

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148996.16 - phase: 0 /pseudo
(332 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
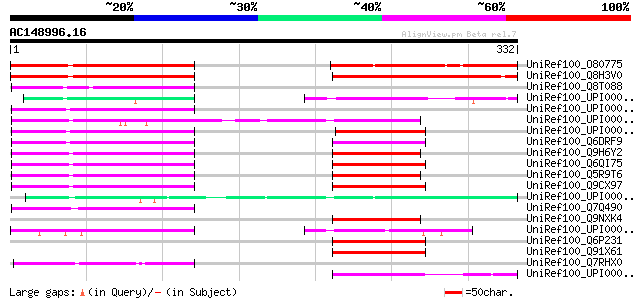
UniRef100_O80775 Expressed protein [Arabidopsis thaliana] 159 1e-37
UniRef100_Q8H3V0 Putative transducin / WD-40 repeat protein [Ory... 137 4e-31
UniRef100_Q8T088 LD21659p [Drosophila melanogaster] 87 8e-16
UniRef100_UPI000043019A UPI000043019A UniRef100 entry 83 1e-14
UniRef100_UPI00002F07DF UPI00002F07DF UniRef100 entry 82 2e-14
UniRef100_UPI000036D2C7 UPI000036D2C7 UniRef100 entry 80 8e-14
UniRef100_UPI0000363F46 UPI0000363F46 UniRef100 entry 78 4e-13
UniRef100_Q6DRF9 AK025355-like [Brachydanio rerio] 78 4e-13
UniRef100_Q9H6Y2 Hypothetical protein FLJ21702 [Homo sapiens] 74 6e-12
UniRef100_Q6QI75 LRRG00133 [Rattus norvegicus] 74 7e-12
UniRef100_Q5R9T6 Hypothetical protein DKFZp469F1823 [Pongo pygma... 73 1e-11
UniRef100_Q9CX97 Mus musculus 16 days embryo lung cDNA, RIKEN fu... 72 2e-11
UniRef100_UPI000049944F UPI000049944F UniRef100 entry 71 4e-11
UniRef100_Q7Q490 ENSANGP00000018332 [Anopheles gambiae str. PEST] 69 2e-10
UniRef100_Q9NXK4 Hypothetical protein FLJ20195 [Homo sapiens] 69 2e-10
UniRef100_UPI00003C1B7C UPI00003C1B7C UniRef100 entry 67 9e-10
UniRef100_Q6P231 2410080P20Rik protein [Mus musculus] 65 3e-09
UniRef100_Q91X61 2410080P20Rik protein [Mus musculus] 64 7e-09
UniRef100_Q7RHX0 Unnamed protein product [Plasmodium yoelii yoelii] 57 9e-07
UniRef100_UPI00003AA9E3 UPI00003AA9E3 UniRef100 entry 55 3e-06
>UniRef100_O80775 Expressed protein [Arabidopsis thaliana]
Length = 353
Score = 159 bits (401), Expect = 1e-37
Identities = 81/121 (66%), Positives = 95/121 (77%), Gaps = 3/121 (2%)
Query: 1 MEINLGKLAFDIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAA 60
MEI+LG AF IDFHPS NLVA+GLIDG LHLYRY SD S VR +V AH ESCRA
Sbjct: 1 MEIDLGANAFGIDFHPSTNLVAAGLIDGHLHLYRYDSD---SSLVRERKVRAHKESCRAV 57
Query: 61 RFINGGRAVLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLINLTESTVASGDDDGCI 120
RFI+ G+ ++T S D SILATDVETG+ +A L+NAHE AVN LIN+TE+T+ASGDD GC+
Sbjct: 58 RFIDDGQRIVTASADCSILATDVETGAQVAHLENAHEDAVNTLINVTETTIASGDDKGCV 117
Query: 121 K 121
K
Sbjct: 118 K 118
Score = 141 bits (356), Expect = 2e-32
Identities = 72/122 (59%), Positives = 90/122 (73%), Gaps = 3/122 (2%)
Query: 211 RIITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRKFLGSIGHDQMLKLWDLD 270
R+ITG +NG+I+LVGILPNRII+PI H +YP+E LA SHD+KFLGS HD MLKLW+L+
Sbjct: 234 RLITGCDNGIISLVGILPNRIIQPIGSH-DYPIEDLALSHDKKFLGSTAHDSMLKLWNLE 292
Query: 271 NILQGSRSTQRNETGVVANDGDSDDGDEMDVDNSASKFSKGNKRKNASNGHAVGDSNNFF 330
IL+GS N +G A D DSD+ D MD+DN SK SKG+KRK S + + +NNFF
Sbjct: 293 EILEGSNVNSGNASG-AAEDSDSDN-DGMDLDNDPSKSSKGSKRKTKSKANTLNATNNFF 350
Query: 331 AD 332
AD
Sbjct: 351 AD 352
>UniRef100_Q8H3V0 Putative transducin / WD-40 repeat protein [Oryza sativa]
Length = 353
Score = 137 bits (345), Expect = 4e-31
Identities = 68/121 (56%), Positives = 89/121 (73%), Gaps = 3/121 (2%)
Query: 1 MEINLGKLAFDIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAA 60
ME ++ FD+ FHPS LVA+ LI G+L+L+RY+++ S P R+ +H ESCRA
Sbjct: 1 MEALHEEMPFDLAFHPSSPLVATSLITGELYLFRYAAE---SQPERLFAAKSHKESCRAV 57
Query: 61 RFINGGRAVLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLINLTESTVASGDDDGCI 120
RF+ G +LTGS D SILA+DVETG IARL++AHE +NRL+ LTE+TVASGDD+GCI
Sbjct: 58 RFVESGNVILTGSADCSILASDVETGKPIARLEDAHENGINRLVCLTETTVASGDDEGCI 117
Query: 121 K 121
K
Sbjct: 118 K 118
Score = 123 bits (308), Expect = 8e-27
Identities = 59/121 (48%), Positives = 86/121 (70%), Gaps = 2/121 (1%)
Query: 212 IITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRKFLGSIGHDQMLKLWDLDN 271
+I+G+ +G+I LVGILPNRII+P+AEHSEYP+E LAFS+DR +LGSI HD+MLKLWDL +
Sbjct: 234 LISGASDGVIRLVGILPNRIIQPLAEHSEYPIEALAFSNDRNYLGSISHDKMLKLWDLQD 293
Query: 272 ILQGSRSTQRNETGVVANDGDSDDGDEMDVDNSASKFSKGNKRKNASNGHAVGDSNNFFA 331
+L + Q ++ G +D DDG ++D+D ++SK S+ K + +++FFA
Sbjct: 294 LLNRQQLVQDDKLGEQDSDDSDDDGMDVDMDPNSSKGSRSTKTSKGQSSDR--PTSDFFA 351
Query: 332 D 332
D
Sbjct: 352 D 352
>UniRef100_Q8T088 LD21659p [Drosophila melanogaster]
Length = 498
Score = 86.7 bits (213), Expect = 8e-16
Identities = 49/120 (40%), Positives = 68/120 (55%), Gaps = 4/120 (3%)
Query: 2 EINLGKLAFDIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAAR 61
EI L DI FHP +++A I GD+HLY Y DN + +R +EVH+ ++CR
Sbjct: 152 EIKLEDFITDICFHPDRDIIALATIIGDVHLYEY--DNEANKLLRTIEVHS--KACRDVE 207
Query: 62 FINGGRAVLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLINLTESTVASGDDDGCIK 121
F GR +LT S D ++ TD+ET + AH+ A+N L L E+ ASGDD G +K
Sbjct: 208 FTEDGRFLLTCSKDKCVMVTDMETEKLKKLYETAHDDAINTLHVLNENLFASGDDAGTVK 267
>UniRef100_UPI000043019A UPI000043019A UniRef100 entry
Length = 384
Score = 82.8 bits (203), Expect = 1e-14
Identities = 50/145 (34%), Positives = 78/145 (53%), Gaps = 29/145 (20%)
Query: 194 AVIDSLILLQTLLIPC*RIITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRK 253
A ID+++ L +I TGSE+GMI ++ +LP++ + +A H E+PVER+ + K
Sbjct: 262 ASIDAIVALTPDIIA-----TGSEDGMIRVIQVLPHKFLGVVATHEEFPVERIRLDRNNK 316
Query: 254 FLGSIGHDQMLKLWDLDNILQGSRSTQRNETGVVANDGDSDDGDEMDVD------NSASK 307
+LGS+ HD+ LKL D++++ + DSD+ DEM+ D + K
Sbjct: 317 WLGSVSHDECLKLTDVEDLFE-----------------DSDEDDEMEEDEPDSDEEKSKK 359
Query: 308 FSKGNKRKNASNGHAVGDSNNFFAD 332
K N K+ S G A D +FFAD
Sbjct: 360 KKKDNGMKDMSRGQAEND-GSFFAD 383
Score = 52.8 bits (125), Expect = 1e-05
Identities = 36/131 (27%), Positives = 52/131 (39%), Gaps = 21/131 (16%)
Query: 10 FDIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAARFINGGRAV 69
FD+ FHP E +V S L+ G + + Y D+ + V + RA G +
Sbjct: 28 FDVAFHPKEPVVFSSLLTGQVCAWSY--DDATGETSSSWSVRPSKRTARALSIEENGDEI 85
Query: 70 LTGSPDFSILAT-------------------DVETGSTIARLDNAHEAAVNRLINLTEST 110
G S+L GS D+AHE +NR+ + +
Sbjct: 86 WMGGKSGSLLCALTPPKVMCPILLMSNCSQLSTRDGSMTRERDSAHECPINRVYCVNRNL 145
Query: 111 VASGDDDGCIK 121
VA+GDDDG IK
Sbjct: 146 VATGDDDGVIK 156
>UniRef100_UPI00002F07DF UPI00002F07DF UniRef100 entry
Length = 615
Score = 82.4 bits (202), Expect = 2e-14
Identities = 43/120 (35%), Positives = 71/120 (58%), Gaps = 2/120 (1%)
Query: 2 EINLGKLAFDIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAAR 61
+I L +A + HPS +++A G +DGD+++Y YS T D + H +SCR R
Sbjct: 19 DIRLEAIANSVALHPSRDILALGDVDGDVYVYSYSC--TEGDNRELWSSGHHVKSCRQVR 76
Query: 62 FINGGRAVLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLINLTESTVASGDDDGCIK 121
F G+ + + S D ++ DVE G ++R+ AH+AA+N L+ + E+ +A+GDD G +K
Sbjct: 77 FSADGQKLYSVSRDKAVHLLDVERGQLLSRIRGAHDAAINSLLLVDENVLATGDDVGGLK 136
>UniRef100_UPI000036D2C7 UPI000036D2C7 UniRef100 entry
Length = 383
Score = 80.1 bits (196), Expect = 8e-14
Identities = 73/310 (23%), Positives = 129/310 (41%), Gaps = 61/310 (19%)
Query: 2 EINLGKLAFDIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAAR 61
+I L A + FHP+ +L+A+G +DGD+ ++ YS + + H ++CRA
Sbjct: 34 DIVLEAPASGLAFHPARDLLAAGDVDGDVFVFSYSCQE--GETKELWSSGHHLKACRAVA 91
Query: 62 FINGGRAVLT-------------------------GSP-------DFSILATDVETGST- 88
F G+ ++T G+P D ++LAT +TG
Sbjct: 92 FSEDGQKLITVSKDKAIHVLDVEQGQLERRVSKAHGAPINSLLLVDENVLATGDDTGGIR 151
Query: 89 ---------IARLDNAHEAAVNRLINLTESTVASGDDDGCIKTLLLHLTQ*KYWPQVETG 139
+ + E + ++ + + + DGC+ + + + + ++G
Sbjct: 152 LWDQRKEGPLMDMRQHEEYIADMALDPAKKLLLTASGDGCLGVFNIKRRRFELLSEPQSG 211
Query: 140 LFLFAIFDEIKSKPNLNFLKMSCCLWF**RMAGKLFADHKLESCYCIHGDASRIAVIDSL 199
++ S + + K C G ++ + A R ID +
Sbjct: 212 --------DLTSVTLMKWGKKVACG----SSEGTIYLFNWNGFGATSDRFALRAECIDCM 259
Query: 200 ILLQTLLIPC*RIITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRKFLGSIG 259
+ + L+ TGS +G+I V ILPNR++ + +H+ PVE LA SH +FL S G
Sbjct: 260 VPVTESLL-----CTGSTDGVIRAVNILPNRVVGSVGQHTGEPVEELALSHCGRFLASSG 314
Query: 260 HDQMLKLWDL 269
HDQ LK WD+
Sbjct: 315 HDQRLKFWDM 324
>UniRef100_UPI0000363F46 UPI0000363F46 UniRef100 entry
Length = 369
Score = 77.8 bits (190), Expect = 4e-13
Identities = 42/120 (35%), Positives = 67/120 (55%), Gaps = 2/120 (1%)
Query: 2 EINLGKLAFDIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAAR 61
+I L +A + HP+ ++V G +DGD+++Y YS T + + H +SCR R
Sbjct: 24 DIRLEAIANTVALHPNRDVVVFGDVDGDVYVYSYSC--TEGENRELWSSGHHMKSCRQVR 81
Query: 62 FINGGRAVLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLINLTESTVASGDDDGCIK 121
F GR + + S D ++ DVE G R+ AHEA +N L+ + E+ +A+GDD G +K
Sbjct: 82 FSADGRKLFSVSRDKAVHLLDVERGQLETRIRGAHEAPINSLLVVDENILATGDDAGSLK 141
Score = 60.8 bits (146), Expect = 5e-08
Identities = 26/59 (44%), Positives = 39/59 (66%)
Query: 214 TGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRKFLGSIGHDQMLKLWDLDNI 272
T S +G I V +LPNR++ I +H+ P+E LA S D +FL S HDQ++K WD+ ++
Sbjct: 259 TASIDGYIRAVNLLPNRVLGCIGQHAGEPIEELAKSWDSRFLASTAHDQLIKFWDISSL 317
>UniRef100_Q6DRF9 AK025355-like [Brachydanio rerio]
Length = 386
Score = 77.8 bits (190), Expect = 4e-13
Identities = 40/120 (33%), Positives = 68/120 (56%), Gaps = 2/120 (1%)
Query: 2 EINLGKLAFDIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAAR 61
+I L + I FHP ++++A+G IDGD++L+ YS T + + H +SCR
Sbjct: 31 DIKLEAIVNTIAFHPKQDILAAGDIDGDIYLFSYSC--TEGENKELWSSGHHLKSCRKVL 88
Query: 62 FINGGRAVLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLINLTESTVASGDDDGCIK 121
F + G+ + + S D +I DVE G R+ AH+ +N ++ + E+ A+GDD+G +K
Sbjct: 89 FSSDGQKLFSVSKDKAIHIMDVEAGKLETRIPKAHKVPINAMLLIDENIFATGDDEGTLK 148
Score = 53.9 bits (128), Expect = 6e-06
Identities = 21/61 (34%), Positives = 36/61 (58%)
Query: 212 IITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRKFLGSIGHDQMLKLWDLDN 271
+ S +G+I + ILPNR++ I +H +E +A D FL S HD+++K WD+ +
Sbjct: 264 LCAASTDGVIRAINILPNRVVGSIGQHVGEAIEEIARCRDTHFLASCAHDELIKFWDISS 323
Query: 272 I 272
+
Sbjct: 324 L 324
>UniRef100_Q9H6Y2 Hypothetical protein FLJ21702 [Homo sapiens]
Length = 383
Score = 73.9 bits (180), Expect = 6e-12
Identities = 41/120 (34%), Positives = 67/120 (55%), Gaps = 2/120 (1%)
Query: 2 EINLGKLAFDIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAAR 61
+I L A + FHP+ +L+A+G +DGD+ ++ YS + + H ++CRA
Sbjct: 34 DIVLEAPASGLAFHPARDLLAAGDVDGDVFVFSYSCQE--GETKELWSSGHHLKACRAVA 91
Query: 62 FINGGRAVLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLINLTESTVASGDDDGCIK 121
F G+ ++T S D +I DVE G R+ AH A +N L+ + E+ +A+GDD G I+
Sbjct: 92 FSEDGQKLITVSKDKAIHVLDVEQGQLERRVSKAHGAPINSLLLVDENVLATGDDTGGIR 151
Score = 68.6 bits (166), Expect = 2e-10
Identities = 30/58 (51%), Positives = 40/58 (68%)
Query: 212 IITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRKFLGSIGHDQMLKLWDL 269
+ TGS +G+I V ILPNR++ + +H+ PVE LA SH +FL S GHDQ LK WD+
Sbjct: 267 LCTGSTDGVIRAVNILPNRVVGSVGQHTGEPVEELALSHCGRFLASSGHDQRLKFWDM 324
>UniRef100_Q6QI75 LRRG00133 [Rattus norvegicus]
Length = 872
Score = 73.6 bits (179), Expect = 7e-12
Identities = 41/120 (34%), Positives = 66/120 (54%), Gaps = 2/120 (1%)
Query: 2 EINLGKLAFDIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAAR 61
+I L A + FHP+ +L+A+G +DGD+ ++ YS + + H +SCRA
Sbjct: 522 DIVLEAPASGLAFHPTRDLLAAGDVDGDVFVFAYSCQE--GETKELWSSGHHLKSCRAVV 579
Query: 62 FINGGRAVLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLINLTESTVASGDDDGCIK 121
F G+ ++T S D +I DVE G R+ AH A +N L+ + E+ + +GDD G I+
Sbjct: 580 FSEDGQKLVTVSKDKAIHVLDVEQGQLERRISKAHSAPINSLLLVDENVLVTGDDTGGIR 639
Score = 67.8 bits (164), Expect = 4e-10
Identities = 30/61 (49%), Positives = 40/61 (65%)
Query: 212 IITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRKFLGSIGHDQMLKLWDLDN 271
+ TGS +G+I V ILPNR++ + +H+ PVE LA SH FL S GHDQ LK WD+
Sbjct: 755 LCTGSTDGIIRAVNILPNRVVGTVGQHAGEPVEELALSHCGHFLASSGHDQRLKFWDMTQ 814
Query: 272 I 272
+
Sbjct: 815 L 815
>UniRef100_Q5R9T6 Hypothetical protein DKFZp469F1823 [Pongo pygmaeus]
Length = 383
Score = 72.8 bits (177), Expect = 1e-11
Identities = 41/120 (34%), Positives = 67/120 (55%), Gaps = 2/120 (1%)
Query: 2 EINLGKLAFDIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAAR 61
+I L A + FHP+ +L+A+G +DGD+ ++ YS + + H ++CRA
Sbjct: 34 DIVLEAPASGLAFHPARDLLAAGDVDGDVFVFSYSCQE--GETKELWSSGHHLKACRAVA 91
Query: 62 FINGGRAVLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLINLTESTVASGDDDGCIK 121
F G+ ++T S D +I DVE G R+ AH A +N L+ + E+ +A+GDD G I+
Sbjct: 92 FSEDGQKLVTVSKDKAIHVLDVEQGRLERRISKAHGAPINSLLLVDENVLATGDDMGGIR 151
Score = 65.9 bits (159), Expect = 2e-09
Identities = 29/58 (50%), Positives = 39/58 (67%)
Query: 212 IITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRKFLGSIGHDQMLKLWDL 269
+ TGS +G+I V ILPNR++ + +H+ PV LA SH +FL S GHDQ LK WD+
Sbjct: 267 LCTGSTDGVIRAVNILPNRVVGSVGQHTGEPVGELALSHCGRFLASSGHDQRLKFWDM 324
>UniRef100_Q9CX97 Mus musculus 16 days embryo lung cDNA, RIKEN full-length enriched
library, clone:8430437C04 product:CDNA: FLJ21702 FIS,
CLONE COL09874 homolog [Mus musculus]
Length = 388
Score = 72.0 bits (175), Expect = 2e-11
Identities = 40/120 (33%), Positives = 66/120 (54%), Gaps = 2/120 (1%)
Query: 2 EINLGKLAFDIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAAR 61
+I L A + FHP+ +L+A+G +DGD+ ++ YS + + H +SCRA
Sbjct: 35 DIVLEAPASGLAFHPTRDLLAAGDVDGDVFVFAYSCQE--GETKELWSSGHHLKSCRAVV 92
Query: 62 FINGGRAVLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLINLTESTVASGDDDGCIK 121
F G+ ++T S D +I DVE G R+ AH A +N ++ + E+ + +GDD G I+
Sbjct: 93 FSEDGQKLVTVSKDKAIHILDVEQGQLERRISKAHSAPINSVLLVDENALVTGDDTGGIR 152
Score = 67.4 bits (163), Expect = 5e-10
Identities = 30/61 (49%), Positives = 40/61 (65%)
Query: 212 IITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRKFLGSIGHDQMLKLWDLDN 271
+ TGS +G+I V ILPNR++ + +H+ PVE LA SH FL S GHDQ LK WD+
Sbjct: 268 LCTGSTDGIIRAVNILPNRVVGTVGQHAGEPVEALALSHCGHFLASSGHDQRLKFWDMTQ 327
Query: 272 I 272
+
Sbjct: 328 L 328
>UniRef100_UPI000049944F UPI000049944F UniRef100 entry
Length = 363
Score = 71.2 bits (173), Expect = 4e-11
Identities = 79/363 (21%), Positives = 144/363 (38%), Gaps = 64/363 (17%)
Query: 11 DIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAARFINGGRAVL 70
DI F P+ ++VA G + G + LY Y + +P + + AH +S R F + G +
Sbjct: 23 DIKFSPTNDVVAVGDMHGLIRLYNYGNIQ---EPNEIFTLKAHEDSTRDVCFNDNGNLLF 79
Query: 71 TGSPDFSILATDVE---------------------------TGSTIARLD---------- 93
+ S D SI TD+ TG R+
Sbjct: 80 STSADGSITITDLNNQKILAKNDKAHKHGIYSLDVKGNIFVTGDESGRIKVWDMRQQKCV 139
Query: 94 ---NAHEAAVNRLINLTESTVASGDDDGCIKTLLLHLTQ*KYWPQVETGLFLFAIFDEIK 150
N H +++L + ++T+ + DGC+ T ++ + + I D +
Sbjct: 140 VVFNEHHDYISQL-TICDNTIFAAGGDGCMSTWNINSKK------------VIGISDNMN 186
Query: 151 SKP-NLNFLKMSCCLWF**RMAGKLFADHKLESCYCIHGDASRIAVIDSLILLQTLLIPC 209
+L+ K L +GKLF Y + I+S+I + I
Sbjct: 187 EDLLSLSITKKDDTLVCG-GQSGKLFVWEWDNWEYPKNALKGHPESIESIIAVNKNTI-- 243
Query: 210 *RIITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRKFLGSIGHDQMLKLWDL 269
ITGS +G+I +V I P++++ + +H ++P+ERLA S D+ L S H + +K WD+
Sbjct: 244 ---ITGSSDGIIRVVQINPHKLLGCVGQH-DFPIERLALSRDKNILASSSHSRHIKFWDV 299
Query: 270 DNILQGSRSTQRNETGVVANDGDSDDGDEMDVDNSASKFSKGNKRKNASNGHAVGDSNNF 329
+ + +++ + + D + + NK+K ++N F
Sbjct: 300 GYLYNDQDDEESDDSKQKTMVIEKPEADIVTPFQEEKVQNAYNKKKKTKGPQQRNETNPF 359
Query: 330 FAD 332
+ D
Sbjct: 360 YKD 362
>UniRef100_Q7Q490 ENSANGP00000018332 [Anopheles gambiae str. PEST]
Length = 477
Score = 68.9 bits (167), Expect = 2e-10
Identities = 38/120 (31%), Positives = 63/120 (51%), Gaps = 4/120 (3%)
Query: 2 EINLGKLAFDIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAAR 61
+I D+ FHP E+++A G GD+ +YRY++D V HT++ R
Sbjct: 150 DIQTEDFVVDLSFHPCEDILAVGTSVGDVLMYRYANDAN----VLAATHELHTKAIRCVE 205
Query: 62 FINGGRAVLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLINLTESTVASGDDDGCIK 121
F+ G+++++ + SI+ D ETG ++AH+ V + L E A+GDDDG +K
Sbjct: 206 FVPDGKSLISTGRERSIVVMDAETGKFKRFWESAHDEPVYSMTVLGEHLFATGDDDGEVK 265
Score = 33.5 bits (75), Expect = 8.3
Identities = 18/75 (24%), Positives = 36/75 (48%), Gaps = 7/75 (9%)
Query: 213 ITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRKFLGSIGHDQMLKLW----- 267
+ E+G++ + ++P R++ + +HS V+ L S + + S HD ++ W
Sbjct: 383 VMAGEDGILRAMHMVPGRVLGIVGQHS-MAVDTLDISGSGELIASSSHDNDVRFWNVKYF 441
Query: 268 -DLDNILQGSRSTQR 281
D D I S+ +R
Sbjct: 442 EDFDGIKYNSKPDKR 456
>UniRef100_Q9NXK4 Hypothetical protein FLJ20195 [Homo sapiens]
Length = 165
Score = 68.6 bits (166), Expect = 2e-10
Identities = 30/58 (51%), Positives = 40/58 (68%)
Query: 212 IITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRKFLGSIGHDQMLKLWDL 269
+ TGS +G+I V ILPNR++ + +H+ PVE LA SH +FL S GHDQ LK WD+
Sbjct: 49 LCTGSTDGVIRAVNILPNRVVGSVGQHTGEPVEELALSHCGRFLASSGHDQRLKFWDM 106
>UniRef100_UPI00003C1B7C UPI00003C1B7C UniRef100 entry
Length = 489
Score = 66.6 bits (161), Expect = 9e-10
Identities = 44/151 (29%), Positives = 71/151 (46%), Gaps = 30/151 (19%)
Query: 1 MEINLGKLAFDIDFHPSE--NLVASGLIDGDLHLYRY----SSDNTNSDPV--------- 45
M+I L A D+ FHP+ NL+A GLI G + L Y SS +++ P+
Sbjct: 1 MDIPLSSDALDLCFHPAAETNLLAVGLISGKIQLINYDDYLSSPSSSRTPLAPPSKKSKP 60
Query: 46 ---------------RVLEVHAHTESCRAARFINGGRAVLTGSPDFSILATDVETGSTIA 90
++ +SCR F G ++ + S D S+ +TD +TG +
Sbjct: 61 STISSAVETPTKLYRKLYTSRPSKKSCRGLHFSTTGSSIFSISKDKSLFSTDTQTGKVVQ 120
Query: 91 RLDNAHEAAVNRLINLTESTVASGDDDGCIK 121
H+AA +R++ + ES V +GDDDG ++
Sbjct: 121 SWIEVHDAAPSRVLPVDESLVVTGDDDGVVR 151
Score = 61.2 bits (147), Expect = 4e-08
Identities = 40/119 (33%), Positives = 63/119 (52%), Gaps = 16/119 (13%)
Query: 194 AVIDSLILLQTLLIPC*RIITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRK 253
A +D+L+ L + +TGS +G++ +V ILP++++ IA H+ PVER+ +
Sbjct: 326 ASVDTLVTLDNETV-----LTGSSDGLVRVVQILPSKLLGVIASHNGLPVERM--KRKQS 378
Query: 254 FLGSIGHDQMLKLWDL-----DNILQGSRSTQR----NETGVVANDGDSDDGDEMDVDN 303
L SIGH +KL DL DN QG ++ G+ +D D DD D+ D D+
Sbjct: 379 VLASIGHTNAVKLTDLSPLLDDNDDQGDDDDEQAGALGIVGLAEDDSDDDDDDDDDDDD 437
>UniRef100_Q6P231 2410080P20Rik protein [Mus musculus]
Length = 226
Score = 65.1 bits (157), Expect = 3e-09
Identities = 29/61 (47%), Positives = 39/61 (63%)
Query: 212 IITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRKFLGSIGHDQMLKLWDLDN 271
+ T S +G+I V ILPNR++ + +H+ PVE LA SH FL S GHDQ LK WD+
Sbjct: 106 LCTSSTDGIIRAVNILPNRVVGTVGQHAGEPVEALALSHCGHFLASSGHDQRLKFWDMTQ 165
Query: 272 I 272
+
Sbjct: 166 L 166
>UniRef100_Q91X61 2410080P20Rik protein [Mus musculus]
Length = 169
Score = 63.5 bits (153), Expect = 7e-09
Identities = 29/61 (47%), Positives = 39/61 (63%)
Query: 212 IITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRKFLGSIGHDQMLKLWDLDN 271
+ TGS +G+I V ILPNR++ + +H+ VE LA SH FL S GHDQ LK WD+
Sbjct: 49 LCTGSTDGIIRAVNILPNRVVGTVGQHAGELVEALALSHCGHFLASSGHDQRLKFWDMTQ 108
Query: 272 I 272
+
Sbjct: 109 L 109
>UniRef100_Q7RHX0 Unnamed protein product [Plasmodium yoelii yoelii]
Length = 322
Score = 56.6 bits (135), Expect = 9e-07
Identities = 34/119 (28%), Positives = 57/119 (47%), Gaps = 4/119 (3%)
Query: 3 INLGKLAFDIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAARF 62
I L FD+DFHP +L+ +GL+DG+L LY+ + + + H +S R F
Sbjct: 8 IKCKSLVFDVDFHPKLDLICTGLLDGNLLLYKLKEEKKKFQ--KKWNIDNHEKSVRFVSF 65
Query: 63 INGGRAVLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLINLTESTVASGDDDGCIK 121
N G+ +L D D+ TG I + H+ ++ ++ +T + D+ G IK
Sbjct: 66 SNNGKNILAAFSDSKCSVFDI-TGD-ILWTNKCHKHPISSILYTGLNTFLTADESGIIK 122
Score = 33.9 bits (76), Expect = 6.3
Identities = 17/59 (28%), Positives = 33/59 (55%), Gaps = 1/59 (1%)
Query: 212 IITGSENGMINLVGILPNRIIEPIAEHSEYP-VERLAFSHDRKFLGSIGHDQMLKLWDL 269
II G+ +G+I PN++ + IA ++ +E+LA ++ + L SI HD + + +
Sbjct: 242 IIFGTSDGLIQTAHFNPNKLGDVIARNNTGDSIEKLAINNKKNLLASISHDYSIDFYQI 300
>UniRef100_UPI00003AA9E3 UPI00003AA9E3 UniRef100 entry
Length = 129
Score = 54.7 bits (130), Expect = 3e-06
Identities = 35/121 (28%), Positives = 53/121 (42%), Gaps = 25/121 (20%)
Query: 212 IITGSENGMINLVGILPNRIIEPIAEHSEYPVERLAFSHDRKFLGSIGHDQMLKLWDLDN 271
I+ +G+I V ILPNR++ + +H P+E+LA + + L S HDQ +K WD+ +
Sbjct: 13 IVQDPLDGVIRAVNILPNRVLGCVGQHLGEPIEQLAVAPGGQLLASCAHDQKVKFWDISS 72
Query: 272 ILQGSRSTQRNETGVVANDGDSDDGDEMDVDNSASKFSKGNKRKNASNGHAVGDSNNFFA 331
+ M VD K +G K A + A G +FFA
Sbjct: 73 L------------------------GSMVVDEYRKKKKRGGPLK-ALSSKAAGSGEDFFA 107
Query: 332 D 332
D
Sbjct: 108 D 108
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.141 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 545,231,281
Number of Sequences: 2790947
Number of extensions: 22154771
Number of successful extensions: 88765
Number of sequences better than 10.0: 569
Number of HSP's better than 10.0 without gapping: 93
Number of HSP's successfully gapped in prelim test: 478
Number of HSP's that attempted gapping in prelim test: 87074
Number of HSP's gapped (non-prelim): 2265
length of query: 332
length of database: 848,049,833
effective HSP length: 128
effective length of query: 204
effective length of database: 490,808,617
effective search space: 100124957868
effective search space used: 100124957868
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 75 (33.5 bits)
Medicago: description of AC148996.16


