN-------RSHISELSLHPDDSECCICLCPYVD 121
Y+F + D+ + Q TG + E L P+D+ECCICL Y D
Sbjct: 150 RYKFRR------TDEPEKQTADETGPFGGIMTECGTNQPIEKVLAPEDAECCICLSAYDD 203
Query: 122 GTELYRLPCTHHFHCECIGRWLRTKATCPLCKFNIR 157
G EL LPC HHFHC CI +WL ATCPLCKFNIR
Sbjct: 204 GAELRELPCGHHFHCACIDKWLHINATCPLCKFNIR 239
>UniRef100_Q7XRX2 OSJNBb0032E06.5 protein [Oryza sativa]
Length = 324
Score = 116 bits (291), Expect = 2e-25
Identities = 65/156 (41%), Positives = 84/156 (53%), Gaps = 17/156 (10%)
Query: 9 LTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQIIALAYALTQPLRIREGASEDDIKSLP 68
L +V L+ + FF +I C+ IIA+ YA++ +EGASEDDI+ +P
Sbjct: 168 LCIVFLAFDVFFVVFCVALACIIGIAVCCCLPCIIAILYAVSD----QEGASEDDIRQIP 223
Query: 69 MYRFCQPNVMIMVDKNKTQLEARTGSHN-------RSHISELSLHPDDSECCICLCPYVD 121
Y+F + D+ + Q TG + E L P+D+ECCICL Y D
Sbjct: 224 RYKFRR------TDEPEKQTADETGPFGGIMTECGTNQPIEKVLAPEDAECCICLSAYDD 277
Query: 122 GTELYRLPCTHHFHCECIGRWLRTKATCPLCKFNIR 157
G EL LPC HHFHC CI +WL ATCPLCKFNIR
Sbjct: 278 GAELRELPCGHHFHCACIDKWLHINATCPLCKFNIR 313
>UniRef100_Q7XIK9 Putative RES protein [Oryza sativa]
Length = 361
Score = 115 bits (288), Expect = 4e-25
Identities = 65/152 (42%), Positives = 83/152 (53%), Gaps = 11/152 (7%)
Query: 9 LTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQIIALAYALTQPLRIREGASEDDIKSLP 68
L++V L+ + FF +I C+ IIA+ YA+T +EGASE+DI +L
Sbjct: 207 LSIVFLAFDVFFVVFCVALACVIGIAVCCCLPCIIAILYAVTD----QEGASEEDINNLS 262
Query: 69 MYRFCQPNVMIMVDKNKTQLEARTGSH----NRSHISELSLHPDDSECCICLCPYVDGTE 124
++F M DK + A G + E L +D+ECCICLCPY DG E
Sbjct: 263 KFKF---RTMGDADKLVAGIAAPVGGVMTECGTNPPVEHFLSAEDAECCICLCPYEDGAE 319
Query: 125 LYRLPCTHHFHCECIGRWLRTKATCPLCKFNI 156
L LPC HHFHC CI +WL ATCPLCKFNI
Sbjct: 320 LRELPCNHHFHCTCIDKWLHINATCPLCKFNI 351
>UniRef100_Q6EPF2 Ring zinc finger protein-like [Oryza sativa]
Length = 399
Score = 115 bits (287), Expect = 5e-25
Identities = 65/156 (41%), Positives = 85/156 (53%), Gaps = 17/156 (10%)
Query: 9 LTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQIIALAYALTQPLRIREGASEDDIKSLP 68
L +V L+ + FF +I C+ IIA+ YA++ +EGASEDDI+ +P
Sbjct: 243 LCIVFLAFDVFFVVFCVALACIIGIAVCCCLPCIIAILYAVSD----QEGASEDDIRQIP 298
Query: 69 MYRFCQPNVMIMVDKNKTQLEARTGSHNRSHIS-------ELSLHPDDSECCICLCPYVD 121
Y+F + +D+ + Q TGS I E L +D+ECCICL Y D
Sbjct: 299 RYKFRR------MDEPEKQSVNMTGSSGGIMIECGTNQPIEKVLAAEDAECCICLSAYDD 352
Query: 122 GTELYRLPCTHHFHCECIGRWLRTKATCPLCKFNIR 157
G EL LPC HHFHC CI +WL ATCPLCKFN+R
Sbjct: 353 GAELRELPCGHHFHCVCIDKWLHINATCPLCKFNVR 388
>UniRef100_Q93Z92 AT4g11680/T5C23_110 [Arabidopsis thaliana]
Length = 390
Score = 113 bits (283), Expect = 2e-24
Identities = 58/148 (39%), Positives = 80/148 (53%), Gaps = 7/148 (4%)
Query: 9 LTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQIIALAYALTQPLRIREGASEDDIKSLP 68
L ++ L + FF +I C+ IIA+ YA+ +EGAS++DI +P
Sbjct: 242 LCIIFLGFDVFFVVFCVALACVIGLAVCCCLPCIIAILYAVAD----QEGASKNDIDQMP 297
Query: 69 MYRFCQPNVMIMVDKNKTQLEARTGSHNRSHISELSLHPDDSECCICLCPYVDGTELYRL 128
+RF + + + + G+ + E SL P+D+ECCICLC Y DG EL L
Sbjct: 298 KFRFTKTGNVEKLSGKARGIMTECGTDSPI---ERSLSPEDAECCICLCEYEDGVELREL 354
Query: 129 PCTHHFHCECIGRWLRTKATCPLCKFNI 156
PC HHFHC CI +WL + CPLCKFNI
Sbjct: 355 PCNHHFHCTCIDKWLHINSRCPLCKFNI 382
>UniRef100_Q9C9X1 Putative RING zinc finger protein; 27623-28978 [Arabidopsis
thaliana]
Length = 343
Score = 113 bits (282), Expect = 2e-24
Identities = 69/159 (43%), Positives = 88/159 (54%), Gaps = 9/159 (5%)
Query: 4 SHSCRLTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQ-IIALAYALTQPLRIREGASED 62
+H LT V L+ + FF A+F + ++ AL C + IIAL YA+ +EGASE
Sbjct: 189 THLYWLTFVFLAFDVFF-AIFCVVLACLIGIALCCCLPCIIALLYAVAG----QEGASEA 243
Query: 63 DIKSLPMYRF-CQPNVMIMVDKNKTQLEARTGSHNRSHISELSLHPDDSECCICLCPYVD 121
D+ LP YRF N D + S N + E L P+D++CCICL Y D
Sbjct: 244 DLSILPKYRFHTMNNDEKQSDGGGKMIPVDAASENLGN--ERVLLPEDADCCICLSSYED 301
Query: 122 GTELYRLPCTHHFHCECIGRWLRTKATCPLCKFNIRIGD 160
G EL LPC HHFH CI +WL+ ATCPLCKFNI G+
Sbjct: 302 GAELVSLPCNHHFHSTCIVKWLKMNATCPLCKFNILKGN 340
>UniRef100_Q8LG38 Putative RING zinc finger protein [Arabidopsis thaliana]
Length = 343
Score = 113 bits (282), Expect = 2e-24
Identities = 69/159 (43%), Positives = 88/159 (54%), Gaps = 9/159 (5%)
Query: 4 SHSCRLTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQ-IIALAYALTQPLRIREGASED 62
+H LT V L+ + FF A+F + ++ AL C + IIAL YA+ +EGASE
Sbjct: 189 THLYWLTFVFLAFDVFF-AIFCVVLACLIGIALCCCLPCIIALLYAVAG----QEGASEA 243
Query: 63 DIKSLPMYRF-CQPNVMIMVDKNKTQLEARTGSHNRSHISELSLHPDDSECCICLCPYVD 121
D+ LP YRF N D + S N + E L P+D++CCICL Y D
Sbjct: 244 DLSILPKYRFHTMNNDEKQSDGGGKMIPVDAASENLGN--ERVLLPEDADCCICLSSYED 301
Query: 122 GTELYRLPCTHHFHCECIGRWLRTKATCPLCKFNIRIGD 160
G EL LPC HHFH CI +WL+ ATCPLCKFNI G+
Sbjct: 302 GAELVSLPCNHHFHSTCIVKWLKMNATCPLCKFNILKGN 340
>UniRef100_Q6Z748 RING zinc finger protein-like [Oryza sativa]
Length = 180
Score = 107 bits (268), Expect = 8e-23
Identities = 64/152 (42%), Positives = 86/152 (56%), Gaps = 7/152 (4%)
Query: 9 LTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQ-IIALAYALTQPLRIREGASEDDIKSL 67
L++V L+ + FF AVF + + AL C + +IA+ YAL +EGAS+ DI L
Sbjct: 31 LSVVFLAFDVFF-AVFCVAMACFIGIALCCCLPCVIAILYALAG----QEGASDADIGFL 85
Query: 68 PMYRFCQPNVMIMVDKNKTQLEARTGSHNRSHISELSLHPDDSECCICLCPYVDGTELYR 127
P YR+ P+ ++ + + S + LH +D+ECCICL Y DG EL
Sbjct: 86 PRYRYSDPSEDGQKGTDEGVMIPVLNNSGTSTSERILLH-EDAECCICLSSYEDGAELSA 144
Query: 128 LPCTHHFHCECIGRWLRTKATCPLCKFNIRIG 159
LPC HHFH CI +WLR ATCPLCK+NI G
Sbjct: 145 LPCNHHFHWTCITKWLRMHATCPLCKYNILKG 176
>UniRef100_Q9LN71 T12C24.29 [Arabidopsis thaliana]
Length = 408
Score = 103 bits (256), Expect = 2e-21
Identities = 59/155 (38%), Positives = 82/155 (52%), Gaps = 16/155 (10%)
Query: 9 LTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQIIALAYALTQPLRIREGASEDDIKSLP 68
L++V L + FF +I C+ IIA+ YA+ +EGAS++DI+ L
Sbjct: 252 LSIVFLGFDVFFVVFCVALACVIGIAVCCCLPCIIAVLYAVAD----QEGASKEDIEQLT 307
Query: 69 MYRFCQPNVMIMVDKNKTQLEARTGSHNRSHIS-------ELSLHPDDSECCICLCPYVD 121
++F + + D NK + G+ E +L +D+ECCICL Y D
Sbjct: 308 KFKFRK-----LGDANKHTNDEAQGTTEGIMTECGTDSPIEHTLLQEDAECCICLSAYED 362
Query: 122 GTELYRLPCTHHFHCECIGRWLRTKATCPLCKFNI 156
GTEL LPC HHFHC C+ +WL ATCPLCK+NI
Sbjct: 363 GTELRELPCGHHFHCSCVDKWLYINATCPLCKYNI 397
>UniRef100_Q8GUU2 RES protein [Arabidopsis thaliana]
Length = 359
Score = 102 bits (255), Expect = 3e-21
Identities = 63/162 (38%), Positives = 89/162 (54%), Gaps = 27/162 (16%)
Query: 9 LTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQ-IIALAYALTQPLRIREGASEDDIKSL 67
L+++ L+++ FF AVF + +V AL C + IIAL YA+ EG SE ++ L
Sbjct: 209 LSVIFLAIDVFF-AVFCVVLACLVGIALCCCLPCIIALLYAVAGT----EGVSEAELGVL 263
Query: 68 PMYRF----------CQPNVMIMVDKNKTQLEARTGSHNRSHISELSLHPDDSECCICLC 117
P+Y+F P M+ + N L +E +L +D++CCICL
Sbjct: 264 PLYKFKAFHSNEKNITGPGKMVPIPINGLCLA-----------TERTLLAEDADCCICLS 312
Query: 118 PYVDGTELYRLPCTHHFHCECIGRWLRTKATCPLCKFNIRIG 159
Y DG EL+ LPC HHFH CI +WL+ +ATCPLCK+NI G
Sbjct: 313 SYEDGAELHALPCNHHFHSTCIVKWLKMRATCPLCKYNILKG 354
>UniRef100_Q93YQ5 Hypothetical protein At3g61180; T20K12.80 [Arabidopsis thaliana]
Length = 379
Score = 100 bits (250), Expect = 1e-20
Identities = 55/129 (42%), Positives = 74/129 (56%), Gaps = 7/129 (5%)
Query: 30 LIVFGALFCIVQIIALAYALTQPLRIREGASEDDIKSLPMYRFC--QPNVMIMVDKNKTQ 87
LI C+ IIA+ YAL +EGA +++I+ L ++F + + + + +TQ
Sbjct: 244 LIGIAVCCCLPCIIAILYALAD----QEGAPDEEIERLLKFKFLTVKNSEKVNGEIRETQ 299
Query: 88 LEARTGSHNRSHISELSLHPDDSECCICLCPYVDGTELYRLPCTHHFHCECIGRWLRTKA 147
TG S +E L +D+EC ICLC Y DG EL LPC HHFH C+ +WLR A
Sbjct: 300 GGIMTGLDTESQ-TERMLLSEDAECSICLCAYEDGVELRELPCRHHFHSLCVDKWLRINA 358
Query: 148 TCPLCKFNI 156
TCPLCKFNI
Sbjct: 359 TCPLCKFNI 367
>UniRef100_Q8LDB8 Hypothetical protein [Arabidopsis thaliana]
Length = 381
Score = 99.8 bits (247), Expect = 2e-20
Identities = 57/152 (37%), Positives = 80/152 (52%), Gaps = 8/152 (5%)
Query: 9 LTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQIIALAYALTQPLRIREGASEDDIKSLP 68
L +V L + FF +I C+ IIA+ YA+ + +EGAS++DI L
Sbjct: 222 LCIVFLGFDVFFVVFCVALACVIGIAVCCCLPCIIAVLYAVAE----QEGASKEDIDQLT 277
Query: 69 MYRFCQPNVMIMVDKNKTQLEARTGSH----NRSHISELSLHPDDSECCICLCPYVDGTE 124
++F + + ++ Q E +G E +L +D+ECCICL Y D TE
Sbjct: 278 KFKFRKVGDTMKHTVDEEQGEGDSGGVMTECGTDSPVEHALPHEDAECCICLSAYEDETE 337
Query: 125 LYRLPCTHHFHCECIGRWLRTKATCPLCKFNI 156
L LPC HHFHC C+ +WL ATCPLCK+NI
Sbjct: 338 LRELPCGHHFHCGCVDKWLYINATCPLCKYNI 369
>UniRef100_Q9CAM6 Hypothetical protein F16M19.7 [Arabidopsis thaliana]
Length = 381
Score = 98.6 bits (244), Expect = 5e-20
Identities = 56/152 (36%), Positives = 80/152 (51%), Gaps = 8/152 (5%)
Query: 9 LTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQIIALAYALTQPLRIREGASEDDIKSLP 68
L +V L + FF +I C+ IIA+ YA+ + +EGAS++DI L
Sbjct: 222 LCIVFLGFDVFFVVFCVALACVIGIAVCCCLPCIIAVLYAVAE----QEGASKEDIDQLT 277
Query: 69 MYRFCQPNVMIMVDKNKTQLEARTGSH----NRSHISELSLHPDDSECCICLCPYVDGTE 124
++F + + ++ Q + +G E +L +D+ECCICL Y D TE
Sbjct: 278 KFKFRKVGDTMKHTVDEEQGQGDSGGVMTECGTDSPVEHALPHEDAECCICLSAYEDETE 337
Query: 125 LYRLPCTHHFHCECIGRWLRTKATCPLCKFNI 156
L LPC HHFHC C+ +WL ATCPLCK+NI
Sbjct: 338 LRELPCGHHFHCGCVDKWLYINATCPLCKYNI 369
>UniRef100_Q93Z65 At1g63170/F16M19_7 [Arabidopsis thaliana]
Length = 381
Score = 98.6 bits (244), Expect = 5e-20
Identities = 56/152 (36%), Positives = 80/152 (51%), Gaps = 8/152 (5%)
Query: 9 LTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQIIALAYALTQPLRIREGASEDDIKSLP 68
L +V L + FF +I C+ IIA+ YA+ + +EGAS++DI L
Sbjct: 222 LCIVFLGFDVFFVVFCVALACVIGIAVCCCLPCIIAVLYAVAE----QEGASKEDIDQLT 277
Query: 69 MYRFCQPNVMIMVDKNKTQLEARTGSH----NRSHISELSLHPDDSECCICLCPYVDGTE 124
++F + + ++ Q + +G E +L +D+ECCICL Y D TE
Sbjct: 278 KFKFRKVGDTMKHTVDEEQGQGDSGGVMTECGTDSPVEHALPHEDAECCICLSAYEDETE 337
Query: 125 LYRLPCTHHFHCECIGRWLRTKATCPLCKFNI 156
L LPC HHFHC C+ +WL ATCPLCK+NI
Sbjct: 338 LRELPCGHHFHCGCVDKWLYINATCPLCKYNI 369
>UniRef100_Q9T0D5 Hypothetical protein AT4g11680 [Arabidopsis thaliana]
Length = 419
Score = 98.2 bits (243), Expect = 7e-20
Identities = 58/177 (32%), Positives = 80/177 (44%), Gaps = 36/177 (20%)
Query: 9 LTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQIIALAYALTQPLRIREGASEDDIKSLP 68
L ++ L + FF +I C+ IIA+ YA+ +EGAS++DI +P
Sbjct: 242 LCIIFLGFDVFFVVFCVALACVIGLAVCCCLPCIIAILYAVAD----QEGASKNDIDQMP 297
Query: 69 MYRFCQPNVMIMVDKNKTQLEARTGSHNRSHISELSLHPDDS------------------ 110
+RF + + + + G+ + E SL P+D+
Sbjct: 298 KFRFTKTGNVEKLSGKARGIMTECGTDSPI---ERSLSPEDAVQSHFHILIKLYILKVCK 354
Query: 111 -----------ECCICLCPYVDGTELYRLPCTHHFHCECIGRWLRTKATCPLCKFNI 156
ECCICLC Y DG EL LPC HHFHC CI +WL + CPLCKFNI
Sbjct: 355 IDNAWLLMILQECCICLCEYEDGVELRELPCNHHFHCTCIDKWLHINSRCPLCKFNI 411
>UniRef100_Q8LES9 Hypothetical protein [Arabidopsis thaliana]
Length = 343
Score = 86.3 bits (212), Expect = 3e-16
Identities = 52/151 (34%), Positives = 81/151 (53%), Gaps = 22/151 (14%)
Query: 5 HSCRLTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQIIA--LAYALTQPLRIREGASED 62
H+ +L ++C+SL + +SF L +F L C+V +I+ L Y + R AS+D
Sbjct: 208 HAPKLHVLCVSLLAWNAICYSFPFLLFLF--LCCLVPLISSLLGYNMNMGSSDR-AASDD 264
Query: 63 DIKSLPMYRFCQPNVMIMVDKNKTQLEARTGSHNRSHISELSLHPDDSECCICLCPYVDG 122
I SLP ++F + +D + + + S+ + DD ECCICL Y D
Sbjct: 265 QISSLPSWKFKR------IDDSASDSD-----------SDSATVTDDPECCICLAKYKDK 307
Query: 123 TELYRLPCTHHFHCECIGRWLRTKATCPLCK 153
E+ +LPC+H FH +C+ +WLR + CPLCK
Sbjct: 308 EEVRKLPCSHKFHSKCVDQWLRIISCCPLCK 338
>UniRef100_Q8LR65 P0421H07.18 protein [Oryza sativa]
Length = 304
Score = 85.1 bits (209), Expect = 6e-16
Identities = 50/148 (33%), Positives = 74/148 (49%), Gaps = 28/148 (18%)
Query: 8 RLTMVCLSLNEFFDAVFSFWIYLIVFGALFCIVQII--ALAYALTQPLRIREGASEDDIK 65
RL +C+ L + V+S + ++F L C V + AL Y + + GAS++ +
Sbjct: 179 RLYALCIGLLAWNAVVYS--LPFLLFLLLCCFVPAVGYALGYNMNSA-SVGRGASDEQLA 235
Query: 66 SLPMYRFCQPNVMIMVDKNKTQLEARTGSHNRSHISELSLHPDDSECCICLCPYVDGTEL 125
+LP +RF +P +R H DD ECCICL Y + E+
Sbjct: 236 ALPQWRFKEP---------------ADAPRDRDH--------DDQECCICLAQYKEKEEV 272
Query: 126 YRLPCTHHFHCECIGRWLRTKATCPLCK 153
+LPCTH FH +C+ RWLR ++CPLCK
Sbjct: 273 RQLPCTHMFHLKCVDRWLRIISSCPLCK 300
>UniRef100_O65539 Hypothetical protein F4D11.200 [Arabidopsis thaliana]
Length = 495
Score = 83.2 bits (204), Expect = 2e-15
Identities = 42/119 (35%), Positives = 63/119 (52%), Gaps = 5/119 (4%)
Query: 38 CIVQIIALAYALTQPLRIREGASEDDIKSLPMYRFCQPNVMIMVDKNKTQLEARTGSHNR 97
CI+ I+ LTQP GA+ + I +LP ++F D N + + G
Sbjct: 294 CIISILGYREDLTQP----RGATPESINALPTHKFKLKKSRSNGDDNGSST-SEGGVVAA 348
Query: 98 SHISELSLHPDDSECCICLCPYVDGTELYRLPCTHHFHCECIGRWLRTKATCPLCKFNI 156
+E ++ +D+ CCICL Y + EL LPC+H FH EC+ +WL+ A+CPLCK +
Sbjct: 349 GTDNERAISGEDAVCCICLAKYANNEELRELPCSHFFHKECVDKWLKINASCPLCKSEV 407
>UniRef100_Q8VY23 Hypothetical protein At4g32600 [Arabidopsis thaliana]
Length = 453
Score = 83.2 bits (204), Expect = 2e-15
Identities = 42/119 (35%), Positives = 63/119 (52%), Gaps = 5/119 (4%)
Query: 38 CIVQIIALAYALTQPLRIREGASEDDIKSLPMYRFCQPNVMIMVDKNKTQLEARTGSHNR 97
CI+ I+ LTQP GA+ + I +LP ++F D N + + G
Sbjct: 294 CIISILGYREDLTQP----RGATPESINALPTHKFKLKKSRSNGDDNGSST-SEGGVVAA 348
Query: 98 SHISELSLHPDDSECCICLCPYVDGTELYRLPCTHHFHCECIGRWLRTKATCPLCKFNI 156
+E ++ +D+ CCICL Y + EL LPC+H FH EC+ +WL+ A+CPLCK +
Sbjct: 349 GTDNERAISGEDAVCCICLAKYANNEELRELPCSHFFHKECVDKWLKINASCPLCKSEV 407
>UniRef100_Q6K2E0 Putative RING zinc finger protein [Oryza sativa]
Length = 414
Score = 82.0 bits (201), Expect = 5e-15
Identities = 43/130 (33%), Positives = 65/130 (49%), Gaps = 1/130 (0%)
Query: 28 IYLIVFGALFCIVQIIALAYALTQPLRIREGASEDDIKSLPMYRFCQPNVMIMVDK-NKT 86
I ++ A+ C + L + L GA+++ I +LP Y+F + VD + +
Sbjct: 253 IPFVMCAAICCCFPCLISLLRLQEDLGHTRGATQELIDALPTYKFKPKRSKMWVDHASSS 312
Query: 87 QLEARTGSHNRSHISELSLHPDDSECCICLCPYVDGTELYRLPCTHHFHCECIGRWLRTK 146
+ + G E + +D+ CCICL Y D EL LPCTH FH +C+ +WL+
Sbjct: 313 ENLSEGGILGPGTKKERIVSAEDAVCCICLTKYGDDDELRELPCTHFFHVQCVDKWLKIN 372
Query: 147 ATCPLCKFNI 156
A CPLCK I
Sbjct: 373 AVCPLCKTEI 382
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.332 0.143 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 267,113,936
Number of Sequences: 2790947
Number of extensions: 9914858
Number of successful extensions: 31674
Number of sequences better than 10.0: 2172
Number of HSP's better than 10.0 without gapping: 1568
Number of HSP's successfully gapped in prelim test: 604
Number of HSP's that attempted gapping in prelim test: 29624
Number of HSP's gapped (non-prelim): 2267
length of query: 163
length of database: 848,049,833
effective HSP length: 117
effective length of query: 46
effective length of database: 521,509,034
effective search space: 23989415564
effective search space used: 23989415564
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 69 (31.2 bits)
Medicago: description of AC149050.11


