

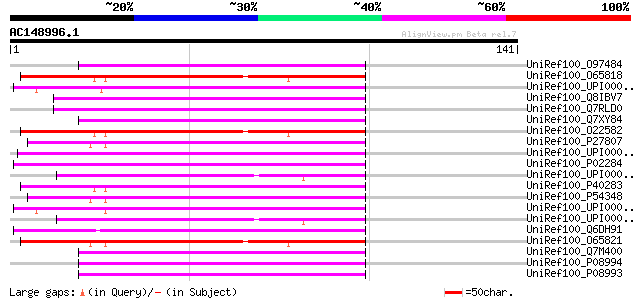
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148996.1 - phase: 0
(141 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O97484 Histone H2B [Euplotes crassus] 57 7e-08
UniRef100_O65818 Histone H2B-2 [Lycopersicon esculentum] 57 9e-08
UniRef100_UPI000029BCE9 UPI000029BCE9 UniRef100 entry 57 1e-07
UniRef100_Q8IBV7 Histone h2b, putative [Plasmodium falciparum] 57 1e-07
UniRef100_Q7RLD0 Histone H2B variant 1 [Plasmodium yoelii yoelii] 57 1e-07
UniRef100_Q7XY84 Histone H2B protein [Griffithsia japonica] 56 1e-07
UniRef100_O22582 Histone H2B [Gossypium hirsutum] 56 1e-07
UniRef100_P27807 Histone H2B [Triticum aestivum] 56 2e-07
UniRef100_UPI000032C773 UPI000032C773 UniRef100 entry 55 3e-07
UniRef100_P02284 Histone H2B, gonadal [Patella granatina] 55 3e-07
UniRef100_UPI000042D23F UPI000042D23F UniRef100 entry 55 3e-07
UniRef100_P40283 Histone H2B [Arabidopsis thaliana] 55 3e-07
UniRef100_P54348 Histone H2B [Zea mays] 55 3e-07
UniRef100_UPI0000318427 UPI0000318427 UniRef100 entry 55 4e-07
UniRef100_UPI000004F97E UPI000004F97E UniRef100 entry 55 4e-07
UniRef100_Q6DH91 Zgc:92591 [Brachydanio rerio] 55 4e-07
UniRef100_O65821 Histone H2B [Lycopersicon esculentum] 55 4e-07
UniRef100_Q7M400 Histone H2B [Tetrahymena pyriformis] 55 4e-07
UniRef100_P08994 Histone H2B.2 [Tetrahymena thermophila] 55 4e-07
UniRef100_P08993 Histone H2B.1 [Tetrahymena thermophila] 55 4e-07
>UniRef100_O97484 Histone H2B [Euplotes crassus]
Length = 113
Score = 57.4 bits (137), Expect = 7e-08
Identities = 30/80 (37%), Positives = 47/80 (58%)
Query: 20 KRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKIVEEVAKNVAALNQKLLV 79
K+RT F +IYRV K+VHP+ ++ +S+ +MN +ND+ KI E +K V + L
Sbjct: 20 KKRTETFSVYIYRVLKQVHPETGVSKKSMSIMNSFINDIFEKIALEASKLVRYNKKHTLS 79
Query: 80 ERDLWVGVKELYPKYLARCA 99
R++ V+ L P LA+ A
Sbjct: 80 SREVQTAVRLLLPGELAKHA 99
>UniRef100_O65818 Histone H2B-2 [Lycopersicon esculentum]
Length = 139
Score = 57.0 bits (136), Expect = 9e-08
Identities = 34/103 (33%), Positives = 63/103 (61%), Gaps = 8/103 (7%)
Query: 4 KKGKNIILEVGSSSGTKRR-----TVE-FKKHIYRVAKKVHPDLDITVESIEMMNDIMND 57
K GK + E GSS K++ ++E +K +I++V K+VHPD+ I+ +S+ +MN +ND
Sbjct: 24 KAGKKLPKESGSSGADKKKKKSKKSIETYKIYIFKVLKQVHPDIGISSKSMGIMNSFIND 83
Query: 58 MLHKIVEEVAKNVAALNQK-LLVERDLWVGVKELYPKYLARCA 99
+ K+ +E + +A +N+K + R++ V+ + P LA+ A
Sbjct: 84 IFEKLAQE-SSRLARINKKPTITSREIQTAVRLVLPGELAKHA 125
>UniRef100_UPI000029BCE9 UPI000029BCE9 UniRef100 entry
Length = 124
Score = 56.6 bits (135), Expect = 1e-07
Identities = 31/102 (30%), Positives = 55/102 (53%), Gaps = 4/102 (3%)
Query: 2 ASKKG-KNIILEVGSSSGTKRRTV---EFKKHIYRVAKKVHPDLDITVESIEMMNDIMND 57
A KKG K + + S G K+R E+ ++Y+V K+VHPD I+ +++ +MN +ND
Sbjct: 8 APKKGSKKAVTKTASKGGKKKRRTRKGEYAIYVYKVLKQVHPDTGISSKAMSIMNSFVND 67
Query: 58 MLHKIVEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
+ +I E ++ + + R++ V+ L P LA+ A
Sbjct: 68 IFERIASEASRLAHYNKRSTITSREIQTAVRLLLPGELAKHA 109
>UniRef100_Q8IBV7 Histone h2b, putative [Plasmodium falciparum]
Length = 123
Score = 56.6 bits (135), Expect = 1e-07
Identities = 27/87 (31%), Positives = 50/87 (57%)
Query: 13 VGSSSGTKRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKIVEEVAKNVAA 72
+G KRRT F +I++V K+VHP+ +T +S+ +MN +ND+ ++V E + +
Sbjct: 21 LGPRHKRKRRTESFSLYIFKVLKQVHPETGVTKKSMNIMNSFINDIFDRLVTEATRLIRY 80
Query: 73 LNQKLLVERDLWVGVKELYPKYLARCA 99
++ L R++ V+ L P L++ A
Sbjct: 81 NKKRTLSSREIQTAVRLLLPGELSKHA 107
>UniRef100_Q7RLD0 Histone H2B variant 1 [Plasmodium yoelii yoelii]
Length = 123
Score = 56.6 bits (135), Expect = 1e-07
Identities = 27/87 (31%), Positives = 50/87 (57%)
Query: 13 VGSSSGTKRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKIVEEVAKNVAA 72
+G KRRT F +I++V K+VHP+ +T +S+ +MN +ND+ ++V E + +
Sbjct: 21 LGPRHKRKRRTESFSLYIFKVLKQVHPETGVTKKSMNIMNSFINDIFDRLVTEATRLIRY 80
Query: 73 LNQKLLVERDLWVGVKELYPKYLARCA 99
++ L R++ V+ L P L++ A
Sbjct: 81 NKKRTLSSREIQTAVRLLLPGELSKHA 107
>UniRef100_Q7XY84 Histone H2B protein [Griffithsia japonica]
Length = 124
Score = 56.2 bits (134), Expect = 1e-07
Identities = 27/80 (33%), Positives = 47/80 (58%)
Query: 20 KRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKIVEEVAKNVAALNQKLLV 79
K+R+ + +IY+V K+VHP+ I+ +++ +MN ++D+ +I E AK N K L
Sbjct: 31 KKRSESYSIYIYKVLKQVHPETGISSKAMSIMNSFVDDLFERIATEAAKLATYNNSKTLT 90
Query: 80 ERDLWVGVKELYPKYLARCA 99
R++ V+ L P LA+ A
Sbjct: 91 SREIQTAVRLLLPGELAKHA 110
>UniRef100_O22582 Histone H2B [Gossypium hirsutum]
Length = 147
Score = 56.2 bits (134), Expect = 1e-07
Identities = 34/103 (33%), Positives = 64/103 (62%), Gaps = 8/103 (7%)
Query: 4 KKGKNIILEVGSSSGTKRR-----TVE-FKKHIYRVAKKVHPDLDITVESIEMMNDIMND 57
K GK + E G+++G K++ +VE +K +I++V K+VHPD+ I+ +++ +MN +ND
Sbjct: 32 KAGKKLPKEGGAAAGDKKKKRVKKSVETYKIYIFKVLKQVHPDIGISSKAMGIMNSFIND 91
Query: 58 MLHKIVEEVAKNVAALNQK-LLVERDLWVGVKELYPKYLARCA 99
+ K+ +E A +A N+K + R++ V+ + P LA+ A
Sbjct: 92 IFEKLAQE-ASRLARYNKKPTITSREIQTAVRLVLPGELAKHA 133
>UniRef100_P27807 Histone H2B [Triticum aestivum]
Length = 152
Score = 55.8 bits (133), Expect = 2e-07
Identities = 31/97 (31%), Positives = 56/97 (56%), Gaps = 3/97 (3%)
Query: 6 GKNIILEVGSSSGTKR--RTVE-FKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKI 62
GK+ E G G K+ ++VE +K +I++V K+VHPD+ I+ +++ +MN +ND+ K+
Sbjct: 42 GKSAAKEGGDKKGKKKAKKSVETYKIYIFKVLKQVHPDIGISSKAMSIMNSFINDIFEKL 101
Query: 63 VEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
E AK + + R++ V+ + P LA+ A
Sbjct: 102 AGEAAKLARYNKKPTITSREIQTSVRLVLPGELAKHA 138
>UniRef100_UPI000032C773 UPI000032C773 UniRef100 entry
Length = 116
Score = 55.5 bits (132), Expect = 3e-07
Identities = 29/97 (29%), Positives = 51/97 (51%)
Query: 3 SKKGKNIILEVGSSSGTKRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKI 62
SKK ++ K +T +K +IY+V K+VHPD I+ +++ +MN +ND+ KI
Sbjct: 6 SKKPAKKTVKGAGGKAKKSKTETYKIYIYKVLKQVHPDTGISSKAMSIMNSFINDIFEKI 65
Query: 63 VEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
E +K + + R++ V+ + P LA+ A
Sbjct: 66 ATEASKLARYNKKPTVTSREIQTAVRLILPGELAKHA 102
>UniRef100_P02284 Histone H2B, gonadal [Patella granatina]
Length = 121
Score = 55.5 bits (132), Expect = 3e-07
Identities = 28/98 (28%), Positives = 50/98 (50%)
Query: 2 ASKKGKNIILEVGSSSGTKRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHK 61
A K GK G +RR + +IY+V K+VHPD ++ +++ +MN +ND+ +
Sbjct: 9 AKKAGKAKAARSGDKKRKRRRKESYSIYIYKVLKQVHPDTGVSSKAMSIMNSFVNDIFER 68
Query: 62 IVEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
I E ++ + + R++ V+ L P LA+ A
Sbjct: 69 IAAEASRLAHYNKRSTITSREIQTAVRLLLPGELAKHA 106
>UniRef100_UPI000042D23F UPI000042D23F UniRef100 entry
Length = 130
Score = 55.1 bits (131), Expect = 3e-07
Identities = 29/87 (33%), Positives = 51/87 (58%), Gaps = 2/87 (2%)
Query: 14 GSSSGTKRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKIVEEVAKNVAAL 73
G+ TK R + +IY+V K+ HPD I+ +++ +MN +ND+ +I E +K +AA
Sbjct: 29 GAKKRTKARKETYSSYIYKVLKQTHPDTGISQKAMSIMNSFVNDIFERIASEASK-LAAY 87
Query: 74 NQKLLVE-RDLWVGVKELYPKYLARCA 99
N+K + R++ V+ + P LA+ A
Sbjct: 88 NKKSTISAREIQTAVRLILPGELAKHA 114
>UniRef100_P40283 Histone H2B [Arabidopsis thaliana]
Length = 150
Score = 55.1 bits (131), Expect = 3e-07
Identities = 30/101 (29%), Positives = 58/101 (56%), Gaps = 5/101 (4%)
Query: 4 KKGKNIILEVGSSSGTKRR----TVE-FKKHIYRVAKKVHPDLDITVESIEMMNDIMNDM 58
K GK + E G+ K++ +VE +K +I++V K+VHPD+ I+ +++ +MN +ND+
Sbjct: 36 KAGKKLPKEAGAGGDKKKKMKKKSVETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDI 95
Query: 59 LHKIVEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
K+ +E +K + + R++ V+ + P LA+ A
Sbjct: 96 FEKLAQEASKLARYNKKPTITSREIQTAVRLVLPGELAKHA 136
>UniRef100_P54348 Histone H2B [Zea mays]
Length = 154
Score = 55.1 bits (131), Expect = 3e-07
Identities = 31/97 (31%), Positives = 56/97 (56%), Gaps = 3/97 (3%)
Query: 6 GKNIILEVGSSSGTKR--RTVE-FKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKI 62
GK+ E G G K+ ++VE +K +I++V K+VHPD+ I+ +++ +MN +ND+ K+
Sbjct: 44 GKSAGKEGGDKKGRKKAKKSVETYKIYIFKVLKQVHPDIGISSKAMSIMNSFINDIFEKL 103
Query: 63 VEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
E AK + + R++ V+ + P LA+ A
Sbjct: 104 AAEAAKLARYNKKPTITSREIQTSVRLVLPGELAKHA 140
>UniRef100_UPI0000318427 UPI0000318427 UniRef100 entry
Length = 124
Score = 54.7 bits (130), Expect = 4e-07
Identities = 30/102 (29%), Positives = 54/102 (52%), Gaps = 4/102 (3%)
Query: 2 ASKKG-KNIILEVGSSSGTKRRTVE---FKKHIYRVAKKVHPDLDITVESIEMMNDIMND 57
A KKG K + + S G K+R + ++Y+V K+VHPD I+ +++ +MN +ND
Sbjct: 8 APKKGSKKAVTKTASKGGKKKRRTRKESYAIYVYKVLKQVHPDTGISSKAMSIMNSFVND 67
Query: 58 MLHKIVEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
+ +I E ++ + + R++ V+ L P LA+ A
Sbjct: 68 IFERIASEASRLAHYNKRSTITSREIQTAVRLLLPGELAKHA 109
>UniRef100_UPI000004F97E UPI000004F97E UniRef100 entry
Length = 130
Score = 54.7 bits (130), Expect = 4e-07
Identities = 29/87 (33%), Positives = 51/87 (58%), Gaps = 2/87 (2%)
Query: 14 GSSSGTKRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKIVEEVAKNVAAL 73
G+ TK R + +IY+V K+ HPD I+ +++ +MN +ND+ +I E +K +AA
Sbjct: 29 GAKKRTKARKETYSSYIYKVLKQTHPDTGISQKAMSIMNSFVNDIFERIATEASK-LAAY 87
Query: 74 NQKLLVE-RDLWVGVKELYPKYLARCA 99
N+K + R++ V+ + P LA+ A
Sbjct: 88 NKKSTISAREIQTAVRLILPGELAKHA 114
>UniRef100_Q6DH91 Zgc:92591 [Brachydanio rerio]
Length = 117
Score = 54.7 bits (130), Expect = 4e-07
Identities = 32/98 (32%), Positives = 53/98 (53%), Gaps = 1/98 (1%)
Query: 2 ASKKGKNIILEVGSSSGTKRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHK 61
A KKGK + GS +KRR + +IY+V K+VHPD I+ ++ +MN +ND+ +
Sbjct: 6 AKKKGKAPGDKKGSKRKSKRRET-YAVYIYKVLKQVHPDTGISSRAMSIMNSFVNDVFER 64
Query: 62 IVEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
I E ++ + + R++ V+ L P LA+ A
Sbjct: 65 IATEASRLAHYNKRSTITSREVQTAVRLLLPGELAKHA 102
>UniRef100_O65821 Histone H2B [Lycopersicon esculentum]
Length = 142
Score = 54.7 bits (130), Expect = 4e-07
Identities = 32/100 (32%), Positives = 62/100 (62%), Gaps = 5/100 (5%)
Query: 4 KKGKNIILEVGSSSGTKR--RTVE-FKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLH 60
K GK + + G+ K+ ++VE +K +I++V K+VHPD+ I+ +S+ +MN +ND+
Sbjct: 30 KAGKKLPKDAGADKKKKKSKKSVETYKIYIFKVLKQVHPDIGISSKSMGIMNSFINDIFE 89
Query: 61 KIVEEVAKNVAALNQK-LLVERDLWVGVKELYPKYLARCA 99
K+ +E + +A +N+K + R++ V+ + P LA+ A
Sbjct: 90 KLAQE-SSRLARINKKPTITSREIQTAVRLVLPGELAKHA 128
>UniRef100_Q7M400 Histone H2B [Tetrahymena pyriformis]
Length = 120
Score = 54.7 bits (130), Expect = 4e-07
Identities = 29/80 (36%), Positives = 48/80 (59%)
Query: 20 KRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKIVEEVAKNVAALNQKLLV 79
K+R+ F +I++V K+VHPD+ I+ +++ +MN +ND +I E +K V ++ L
Sbjct: 25 KKRSETFAIYIFKVLKQVHPDVGISKKAMNIMNSFINDSFERIALESSKLVRFNKRRTLS 84
Query: 80 ERDLWVGVKELYPKYLARCA 99
R++ VK L P LAR A
Sbjct: 85 SREVQTAVKLLLPGELARHA 104
>UniRef100_P08994 Histone H2B.2 [Tetrahymena thermophila]
Length = 121
Score = 54.7 bits (130), Expect = 4e-07
Identities = 29/80 (36%), Positives = 48/80 (59%)
Query: 20 KRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKIVEEVAKNVAALNQKLLV 79
K+R+ F +I++V K+VHPD+ I+ +++ +MN +ND +I E +K V ++ L
Sbjct: 26 KKRSETFAIYIFKVLKQVHPDVGISKKAMNIMNSFINDSFERIALESSKLVRFNKRRTLS 85
Query: 80 ERDLWVGVKELYPKYLARCA 99
R++ VK L P LAR A
Sbjct: 86 SREVQTAVKLLLPGELARHA 105
>UniRef100_P08993 Histone H2B.1 [Tetrahymena thermophila]
Length = 121
Score = 54.7 bits (130), Expect = 4e-07
Identities = 29/80 (36%), Positives = 48/80 (59%)
Query: 20 KRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKIVEEVAKNVAALNQKLLV 79
K+R+ F +I++V K+VHPD+ I+ +++ +MN +ND +I E +K V ++ L
Sbjct: 26 KKRSETFAIYIFKVLKQVHPDVGISKKAMNIMNSFINDSFERIALESSKLVRFNKRRTLS 85
Query: 80 ERDLWVGVKELYPKYLARCA 99
R++ VK L P LAR A
Sbjct: 86 SREVQTAVKLLLPGELARHA 105
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.134 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 213,570,597
Number of Sequences: 2790947
Number of extensions: 7711855
Number of successful extensions: 27457
Number of sequences better than 10.0: 392
Number of HSP's better than 10.0 without gapping: 305
Number of HSP's successfully gapped in prelim test: 87
Number of HSP's that attempted gapping in prelim test: 27142
Number of HSP's gapped (non-prelim): 393
length of query: 141
length of database: 848,049,833
effective HSP length: 117
effective length of query: 24
effective length of database: 521,509,034
effective search space: 12516216816
effective search space used: 12516216816
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC148996.1


