

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148996.5 + phase: 0
(196 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
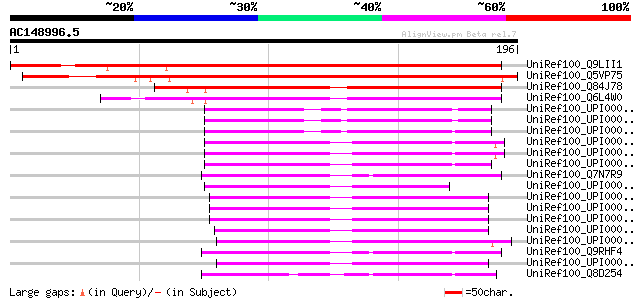
UniRef100_Q9LII1 Similarity to unknown protein [Arabidopsis thal... 215 6e-55
UniRef100_Q5VP75 Single-strand DNA binding protein-like [Oryza s... 206 3e-52
UniRef100_Q84J78 Single-stranded DNA-binding protein, mitochondr... 99 6e-20
UniRef100_Q6L4W0 Putative single-strand DNA binding protein [Ory... 97 3e-19
UniRef100_UPI00002C9AFE UPI00002C9AFE UniRef100 entry 60 3e-08
UniRef100_UPI0000307E7F UPI0000307E7F UniRef100 entry 60 4e-08
UniRef100_UPI0000270BD0 UPI0000270BD0 UniRef100 entry 57 2e-07
UniRef100_UPI0000319EBE UPI0000319EBE UniRef100 entry 56 5e-07
UniRef100_UPI0000305EE2 UPI0000305EE2 UniRef100 entry 56 6e-07
UniRef100_UPI00002CD5CB UPI00002CD5CB UniRef100 entry 56 6e-07
UniRef100_Q7N7R9 Single-strand binding protein [Photorhabdus lum... 55 8e-07
UniRef100_UPI00003302C4 UPI00003302C4 UniRef100 entry 55 1e-06
UniRef100_UPI0000256578 UPI0000256578 UniRef100 entry 55 1e-06
UniRef100_UPI000032B92D UPI000032B92D UniRef100 entry 55 1e-06
UniRef100_UPI0000291169 UPI0000291169 UniRef100 entry 55 1e-06
UniRef100_UPI00002D73DB UPI00002D73DB UniRef100 entry 54 2e-06
UniRef100_UPI00002D0DAD UPI00002D0DAD UniRef100 entry 54 2e-06
UniRef100_Q9RHF4 Single-strand binding protein 2 [Salmonella typhi] 54 2e-06
UniRef100_UPI000028DFA1 UPI000028DFA1 UniRef100 entry 54 2e-06
UniRef100_Q8D254 Single-strand binding protein [Wigglesworthia g... 54 2e-06
>UniRef100_Q9LII1 Similarity to unknown protein [Arabidopsis thaliana]
Length = 217
Score = 215 bits (547), Expect = 6e-55
Identities = 112/199 (56%), Positives = 142/199 (71%), Gaps = 14/199 (7%)
Query: 1 MAKFSISRTSTLYRSLLSTPPPPLHHHRHIPLRFCT---TTTSLSDYDDADSAAPSPSPE 57
MA + + LYRSLLS P + FCT T+ SD+D+ +S + +
Sbjct: 1 MANSMATLSRRLYRSLLSNP-----RISQASMSFCTNNITSPEDSDFDELESPIEPKASD 55
Query: 58 QT------ERTFFDRPLENGLDPGVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIR 111
ER +RPLENGLD G+++AILVG+ GQ PLQKKLKSG VTL S+GTGGIR
Sbjct: 56 PVSRFSGEERVMEERPLENGLDSGIFKAILVGQVGQLPLQKKLKSGRTVTLFSVGTGGIR 115
Query: 112 NNRRPLDHENPREYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDP 171
NNRRPL +E+PREYA+R AVQWHRV+VYPERL +L++K V PG+ +Y+EGNLETK+F+DP
Sbjct: 116 NNRRPLINEDPREYASRSAVQWHRVSVYPERLADLVLKNVEPGTVIYLEGNLETKIFTDP 175
Query: 172 VTGLVRRVREVAVRRNGNL 190
VTGLVRR+REVA+RRNG +
Sbjct: 176 VTGLVRRIREVAIRRNGRV 194
>UniRef100_Q5VP75 Single-strand DNA binding protein-like [Oryza sativa]
Length = 224
Score = 206 bits (524), Expect = 3e-52
Identities = 115/214 (53%), Positives = 139/214 (64%), Gaps = 33/214 (15%)
Query: 6 ISRTSTLYRSLLSTPPPPLHHHRHIPLRFCTTTTSLSDYDDA---DSAAPS---PSPEQT 59
++R L R +LS+P P F TT +S S + DS A S P PEQ
Sbjct: 9 LARRLLLTRRVLSSPLRP----------FSTTDSSSSSSSSSSSDDSRAGSDAGPDPEQQ 58
Query: 60 E--------------RTFFDRPLENGLDPGVYRAILVGKAGQKPLQKKLKSGTVVTLLSI 105
+ R RPLENGLDPG+Y+AI+VGK GQ+P+QK+L+SG V L S+
Sbjct: 59 QPPPAGQDQQAAARPRAGDTRPLENGLDPGIYKAIMVGKVGQEPIQKRLRSGRTVVLFSL 118
Query: 106 GTGGIRNNRRPLDHENPREYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLET 165
GTGGIRNNRRPLD E P +YA RC+VQWHRV +YPERLG+L +K V GS LY+EGNLET
Sbjct: 119 GTGGIRNNRRPLDREEPHQYAERCSVQWHRVCIYPERLGSLALKHVKTGSVLYLEGNLET 178
Query: 166 KVFSDPVTGLVRRVREVAVRRNGN---LANSSNA 196
KVFSDP+TGLVRR+RE+AVR NG L N NA
Sbjct: 179 KVFSDPITGLVRRIREIAVRSNGRLLFLGNDCNA 212
>UniRef100_Q84J78 Single-stranded DNA-binding protein, mitochondrial precursor
[Arabidopsis thaliana]
Length = 201
Score = 99.0 bits (245), Expect = 6e-20
Identities = 53/143 (37%), Positives = 86/143 (60%), Gaps = 15/143 (10%)
Query: 57 EQTERTFFDRP--LENGLDP-------GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGT 107
E E +RP +G+DP GV+RAI+ GK GQ PLQK L++G VT+ ++GT
Sbjct: 43 EDFEENVTERPELQPHGVDPRKGWGFRGVHRAIICGKVGQAPLQKILRNGRTVTIFTVGT 102
Query: 108 GGIRNNRRPLDHENPREYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLETKV 167
GG+ + R P+ QWHR+ V+ E LG+ ++ + S++YVEG++ET+V
Sbjct: 103 GGMFDQRLVGATNQPKP------AQWHRIAVHNEVLGSYAVQKLAKNSSVYVEGDIETRV 156
Query: 168 FSDPVTGLVRRVREVAVRRNGNL 190
++D ++ V+ + E+ VRR+G +
Sbjct: 157 YNDSISSEVKSIPEICVRRDGKI 179
>UniRef100_Q6L4W0 Putative single-strand DNA binding protein [Oryza sativa]
Length = 206
Score = 96.7 bits (239), Expect = 3e-19
Identities = 55/163 (33%), Positives = 94/163 (56%), Gaps = 19/163 (11%)
Query: 36 TTTTSLSDYDDADSAAPSPSPEQTERTFFDRPLE-NGLDP-------GVYRAILVGKAGQ 87
++T S SD D+ S + T R LE +DP GV+RAI+ GK GQ
Sbjct: 33 SSTVSFSDIDEK-----SEMGGDDDYTDSRRELEPQSVDPKKGWGFRGVHRAIICGKVGQ 87
Query: 88 KPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHRVTVYPERLGNLL 147
P+QK L++G VT+ ++GTGG+ + R D + P+ QWHR+ ++ ++LG
Sbjct: 88 VPVQKILRNGRTVTVFTVGTGGMFDQRVVGDADLPKP------AQWHRIAIHNDQLGAFA 141
Query: 148 MKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRRNGNL 190
++ ++ S +YVEG++ET+V++D + V+ + E+ +RR+G +
Sbjct: 142 VQKLVKNSAVYVEGDIETRVYNDSINDQVKNIPEICLRRDGKI 184
>UniRef100_UPI00002C9AFE UPI00002C9AFE UniRef100 entry
Length = 160
Score = 60.1 bits (144), Expect = 3e-08
Identities = 35/111 (31%), Positives = 59/111 (52%), Gaps = 10/111 (9%)
Query: 76 VYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHR 135
+ + ILVG G+ P + ++ G + LS+ T +R N E R +WHR
Sbjct: 5 INKVILVGNLGRDPEVRSMQDGGKIVQLSLATSETWKDR------NSGERRER--TEWHR 56
Query: 136 VTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRR 186
V ++ E LGN+ + + GST+Y+EG L+T+ + +P G + EV ++R
Sbjct: 57 VVIFNEALGNIAEQYLRKGSTIYIEGQLQTRKWGEP--GQEKYTTEVVLQR 105
>UniRef100_UPI0000307E7F UPI0000307E7F UniRef100 entry
Length = 160
Score = 59.7 bits (143), Expect = 4e-08
Identities = 35/111 (31%), Positives = 59/111 (52%), Gaps = 10/111 (9%)
Query: 76 VYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHR 135
+ + ILVG G+ P + ++ G + LS+ T +R N E R +WHR
Sbjct: 5 INKVILVGNLGRDPEVRSMQDGGKIVQLSLATSETWKDR------NSGERRER--TEWHR 56
Query: 136 VTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRR 186
V ++ E LGN+ + + GST+Y+EG L+T+ + +P G + EV ++R
Sbjct: 57 VVIFNEALGNIAEQYLRKGSTVYIEGQLQTRKWGEP--GQEKYTTEVVLQR 105
>UniRef100_UPI0000270BD0 UPI0000270BD0 UniRef100 entry
Length = 131
Score = 57.4 bits (137), Expect = 2e-07
Identities = 34/111 (30%), Positives = 59/111 (52%), Gaps = 9/111 (8%)
Query: 76 VYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHR 135
+ + ILVG G+ P + ++ G V LSI T +R N E R +WHR
Sbjct: 5 INKVILVGNLGRDPESRTMQDGNPVVNLSIATSESWRDR------NSGERRER--TEWHR 56
Query: 136 VTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRR 186
V ++ ++L + K + GS +Y+EG L+T+ ++D +G+ + EV ++R
Sbjct: 57 VVIFNDKLAEIAQKYLRKGSKIYLEGQLQTRKWTDQ-SGMEKYTTEVVLQR 106
>UniRef100_UPI0000319EBE UPI0000319EBE UniRef100 entry
Length = 159
Score = 56.2 bits (134), Expect = 5e-07
Identities = 36/117 (30%), Positives = 58/117 (48%), Gaps = 10/117 (8%)
Query: 76 VYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHR 135
V + IL+G GQ P+ + + G + LSI T +R + E WHR
Sbjct: 5 VNKVILLGNLGQDPVVRTSQDGNKIVQLSIATSERWKDRMSGEQRERTE--------WHR 56
Query: 136 VTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRR-NGNLA 191
V ++ E L N+ + + GST+Y+EG L+T+ + D G+ + E+ + R G LA
Sbjct: 57 VVIFNEGLSNIAEQYLKKGSTIYIEGQLQTRKWQDQ-NGVDKYTTEIVLGRFRGELA 112
>UniRef100_UPI0000305EE2 UPI0000305EE2 UniRef100 entry
Length = 146
Score = 55.8 bits (133), Expect = 6e-07
Identities = 36/117 (30%), Positives = 58/117 (48%), Gaps = 10/117 (8%)
Query: 76 VYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHR 135
V + IL+G GQ P+ + + G + LSI T +R + E WHR
Sbjct: 5 VNKVILLGNLGQDPVVRTSQDGNKIVQLSIATSERWKDRMSGEQRERTE--------WHR 56
Query: 136 VTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRR-NGNLA 191
V ++ E L N+ + + GST+Y+EG L+T+ + D G+ + E+ + R G LA
Sbjct: 57 VVIFNEGLSNIAEQYLKKGSTVYIEGQLQTRKWQDQ-NGVDKYTTEIVLGRFRGELA 112
>UniRef100_UPI00002CD5CB UPI00002CD5CB UniRef100 entry
Length = 140
Score = 55.8 bits (133), Expect = 6e-07
Identities = 32/111 (28%), Positives = 57/111 (50%), Gaps = 9/111 (8%)
Query: 76 VYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHR 135
+ + ILVG G+ P + ++ G V LSI T +R + E WHR
Sbjct: 5 INKVILVGNLGRDPESRTMQDGNSVVNLSIATSESWRDRTSGERRERTE--------WHR 56
Query: 136 VTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRR 186
V ++ ++L + K + GS +Y+EG L+T+ ++D +G+ + EV ++R
Sbjct: 57 VVIFNDKLAEIAQKYLRKGSKVYLEGQLQTRKWTDQ-SGMEKYTTEVVLQR 106
>UniRef100_Q7N7R9 Single-strand binding protein [Photorhabdus luminescens]
Length = 154
Score = 55.5 bits (132), Expect = 8e-07
Identities = 33/116 (28%), Positives = 58/116 (49%), Gaps = 9/116 (7%)
Query: 75 GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWH 134
GV + IL+G+ GQ+P + G V +LS+ T +R+ + + E WH
Sbjct: 5 GVNKVILIGRLGQEPEVRYRPEGDTVAVLSLATSETWRDRQTGEQKERTE--------WH 56
Query: 135 RVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRRNGNL 190
RV ++ +L + + + GS +Y+EG L T+ + D +G R EV + + G +
Sbjct: 57 RVVIF-GKLAEIAGEYLKKGSQVYIEGQLRTRKWFDKASGQDRYSTEVVLNQQGTM 111
>UniRef100_UPI00003302C4 UPI00003302C4 UniRef100 entry
Length = 149
Score = 55.1 bits (131), Expect = 1e-06
Identities = 30/95 (31%), Positives = 48/95 (49%), Gaps = 8/95 (8%)
Query: 76 VYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHR 135
+ + +L+G G+ P K + +G + LSI T +R + E WHR
Sbjct: 5 INKVVLIGNLGRDPEVKTMPNGNKIVNLSIATSERWKDRATGEQREKTE--------WHR 56
Query: 136 VTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSD 170
V ++ E LGN+ + + GST YVEG L T+ ++D
Sbjct: 57 VVIFNEALGNVAEQYLRKGSTCYVEGQLRTRKWND 91
>UniRef100_UPI0000256578 UPI0000256578 UniRef100 entry
Length = 152
Score = 55.1 bits (131), Expect = 1e-06
Identities = 30/108 (27%), Positives = 56/108 (51%), Gaps = 8/108 (7%)
Query: 78 RAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHRVT 137
+ +L+G+ G P K++ +G V LS+ T ++ + + E WHRV
Sbjct: 7 KVLLIGRLGADPEIKQMVNGKSVARLSLATSNTWKDKNTGEKKEKTE--------WHRVV 58
Query: 138 VYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVR 185
++ E L N++ + V G+ +Y+EG L T+ + D +G+ R EV ++
Sbjct: 59 IFNEGLVNVVQQYVKKGAQVYIEGQLTTRKWKDEKSGIDRYSTEVVLQ 106
>UniRef100_UPI000032B92D UPI000032B92D UniRef100 entry
Length = 152
Score = 54.7 bits (130), Expect = 1e-06
Identities = 29/108 (26%), Positives = 56/108 (51%), Gaps = 8/108 (7%)
Query: 78 RAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHRVT 137
+ +L+G+ G P K++ +G V +S+ T ++ + + E WHRV
Sbjct: 7 KVLLIGRLGADPEIKQMTNGKSVARISLATSNTWKDKNTGEKKEKTE--------WHRVV 58
Query: 138 VYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVR 185
++ E L N++ + V G+ +Y+EG L T+ + D +G+ R EV ++
Sbjct: 59 IFNEGLVNVVQQYVKKGAQVYIEGQLTTRKWKDEKSGMDRYSTEVVLQ 106
>UniRef100_UPI0000291169 UPI0000291169 UniRef100 entry
Length = 152
Score = 54.7 bits (130), Expect = 1e-06
Identities = 31/108 (28%), Positives = 56/108 (51%), Gaps = 8/108 (7%)
Query: 78 RAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHRVT 137
+ +L+G+ G P K++ +G V LS+ T ++ + + E WHRV
Sbjct: 7 KVLLIGRLGADPDIKQMVNGKNVARLSLATSNTWKDKNTGEKKEKTE--------WHRVV 58
Query: 138 VYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVR 185
++ E L N++ + V G+ +Y+EG L T+ + D +GL R EV ++
Sbjct: 59 IFNEGLVNVVKQYVKKGTQVYIEGQLTTRKWKDEKSGLDRYSTEVVLQ 106
>UniRef100_UPI00002D73DB UPI00002D73DB UniRef100 entry
Length = 144
Score = 54.3 bits (129), Expect = 2e-06
Identities = 31/106 (29%), Positives = 55/106 (51%), Gaps = 8/106 (7%)
Query: 80 ILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHRVTVY 139
+L+G+ G P K++ +G V LSI T ++ + + E WHRV ++
Sbjct: 1 LLIGRLGGDPEIKQMVNGKNVARLSIATSNTWKDKNTGEKKEKTE--------WHRVVIF 52
Query: 140 PERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVR 185
E L N++ + V G+ +Y+EG L T+ + D +G+ R EV ++
Sbjct: 53 NEGLVNVVQQYVKKGTQVYIEGQLTTRKWKDEKSGIDRYSTEVVLQ 98
>UniRef100_UPI00002D0DAD UPI00002D0DAD UniRef100 entry
Length = 142
Score = 54.3 bits (129), Expect = 2e-06
Identities = 35/125 (28%), Positives = 60/125 (48%), Gaps = 19/125 (15%)
Query: 81 LVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHRVTVYP 140
L+G+ G P K++ +G V LSI T ++ + + E WHRV ++
Sbjct: 10 LIGRLGADPEIKQMVNGKSVARLSIATSQSWKDKASGERKEKTE--------WHRVVIFN 61
Query: 141 ERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVR-----------RNGN 189
E L N++ + V G+ +Y+EG L T+ + D TG + E+ ++ R+GN
Sbjct: 62 EGLVNVVQQYVKKGANVYIEGQLSTRKWKDEATGQDKYSTEILLQGYNSSLTMLDSRSGN 121
Query: 190 LANSS 194
NS+
Sbjct: 122 NGNSN 126
>UniRef100_Q9RHF4 Single-strand binding protein 2 [Salmonella typhi]
Length = 178
Score = 54.3 bits (129), Expect = 2e-06
Identities = 35/116 (30%), Positives = 60/116 (51%), Gaps = 10/116 (8%)
Query: 75 GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWH 134
GV + ILVG GQ P + + +G V LS+ T +++ D + E WH
Sbjct: 5 GVNKVILVGNLGQDPEVRYMPNGGAVVNLSLATSDTWTDKQTGDKKERTE--------WH 56
Query: 135 RVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRRNGNL 190
RV +Y +L + + + GS +Y+EG L T+ ++D +G+ + EV V ++G +
Sbjct: 57 RVVLY-GKLAEIASEYLRKGSQVYIEGALRTRKWTDQ-SGVEKYTTEVVVSQSGTM 110
>UniRef100_UPI000028DFA1 UPI000028DFA1 UniRef100 entry
Length = 152
Score = 53.9 bits (128), Expect = 2e-06
Identities = 30/105 (28%), Positives = 53/105 (49%), Gaps = 8/105 (7%)
Query: 81 LVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHRVTVYP 140
L+G+ G P K++ +G V LSI T ++ + + E WHR+ ++
Sbjct: 10 LIGRLGADPEIKQMVNGKSVARLSIATSQSWKDKSSGERKEKTE--------WHRIVIFN 61
Query: 141 ERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVR 185
E L N++ + V GS +Y+EG L T+ + D TG + E+ ++
Sbjct: 62 EGLVNVVQQYVKKGSNVYIEGQLSTRKWKDEATGQDKYSTEIVLQ 106
>UniRef100_Q8D254 Single-strand binding protein [Wigglesworthia glossinidia
brevipalpis]
Length = 163
Score = 53.9 bits (128), Expect = 2e-06
Identities = 35/114 (30%), Positives = 59/114 (51%), Gaps = 10/114 (8%)
Query: 75 GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWH 134
G+ + IL+G GQ P + + +G +T LS+ T N R + +E +WH
Sbjct: 5 GINKVILIGNLGQDPEVRYIPNGNAITNLSLATS---ENWRDKNTGESKE-----KTEWH 56
Query: 135 RVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRRNG 188
RV ++ +L + + + GS +Y+EGNL+T+ + D G R + E+ V NG
Sbjct: 57 RVILF-GKLAEVAGEYLKKGSQVYIEGNLQTRKWQDQ-NGQDRYITEIVVGING 108
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.134 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 346,999,861
Number of Sequences: 2790947
Number of extensions: 15147826
Number of successful extensions: 39666
Number of sequences better than 10.0: 447
Number of HSP's better than 10.0 without gapping: 238
Number of HSP's successfully gapped in prelim test: 210
Number of HSP's that attempted gapping in prelim test: 39233
Number of HSP's gapped (non-prelim): 464
length of query: 196
length of database: 848,049,833
effective HSP length: 121
effective length of query: 75
effective length of database: 510,345,246
effective search space: 38275893450
effective search space used: 38275893450
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 71 (32.0 bits)
Medicago: description of AC148996.5


