

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148995.13 - phase: 0
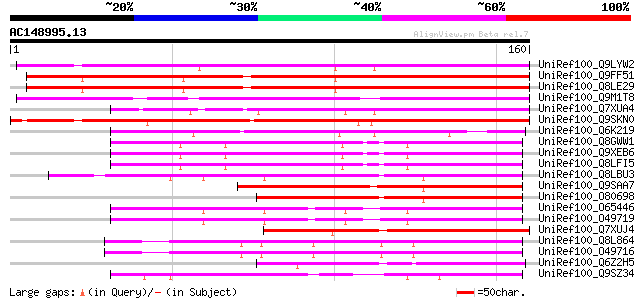
(160 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LYW2 Hypothetical protein F15A17_260 [Arabidopsis th... 132 4e-30
UniRef100_Q9FF51 Emb|CAB72159.1 [Arabidopsis thaliana] 127 1e-28
UniRef100_Q8LE29 Hypothetical protein [Arabidopsis thaliana] 119 2e-26
UniRef100_Q9M1T8 Hypothetical protein T14D3.150 [Arabidopsis tha... 119 3e-26
UniRef100_Q7XUA4 OSJNBa0019D11.12 protein [Oryza sativa] 108 6e-23
UniRef100_Q9SKN0 Hypothetical protein At2g28400 [Arabidopsis tha... 103 1e-21
UniRef100_Q6K219 Hypothetical protein B1469H02.18 [Oryza sativa] 95 7e-19
UniRef100_Q8GWW1 Hypothetical protein At4g04630/F4H6_15 [Arabido... 89 3e-17
UniRef100_Q9XEB6 Hypothetical protein F4H6.15 [Arabidopsis thali... 89 3e-17
UniRef100_Q8LFI5 Hypothetical protein [Arabidopsis thaliana] 89 3e-17
UniRef100_Q8LBU3 Hypothetical protein [Arabidopsis thaliana] 85 7e-16
UniRef100_Q9SAA7 F25C20.15 protein [Arabidopsis thaliana] 84 2e-15
UniRef100_O80698 F8K4.12 [Arabidopsis thaliana] 84 2e-15
UniRef100_O65446 Hypothetical protein AT4g21970 [Arabidopsis tha... 80 2e-14
UniRef100_O49719 Hypothetical protein T8O5.180 [Arabidopsis thal... 80 2e-14
UniRef100_Q7XUJ4 OSJNBb0103I08.8 protein [Oryza sativa] 76 2e-13
UniRef100_Q8L864 Hypothetical protein At4g21930 [Arabidopsis tha... 75 7e-13
UniRef100_O49716 Hypothetical protein T8O5.140 [Arabidopsis thal... 75 7e-13
UniRef100_Q6Z2H5 Hypothetical protein P0650C03.15 [Oryza sativa] 73 3e-12
UniRef100_Q9SZ34 Hypothetical protein F10M23.290 [Arabidopsis th... 72 5e-12
>UniRef100_Q9LYW2 Hypothetical protein F15A17_260 [Arabidopsis thaliana]
Length = 166
Score = 132 bits (331), Expect = 4e-30
Identities = 74/167 (44%), Positives = 99/167 (58%), Gaps = 11/167 (6%)
Query: 3 SSRKSFLSRTSYIFPETNFNQKSSQGKELEFDEADVWNMSYSNSNTNIEPKKGVP--GLK 60
+SRK F + YI+PE + S EFDE+D+ N+ + + K+ + L+
Sbjct: 2 ASRKLFFVKPKYIYPEPK--PEMSDENVFEFDESDIHNLGDHQLPNSFDAKRSISISRLR 59
Query: 61 RVSRKMEANNKVNPLASSSLPMNIPDWSKILKEEYKKKK---ESSDDEDEGDYD----GV 113
R K + + SLP+NIPDWSKILK EY+ + SDD+DE D D G
Sbjct: 60 RKPTKTGDSGNREITKTGSLPVNIPDWSKILKSEYRGHAIPDDDSDDDDEDDDDSNDGGR 119
Query: 114 VQLPPHEYLARTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGFED 160
+PPHEYLAR RG+S +VHEG G T KGRDLR +RNAIW+K+GF+D
Sbjct: 120 RMIPPHEYLARRRGSSFTVHEGIGGTAKGRDLRRLRNAIWEKIGFQD 166
>UniRef100_Q9FF51 Emb|CAB72159.1 [Arabidopsis thaliana]
Length = 163
Score = 127 bits (318), Expect = 1e-28
Identities = 75/161 (46%), Positives = 104/161 (64%), Gaps = 8/161 (4%)
Query: 6 KSFLSRTSYIFPETNF-NQKSSQGKELEFDEADVWNMSYSNSNTNIEP-KKGVPGLKRVS 63
KS+ +R SY F T+ + ++ LEFDE+D++N +S+S V K+ S
Sbjct: 5 KSYYARPSYRFLGTDQPSYFTASDSGLEFDESDLFNPIHSDSPDFCRKISSSVRSGKKSS 64
Query: 64 RKMEANNKVNPLASSSLPMNIPDWSKILKEEYKKKK----ESSDDEDEGDYDGVVQLPPH 119
+ A + A+SSLP+N+PDWSKIL+ EY+ + E +DD+D+ + DG LPPH
Sbjct: 65 NRPSAASSA--AAASSLPVNVPDWSKILRGEYRDNRRRSIEDNDDDDDDNEDGGDWLPPH 122
Query: 120 EYLARTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGFED 160
E+LA+TR AS SVHEG GRTLKGRDL VRNAI++K GF+D
Sbjct: 123 EFLAKTRMASFSVHEGVGRTLKGRDLSRVRNAIFEKFGFQD 163
>UniRef100_Q8LE29 Hypothetical protein [Arabidopsis thaliana]
Length = 163
Score = 119 bits (299), Expect = 2e-26
Identities = 72/161 (44%), Positives = 100/161 (61%), Gaps = 8/161 (4%)
Query: 6 KSFLSRTSYIFPETNF-NQKSSQGKELEFDEADVWNMSYSNSNTNIEP-KKGVPGLKRVS 63
KS+ +R SY F T+ + ++ LEFDE+D++N +S+S V K+ S
Sbjct: 5 KSYYARPSYRFLGTDQPSYFTASDSGLEFDESDLFNPIHSDSPDFCRKISSSVRSGKKSS 64
Query: 64 RKMEANNKVNPLASSSLPMNIPDWSKILKEEYKKKK----ESSDDEDEGDYDGVVQLPPH 119
+ A + A+SSLP+N+PDWSK L+ EY+ + E +DD+D+ + DG LPPH
Sbjct: 65 NRPSAASSA--AAASSLPVNVPDWSKXLRGEYRDNRRRSIEDNDDDDDDNEDGGDWLPPH 122
Query: 120 EYLARTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGFED 160
E+LA+TR AS SVHEG GRTLKGRDL VR A + K GF+D
Sbjct: 123 EFLAKTRMASFSVHEGVGRTLKGRDLSRVRXAXFXKFGFQD 163
>UniRef100_Q9M1T8 Hypothetical protein T14D3.150 [Arabidopsis thaliana]
Length = 148
Score = 119 bits (298), Expect = 3e-26
Identities = 72/158 (45%), Positives = 95/158 (59%), Gaps = 13/158 (8%)
Query: 3 SSRKSFLSRTSYIFPETNFNQKSSQGKELEFDEADVWNMSYSNSNTNIEPKKGVPGLKRV 62
++RKS+ R S+ F T+ + E EFDE+D+ YSN + + E ++ L
Sbjct: 4 ATRKSYYQRPSHRFLPTDRTYNVTGDSEFEFDESDL----YSNRSDSPEFRRK---LITS 56
Query: 63 SRKMEANNKVNPLASSSLPMNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQLPPHEYL 122
+RK +SSLPMN+ +WSKIL +E +K E+ DD EG +LPPHEYL
Sbjct: 57 NRKSSPATVTTTTVASSLPMNVQNWSKILGKENRKSIENDDDGGEG------KLPPHEYL 110
Query: 123 ARTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGFED 160
A+TR AS SVHEG GRTLKGRD+ VRNAI +K GF D
Sbjct: 111 AKTRMASFSVHEGIGRTLKGRDMSRVRNAILEKTGFLD 148
>UniRef100_Q7XUA4 OSJNBa0019D11.12 protein [Oryza sativa]
Length = 174
Score = 108 bits (269), Expect = 6e-23
Identities = 69/144 (47%), Positives = 84/144 (57%), Gaps = 19/144 (13%)
Query: 32 EFDEADVWNMSYSNSNTNIEPKK-GVPGLKRVSRKMEANNKVNPL--ASSSLPMNIPDWS 88
EFDE+DVW SY + P + G G R A K PL A+ SLP+NIPDW
Sbjct: 35 EFDESDVWG-SYGAAGVESSPAELGARG--RAIPSARAGRKA-PLDRAAGSLPVNIPDWQ 90
Query: 89 KILKEEYKKKKESS-------DDEDEGDYD-----GVVQLPPHEYLARTRGASLSVHEGK 136
KIL EY+ + ++ D +D+ +Y G V +PPHE R R ASLSVHEG
Sbjct: 91 KILGVEYRDHQAAAAEWELQGDGDDDYEYGKVAGVGGVVIPPHELAWRGRAASLSVHEGI 150
Query: 137 GRTLKGRDLRSVRNAIWKKVGFED 160
GRTLKGRDL VR+A+WKK GFED
Sbjct: 151 GRTLKGRDLSRVRDAVWKKTGFED 174
>UniRef100_Q9SKN0 Hypothetical protein At2g28400 [Arabidopsis thaliana]
Length = 162
Score = 103 bits (258), Expect = 1e-21
Identities = 69/166 (41%), Positives = 101/166 (60%), Gaps = 10/166 (6%)
Query: 1 MASSRKSFLSRTSYIFPETNFNQKSSQGKELEFDEADVWNM-SYSNSNTNIEPKKGVPGL 59
MA+S K + R S+ F T+ Q + + E DE D++N S S+S+ + G
Sbjct: 1 MATS-KCYYPRPSHRFFTTD--QHVTATSDFELDEWDLFNTGSDSSSSFSFSDLTITSGR 57
Query: 60 KRVSRKMEANNKVNPLASSSLPMNIPDWSKILKEEYKKKKESSDDED-EGDY----DGVV 114
+R++ + A+SSLP+N+PDWSKIL +E +++++ S++E+ +GD +G
Sbjct: 58 TGTNRQIHGGSDSGK-AASSLPVNVPDWSKILGDESRRQRKISNEEEVDGDEILCGEGTR 116
Query: 115 QLPPHEYLARTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGFED 160
++PPHE LA R AS SVHEG GRTLKGRDL VRN I+K G ED
Sbjct: 117 RVPPHELLANRRMASFSVHEGAGRTLKGRDLSRVRNTIFKIRGIED 162
>UniRef100_Q6K219 Hypothetical protein B1469H02.18 [Oryza sativa]
Length = 170
Score = 94.7 bits (234), Expect = 7e-19
Identities = 62/140 (44%), Positives = 75/140 (53%), Gaps = 19/140 (13%)
Query: 32 EFDEADVWNMSYSNSNTNIEPKKG---VPGLKRVSRKMEANNKVNPLASSSLPMNIPDWS 88
EFDE+D+W + P+ VP + RK A P A SLP+NIPDWS
Sbjct: 37 EFDESDIWGSFEPAAEVAESPRAARHQVPAARPPGRKAAAAAS-KPAAHGSLPVNIPDWS 95
Query: 89 KILKEEYKKKKE---SSDDEDEGDYD---GVVQLPPHEYLARTRGASLSVHE---GKGRT 139
KIL +EY+ +DD D+ D D V LPPHE R R ASLSVHE G GRT
Sbjct: 96 KILGDEYRGHHAGDWEADDVDDDDIDAASAVAVLPPHELAWRRRAASLSVHEDGMGIGRT 155
Query: 140 LKGRDLRSVRNAIWKKVGFE 159
LK VR+A+WKK GF+
Sbjct: 156 LK------VRDAVWKKTGFQ 169
>UniRef100_Q8GWW1 Hypothetical protein At4g04630/F4H6_15 [Arabidopsis thaliana]
Length = 168
Score = 89.4 bits (220), Expect = 3e-17
Identities = 57/143 (39%), Positives = 80/143 (55%), Gaps = 18/143 (12%)
Query: 32 EFDEADVWNMSYSNSNTNIE---PKKGVPGLKRVSRK---MEANNKVNPLASSSLPMNIP 85
EF E DVW++ ++ E PK S + + +V+ + SS PMN+P
Sbjct: 26 EFQEEDVWSVLREGETSSPEIKIPKSHFSSSSSSSSSPWNIRRSKEVSGVKQSSAPMNVP 85
Query: 86 DWSKILKEEYKKKKES------SDDEDEGDYDGVVQLPPHEY----LARTRGASLSVHEG 135
DWSK+ + ++ S +DD+DE D DG + +PPHE+ LART+ +S S+ EG
Sbjct: 86 DWSKVYGDSKSNRRSSHLHSHAADDDDEDD-DGCM-VPPHEWVARKLARTQISSFSMCEG 143
Query: 136 KGRTLKGRDLRSVRNAIWKKVGF 158
GRTLKGRDL VRNA+ K GF
Sbjct: 144 VGRTLKGRDLSKVRNAVLSKTGF 166
>UniRef100_Q9XEB6 Hypothetical protein F4H6.15 [Arabidopsis thaliana]
Length = 202
Score = 89.4 bits (220), Expect = 3e-17
Identities = 57/143 (39%), Positives = 80/143 (55%), Gaps = 18/143 (12%)
Query: 32 EFDEADVWNMSYSNSNTNIE---PKKGVPGLKRVSRK---MEANNKVNPLASSSLPMNIP 85
EF E DVW++ ++ E PK S + + +V+ + SS PMN+P
Sbjct: 26 EFQEEDVWSVLREGETSSPEMKIPKSHFSSSSSSSSSPWNIRRSKEVSGVKQSSAPMNVP 85
Query: 86 DWSKILKEEYKKKKES------SDDEDEGDYDGVVQLPPHEY----LARTRGASLSVHEG 135
DWSK+ + ++ S +DD+DE D DG + +PPHE+ LART+ +S S+ EG
Sbjct: 86 DWSKVYGDSKSNRRSSHLHSHAADDDDEDD-DGCM-VPPHEWVARKLARTQISSFSMCEG 143
Query: 136 KGRTLKGRDLRSVRNAIWKKVGF 158
GRTLKGRDL VRNA+ K GF
Sbjct: 144 VGRTLKGRDLSKVRNAVLSKTGF 166
>UniRef100_Q8LFI5 Hypothetical protein [Arabidopsis thaliana]
Length = 168
Score = 89.4 bits (220), Expect = 3e-17
Identities = 57/143 (39%), Positives = 80/143 (55%), Gaps = 18/143 (12%)
Query: 32 EFDEADVWNMSYSNSNTNIE---PKKGVPGLKRVSRK---MEANNKVNPLASSSLPMNIP 85
EF E DVW++ ++ E PK S + + +V+ + SS PMN+P
Sbjct: 26 EFQEEDVWSVLREGETSSPEMKIPKSHFSSSSSSSSSPWNIRRSKEVSGVKQSSAPMNVP 85
Query: 86 DWSKILKEEYKKKKES------SDDEDEGDYDGVVQLPPHEY----LARTRGASLSVHEG 135
DWSK+ + ++ S +DD+DE D DG + +PPHE+ LART+ +S S+ EG
Sbjct: 86 DWSKVYGDSKSNRRSSHLHSHAADDDDEDD-DGCM-VPPHEWVARKLARTQISSFSMCEG 143
Query: 136 KGRTLKGRDLRSVRNAIWKKVGF 158
GRTLKGRDL VRNA+ K GF
Sbjct: 144 VGRTLKGRDLSKVRNAVLSKTGF 166
>UniRef100_Q8LBU3 Hypothetical protein [Arabidopsis thaliana]
Length = 203
Score = 84.7 bits (208), Expect = 7e-16
Identities = 60/187 (32%), Positives = 91/187 (48%), Gaps = 45/187 (24%)
Query: 13 SYIFPETNFNQKSSQGKELEFDEADVWNMSYSNSNT----------NIEPKKGVPG-LKR 61
SY + +++ N + ELE E D+W+ + + +T N+ G G +
Sbjct: 19 SYTYGDSHGNSVTD---ELELGEEDIWSPAVIHDDTTENEESYGTWNLRATLGKNGRVGG 75
Query: 62 VSRKMEANNKVNPLAS----------------------SSLPMNIPDWSKILKEEYKKKK 99
+S E + PL+S SS P+N+PDWSKI + + +
Sbjct: 76 LSLAFEGSLVAPPLSSPMIVQKIHGGGGEGEEDRRKLASSAPVNVPDWSKIYRVDSVESI 135
Query: 100 ESSDDEDEGDYDGVVQLPPHEYLARTR--------GASLSVHEGKGRTLKGRDLRSVRNA 151
DDED+ D + + +PPHEYLA+++ G SV +G GRTLKGR+LR VR+A
Sbjct: 136 HELDDEDDEDEESGM-MPPHEYLAKSQARRSRKIGGGGASVFDGVGRTLKGRELRRVRDA 194
Query: 152 IWKKVGF 158
IW + GF
Sbjct: 195 IWSQTGF 201
>UniRef100_Q9SAA7 F25C20.15 protein [Arabidopsis thaliana]
Length = 201
Score = 83.6 bits (205), Expect = 2e-15
Identities = 44/95 (46%), Positives = 60/95 (62%), Gaps = 9/95 (9%)
Query: 71 KVNPLASSSLPMNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQLPPHEYLARTR---- 126
+V +SS P+N+PDWSKI + + SD+E+E D + +PPHEYLA+++
Sbjct: 107 RVERQLASSAPVNVPDWSKIYRVNSVESIHESDEEEEEDSG--MMMPPHEYLAKSQQRRS 164
Query: 127 ---GASLSVHEGKGRTLKGRDLRSVRNAIWKKVGF 158
G SV EG GRTLKGR+LR VR+AIW + GF
Sbjct: 165 RKSGGGGSVFEGVGRTLKGRELRRVRDAIWSQTGF 199
>UniRef100_O80698 F8K4.12 [Arabidopsis thaliana]
Length = 203
Score = 83.6 bits (205), Expect = 2e-15
Identities = 42/90 (46%), Positives = 59/90 (64%), Gaps = 9/90 (10%)
Query: 77 SSSLPMNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQLPPHEYLARTR--------GA 128
+SS P+N+PDWSKI + + + DDED+ D + + +PPHEYLA+++ G
Sbjct: 113 ASSAPVNVPDWSKIYRVDSVESIHELDDEDDEDEESGM-MPPHEYLAKSQARRSRKIGGG 171
Query: 129 SLSVHEGKGRTLKGRDLRSVRNAIWKKVGF 158
SV +G GRTLKGR+LR VR+AIW + GF
Sbjct: 172 GASVFDGVGRTLKGRELRRVRDAIWSQTGF 201
>UniRef100_O65446 Hypothetical protein AT4g21970 [Arabidopsis thaliana]
Length = 181
Score = 80.1 bits (196), Expect = 2e-14
Identities = 58/145 (40%), Positives = 74/145 (51%), Gaps = 25/145 (17%)
Query: 32 EFDEADVWNMSYSNSNTNIEPKKGVPG------LKRVSRKMEANNKVNPLAS-SSLPMNI 84
EF E +VW++ + E K +R + N+V+ A SS PMNI
Sbjct: 6 EFQEEEVWSVLRESETQGPEMKMSKSNNLFSASSSSSARYIPKGNEVSGGAKQSSAPMNI 65
Query: 85 PDWSKILKEEYKKKKESS-------DDEDEGDYDGVVQLPPHEY----LARTRGASLSVH 133
PDWSK+ Y KK SS DD+DEG +PPHE LART+ +S S+
Sbjct: 66 PDWSKVYG--YSKKNTSSHLHSWAIDDDDEGS-----MVPPHELVAKRLARTQISSFSMC 118
Query: 134 EGKGRTLKGRDLRSVRNAIWKKVGF 158
EG GRTLKGRDL RNA+ + GF
Sbjct: 119 EGIGRTLKGRDLSKTRNAVLTRTGF 143
>UniRef100_O49719 Hypothetical protein T8O5.180 [Arabidopsis thaliana]
Length = 241
Score = 80.1 bits (196), Expect = 2e-14
Identities = 58/145 (40%), Positives = 74/145 (51%), Gaps = 25/145 (17%)
Query: 32 EFDEADVWNMSYSNSNTNIEPKKGVPG------LKRVSRKMEANNKVNPLAS-SSLPMNI 84
EF E +VW++ + E K +R + N+V+ A SS PMNI
Sbjct: 6 EFQEEEVWSVLRESETQGPEMKMSKSNNLFSASSSSSARYIPKGNEVSGGAKQSSAPMNI 65
Query: 85 PDWSKILKEEYKKKKESS-------DDEDEGDYDGVVQLPPHEY----LARTRGASLSVH 133
PDWSK+ Y KK SS DD+DEG +PPHE LART+ +S S+
Sbjct: 66 PDWSKVYG--YSKKNTSSHLHSWAIDDDDEGS-----MVPPHELVAKRLARTQISSFSMC 118
Query: 134 EGKGRTLKGRDLRSVRNAIWKKVGF 158
EG GRTLKGRDL RNA+ + GF
Sbjct: 119 EGIGRTLKGRDLSKTRNAVLTRTGF 143
>UniRef100_Q7XUJ4 OSJNBb0103I08.8 protein [Oryza sativa]
Length = 209
Score = 76.3 bits (186), Expect = 2e-13
Identities = 40/85 (47%), Positives = 54/85 (63%), Gaps = 5/85 (5%)
Query: 79 SLPMNIPDWSKILKEEYKKK---KESSDDEDEGDYDGVVQLPPHEYLARTRGASLSVHEG 135
S P+ +P W K + +++ E+ +DE++ D + VV PPHE AR A+ SV EG
Sbjct: 126 SAPVAVPAWPKATDSDRRRRGVQHEALNDEEDDDDELVV--PPHEMAARRAAAAASVMEG 183
Query: 136 KGRTLKGRDLRSVRNAIWKKVGFED 160
GRTLKGRDLR VRNA+W+ GF D
Sbjct: 184 AGRTLKGRDLRRVRNAVWRTTGFLD 208
>UniRef100_Q8L864 Hypothetical protein At4g21930 [Arabidopsis thaliana]
Length = 183
Score = 74.7 bits (182), Expect = 7e-13
Identities = 54/156 (34%), Positives = 76/156 (48%), Gaps = 35/156 (22%)
Query: 30 ELEFDEADVWNMSYSNSNTNIEPKKGVPGLKRVSRKMEANN--------KVNPLA----- 76
ELE E DVW++ +EP + +R +EA+ +V+ L
Sbjct: 34 ELELMEEDVWSV--------VEPDEPKELGAWNARSLEASGSEWRRKGGRVSDLTVPSDG 85
Query: 77 ------SSSLPMNIPDWSKILK-EEYKKKKESSDDEDEGDYDGV----VQLPPHEYLA-- 123
++S P+ +PDWSKILK E K +++D D D +PPHEY+A
Sbjct: 86 QRKRHVATSAPVKVPDWSKILKVESVKSMHNNNNDNDNADVADCDWESAMVPPHEYVAAR 145
Query: 124 -RTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGF 158
R SV G GRTLKGRD+R VR+A+W + GF
Sbjct: 146 SRNGDGGSSVFLGVGRTLKGRDMRRVRDAVWSQTGF 181
>UniRef100_O49716 Hypothetical protein T8O5.140 [Arabidopsis thaliana]
Length = 193
Score = 74.7 bits (182), Expect = 7e-13
Identities = 54/156 (34%), Positives = 76/156 (48%), Gaps = 35/156 (22%)
Query: 30 ELEFDEADVWNMSYSNSNTNIEPKKGVPGLKRVSRKMEANN--------KVNPLA----- 76
ELE E DVW++ +EP + +R +EA+ +V+ L
Sbjct: 44 ELELMEEDVWSV--------VEPDEPKELGAWNARSLEASGSEWRRKGGRVSDLTVPSDG 95
Query: 77 ------SSSLPMNIPDWSKILK-EEYKKKKESSDDEDEGDYDGV----VQLPPHEYLA-- 123
++S P+ +PDWSKILK E K +++D D D +PPHEY+A
Sbjct: 96 QRKRHVATSAPVKVPDWSKILKVESVKSMHNNNNDNDNADVADCDWESAMVPPHEYVAAR 155
Query: 124 -RTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGF 158
R SV G GRTLKGRD+R VR+A+W + GF
Sbjct: 156 SRNGDGGSSVFLGVGRTLKGRDMRRVRDAVWSQTGF 191
>UniRef100_Q6Z2H5 Hypothetical protein P0650C03.15 [Oryza sativa]
Length = 196
Score = 72.8 bits (177), Expect = 3e-12
Identities = 41/90 (45%), Positives = 54/90 (59%), Gaps = 10/90 (11%)
Query: 77 SSSLPMNIPDW-------SKILKEEYKKKKESSDDEDEGDYDGVVQLPPHEYLARTRGAS 129
++S P+ +P W + E +K++E DD +GD DG V PPH YLAR R A
Sbjct: 109 AASAPVQVPAWPGRYADPDQAAFAEEEKRREEEDDAGDGDGDGWV--PPHVYLAR-RQAR 165
Query: 130 LSVHEGKGRTLKGRDLRSVRNAIWKKVGFE 159
SV EG GRTLKGRD VR+A+W + GF+
Sbjct: 166 SSVVEGVGRTLKGRDASRVRDAVWSRTGFD 195
>UniRef100_Q9SZ34 Hypothetical protein F10M23.290 [Arabidopsis thaliana]
Length = 144
Score = 72.0 bits (175), Expect = 5e-12
Identities = 52/140 (37%), Positives = 73/140 (52%), Gaps = 25/140 (17%)
Query: 32 EFDEADVWN---------MSYSNSNT--NIEPKKGVPGLKRVSRKMEANNKVNPLASSSL 80
+F E DVW+ M SN T + + +P R+ + + + P+ S+
Sbjct: 15 DFQEDDVWDVLDGYQSPFMISSNHTTKPSFSTQTLLPSEPRMIPERQRTEGMAPMRQQSV 74
Query: 81 PMNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQLPPHEY-LARTRGASLS-VHEGKGR 138
P+N+PDWS + + KK K+ DDE+ + P EY L R+R +S S V EG GR
Sbjct: 75 PVNVPDWSMVQR---KKTKKVVDDEN---------VSPEEYFLRRSRSSSSSSVMEGVGR 122
Query: 139 TLKGRDLRSVRNAIWKKVGF 158
LKGRDL VRNAI K+ GF
Sbjct: 123 KLKGRDLSKVRNAILKQTGF 142
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.308 0.128 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 286,157,578
Number of Sequences: 2790947
Number of extensions: 12245877
Number of successful extensions: 42545
Number of sequences better than 10.0: 183
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 137
Number of HSP's that attempted gapping in prelim test: 42383
Number of HSP's gapped (non-prelim): 231
length of query: 160
length of database: 848,049,833
effective HSP length: 117
effective length of query: 43
effective length of database: 521,509,034
effective search space: 22424888462
effective search space used: 22424888462
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 69 (31.2 bits)
Medicago: description of AC148995.13


