

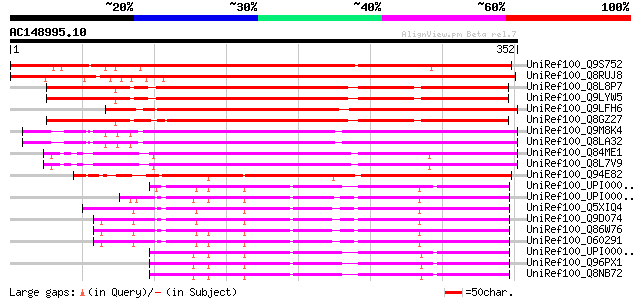
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148995.10 - phase: 0
(352 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9S752 Hypothetical protein F11F8.36 [Arabidopsis thal... 417 e-115
UniRef100_Q8RUJ8 Putative hydroxyproline-rich glycoprotein [Oryz... 417 e-115
UniRef100_Q8L8P7 Putative RING zinc finger protein [Arabidopsis ... 327 3e-88
UniRef100_Q9LYW5 Hypothetical protein F15A17_230 [Arabidopsis th... 325 9e-88
UniRef100_Q9LFH6 Hypothetical protein F4P12_110 [Arabidopsis tha... 322 1e-86
UniRef100_Q8GZ27 Hypothetical protein At5g03200/F15A17_230 [Arab... 321 2e-86
UniRef100_Q9M8K4 Putative RING zinc finger protein [Arabidopsis ... 280 6e-74
UniRef100_Q8LA32 Putative RING zinc finger protein [Arabidopsis ... 280 6e-74
UniRef100_Q84ME1 At5g19080/T16G12_120 [Arabidopsis thaliana] 270 6e-71
UniRef100_Q8L7V9 AT5g19080/T16G12_120 [Arabidopsis thaliana] 266 8e-70
UniRef100_Q94E82 Mahogunin, ring finger 1-like protein [Oryza sa... 254 3e-66
UniRef100_UPI0000248DFA UPI0000248DFA UniRef100 entry 151 2e-35
UniRef100_UPI00003AAE96 UPI00003AAE96 UniRef100 entry 149 1e-34
UniRef100_Q5XIQ4 Hypothetical protein [Rattus norvegicus] 142 1e-32
UniRef100_Q9D074 Mus musculus 10 days embryo whole body cDNA, RI... 141 2e-32
UniRef100_Q86W76 Mahogunin, ring finger 1 [Homo sapiens] 141 2e-32
UniRef100_O60291 KIAA0544 protein [Homo sapiens] 141 2e-32
UniRef100_UPI000036AEB7 UPI000036AEB7 UniRef100 entry 139 9e-32
UniRef100_Q96PX1 KIAA1917 protein [Homo sapiens] 139 9e-32
UniRef100_Q8NB72 Hypothetical protein FLJ34148 [Homo sapiens] 139 9e-32
>UniRef100_Q9S752 Hypothetical protein F11F8.36 [Arabidopsis thaliana]
Length = 388
Score = 417 bits (1072), Expect = e-115
Identities = 218/371 (58%), Positives = 263/371 (70%), Gaps = 25/371 (6%)
Query: 1 MGNISSTGAHNRRRHATASRRTHPPPPPP----VTPQP---EIAPHQFVYPGAAPYPNPP 53
MGNISS+G RRR PPPPPP + P P EI + V+ PYPNP
Sbjct: 1 MGNISSSGGEGRRRRRRNHTAAPPPPPPPPSSSLPPPPLPTEIQANPIVFAAVTPYPNPN 60
Query: 54 MHYPQYHYPGYY---PPPIPMP----HHHHPHPHPHPHMDPAW----VSRYYPCGPVVNQ 102
+ P Y YP Y PPP MP HH H PHP+ + +W ++RY G ++ Q
Sbjct: 61 PN-PVYQYPASYYHHPPPGAMPLPPYDHHLQHHPPHPYHNHSWAPVAMARYPYAGHMMAQ 119
Query: 103 PAPFVEHQKAVTIRNDVNIKKETIVISPDEENPGFFLVSFTFDAAVSGSITIFFFAKEDE 162
P P+VEHQKAVTIRNDVN+KKE++ + PD +NPG FLVSFTFDA VSG I++ FFAKE E
Sbjct: 120 PTPYVEHQKAVTIRNDVNLKKESLRLEPDPDNPGRFLVSFTFDATVSGRISVIFFAKESE 179
Query: 163 GCILTPTKETHLAPVTVQFQQGLGQKFRQQAGTGINFSMFEESDLVK-VGDVDVYPLAVK 221
C LT TKE L P+T+ F++GLGQKF+Q +G+GI+FS+FE+ +L K D ++YPLAVK
Sbjct: 180 DCKLTATKEDILPPITLDFEKGLGQKFKQSSGSGIDFSVFEDVELFKAAADTEIYPLAVK 239
Query: 222 ADASSDNHDGSNETETSSKPNSQITQAVFEKEKGEFRVKVVKQILSVNGMRYELQEIYGI 281
A+A+ + E + SK N+QITQAV+EK+KGE +++VVKQIL VNG RYELQEIYGI
Sbjct: 240 AEAAPSGGENEEEERSGSK-NAQITQAVYEKDKGEIKIRVVKQILWVNGTRYELQEIYGI 298
Query: 282 GNSVESDVDD----NEQGKECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQTNRCPICR 337
GN+VE D D N+ GKECVICLSEPRDT V PCRHMCMCSGCAKVLRFQTNRCPICR
Sbjct: 299 GNTVEGDDDSADDANDPGKECVICLSEPRDTTVLPCRHMCMCSGCAKVLRFQTNRCPICR 358
Query: 338 QPVERLLEIKV 348
QPVERLLEIKV
Sbjct: 359 QPVERLLEIKV 369
>UniRef100_Q8RUJ8 Putative hydroxyproline-rich glycoprotein [Oryza sativa]
Length = 425
Score = 417 bits (1071), Expect = e-115
Identities = 221/398 (55%), Positives = 270/398 (67%), Gaps = 49/398 (12%)
Query: 1 MGNISSTGAHNRRRHATASRRTH----PPPPPPVTPQPEIAPHQFVYPGAAPYP------ 50
MGN+ S+G H RR + H PPPPP Q E+AP+++V+ A+PYP
Sbjct: 1 MGNMGSSGGHRRRNNGHGRHHHHGQPTAPPPPPQQQQQEVAPNRYVFAAASPYPPQYPNP 60
Query: 51 NPPMHYPQYHYPGYYPPP-------IPMPHHHH-----------PHPHPHP-----HMDP 87
N P +YPQY +YPPP +P P+ HH P P P P P
Sbjct: 61 NLPQYYPQYG--NFYPPPPPSMPGPLPAPYDHHHRGGGPAQPPPPPPPPQPIHAAGEFPP 118
Query: 88 AWVSRY---------YPCGPVVNQPAP---FVEHQKAVTIRNDVNIKKETIVISPDEENP 135
A + ++ + GP PAP +VEHQKAVTIRNDVN+KKET+ + PD+E P
Sbjct: 119 AMLQQHPHYHGWGGNFSYGPPTQPPAPAPPYVEHQKAVTIRNDVNLKKETLRVEPDDECP 178
Query: 136 GFFLVSFTFDAAVSGSITIFFFAKEDEGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGT 195
G FL++FTFDA V+GS+T++FFAKE+ C LT TKE L PVTV F++GLGQKFRQ +GT
Sbjct: 179 GRFLITFTFDATVAGSMTVYFFAKEELNCNLTATKEDLLKPVTVTFKEGLGQKFRQPSGT 238
Query: 196 GINFSMFEESDLVKVGDVDVYPLAVKADAS-SDNHDGSNETETSSKPNSQITQAVFE-KE 253
GI+FS+FE+++L K G++DVYPLAVKA+ + S E + S PNSQITQAVFE KE
Sbjct: 239 GIDFSLFEDAELFKEGEMDVYPLAVKAETTFSIGQFSEGEEQKSQTPNSQITQAVFERKE 298
Query: 254 KGEFRVKVVKQILSVNGMRYELQEIYGIGNSVESDVDDNEQGKECVICLSEPRDTIVHPC 313
G++ V+VVKQIL VNG RYELQEIYGIGNSVE D + N+ GKECVICLSEPRDT V PC
Sbjct: 299 NGDYHVRVVKQILWVNGTRYELQEIYGIGNSVEGDTEGNDPGKECVICLSEPRDTTVLPC 358
Query: 314 RHMCMCSGCAKVLRFQTNRCPICRQPVERLLEIKVGTE 351
RHMCMCS CAKVLR+QTNRCPICRQPVERLLEIKV +
Sbjct: 359 RHMCMCSECAKVLRYQTNRCPICRQPVERLLEIKVNNK 396
>UniRef100_Q8L8P7 Putative RING zinc finger protein [Arabidopsis thaliana]
Length = 337
Score = 327 bits (838), Expect = 3e-88
Identities = 175/331 (52%), Positives = 214/331 (63%), Gaps = 27/331 (8%)
Query: 26 PPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMP----------HHH 75
PP T E+ P++FV+ A PY NP +Y + PPP+ P HH+
Sbjct: 20 PPAMETAPLELPPNRFVFAAAPPYLNPNPNYVDQYPGNCLPPPVTEPPMLPYNFNHLHHY 79
Query: 76 HPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEHQKAVTIRNDVNIKKETIVISPDEENP 135
P+ + PH P + YP P P HQKAVTIRNDVN+KK+T+ + PD ENP
Sbjct: 80 PPNSYQLPH--PLFHGGRYPI-----LPPPTYVHQKAVTIRNDVNLKKKTLTLIPDPENP 132
Query: 136 GFFLVSFTFDAAVSGSITIFFFAKEDEGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGT 195
LVSFTFDA++ G IT+ FFA ED C L TKE L P+T F +GLGQKF Q +GT
Sbjct: 133 NRLLVSFTFDASMPGRITVVFFATEDAECNLRATKEDTLPPITFDFGEGLGQKFIQSSGT 192
Query: 196 GINFSMFEESDLVKVGDVDVYPLAVKADASSDNHDGSNETETSSKPNSQITQAVFEKEKG 255
GI+ + F++S+L K D DV+PLAVKA+A+ S T N QITQ V+ KEKG
Sbjct: 193 GIDLTAFKDSELFKEVDTDVFPLAVKAEATPAEEGKSGST------NVQITQVVYTKEKG 246
Query: 256 EFRVKVVKQILSVNGMRYELQEIYGIGNSVESDVDDNEQGKECVICLSEPRDTIVHPCRH 315
E +++VVKQIL VN RYEL EIYGI E+ VD +++GKECV+CLSEPRDT V PCRH
Sbjct: 247 EIKIEVVKQILWVNKRRYELLEIYGI----ENTVDGSDEGKECVVCLSEPRDTTVLPCRH 302
Query: 316 MCMCSGCAKVLRFQTNRCPICRQPVERLLEI 346
MCMCSGCAK LRFQTN CP+CRQPVE LLEI
Sbjct: 303 MCMCSGCAKALRFQTNLCPVCRQPVEMLLEI 333
>UniRef100_Q9LYW5 Hypothetical protein F15A17_230 [Arabidopsis thaliana]
Length = 337
Score = 325 bits (834), Expect = 9e-88
Identities = 174/331 (52%), Positives = 213/331 (63%), Gaps = 27/331 (8%)
Query: 26 PPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMP----------HHH 75
PP T E+ P++FV+ PY NP +Y + PPP+ P HH+
Sbjct: 20 PPAMETAPLELPPNRFVFAAVPPYLNPNPNYVDQYPGNCLPPPVTEPPMLPYNFNHLHHY 79
Query: 76 HPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEHQKAVTIRNDVNIKKETIVISPDEENP 135
P+ + PH P + YP P P HQKAVTIRNDVN+KK+T+ + PD ENP
Sbjct: 80 PPNSYQLPH--PLFHGGRYPI-----LPPPTYVHQKAVTIRNDVNLKKKTLTLIPDPENP 132
Query: 136 GFFLVSFTFDAAVSGSITIFFFAKEDEGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGT 195
LVSFTFDA++ G IT+ FFA ED C L TKE L P+T F +GLGQKF Q +GT
Sbjct: 133 NRLLVSFTFDASMPGRITVVFFATEDAECNLRATKEDTLPPITFDFGEGLGQKFIQSSGT 192
Query: 196 GINFSMFEESDLVKVGDVDVYPLAVKADASSDNHDGSNETETSSKPNSQITQAVFEKEKG 255
GI+ + F++S+L K D DV+PLAVKA+A+ S T N QITQ V+ KEKG
Sbjct: 193 GIDLTAFKDSELFKEVDTDVFPLAVKAEATPAEEGKSGST------NVQITQVVYTKEKG 246
Query: 256 EFRVKVVKQILSVNGMRYELQEIYGIGNSVESDVDDNEQGKECVICLSEPRDTIVHPCRH 315
E +++VVKQIL VN RYEL EIYGI E+ VD +++GKECV+CLSEPRDT V PCRH
Sbjct: 247 EIKIEVVKQILWVNKRRYELLEIYGI----ENTVDGSDEGKECVVCLSEPRDTTVLPCRH 302
Query: 316 MCMCSGCAKVLRFQTNRCPICRQPVERLLEI 346
MCMCSGCAK LRFQTN CP+CRQPVE LLEI
Sbjct: 303 MCMCSGCAKALRFQTNLCPVCRQPVEMLLEI 333
>UniRef100_Q9LFH6 Hypothetical protein F4P12_110 [Arabidopsis thaliana]
Length = 299
Score = 322 bits (825), Expect = 1e-86
Identities = 168/289 (58%), Positives = 207/289 (71%), Gaps = 14/289 (4%)
Query: 67 PPIPMPHHHHPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEHQKAVTIRNDVNIKKETI 126
PP P P+ + +P P+ D A YP G + + P +VEHQ+AVTIRND+N+KKET+
Sbjct: 20 PPYPNPNAQYQGNYPSPYQDCA----RYPYGEMAS-PVQYVEHQEAVTIRNDINLKKETL 74
Query: 127 VISPDEENPGFFLVSFTFDAAVSGSITIFFFAKEDEGCILTPTKETHLAPVTVQFQQGLG 186
+ PDE+NPG FL+SFTFDA+V GSIT+ FFAKE + C L TKE V F +GL
Sbjct: 75 RLEPDEQNPGKFLLSFTFDASVPGSITVMFFAKEGKDCNLIATKEDLFPSTQVSFAKGLE 134
Query: 187 QKFRQQAGTGINFSMFEESDLVKVGDVDVYPLAVKAD-ASSDNHDGSNETETSSKPNSQI 245
Q+F+Q GTGI+FS E+DLV+ + DVY +AVKA+ S D+H S PN QI
Sbjct: 135 QRFKQACGTGIDFSDMSEADLVEANETDVYHVAVKAEVVSEDDH------PESGTPNRQI 188
Query: 246 TQAVFEKE-KGEFRVKVVKQILSVNGMRYELQEIYGIGNSVESDVDD-NEQGKECVICLS 303
T V EK+ KGE++ +VVKQIL VNG RY LQEIYGIGN+V+ + +D NE+GKECVICLS
Sbjct: 189 THVVLEKDHKGEYKARVVKQILWVNGNRYVLQEIYGIGNTVDDNGEDANERGKECVICLS 248
Query: 304 EPRDTIVHPCRHMCMCSGCAKVLRFQTNRCPICRQPVERLLEIKVGTEE 352
EPRDT V PCRHMCMCSGCAK+LRFQTN CPICRQPV+RLLEI V +
Sbjct: 249 EPRDTTVLPCRHMCMCSGCAKLLRFQTNLCPICRQPVDRLLEITVNNND 297
>UniRef100_Q8GZ27 Hypothetical protein At5g03200/F15A17_230 [Arabidopsis thaliana]
Length = 337
Score = 321 bits (823), Expect = 2e-86
Identities = 173/331 (52%), Positives = 215/331 (64%), Gaps = 27/331 (8%)
Query: 26 PPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMP----------HHH 75
PP T ++ P++FV+ PY NP +Y + PPP+ P HH+
Sbjct: 20 PPAMETAPLDLPPNRFVFAAVPPYLNPNPNYVDQYPGNCLPPPVTEPPMLPYNFNHLHHY 79
Query: 76 HPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEHQKAVTIRNDVNIKKETIVISPDEENP 135
P+ + PH P + YP P P +V+ QKAVTIRNDVN+KK+T+ + PD ENP
Sbjct: 80 PPNSYQLPH--PLFHGGRYPILP----PPTYVD-QKAVTIRNDVNLKKKTLTLIPDPENP 132
Query: 136 GFFLVSFTFDAAVSGSITIFFFAKEDEGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGT 195
LVSFTFDA++ G IT+ FFA ED C L TKE L P+T F +GLGQKF Q +GT
Sbjct: 133 NRLLVSFTFDASMPGRITVVFFATEDAECNLRATKEDTLPPITFDFGEGLGQKFIQSSGT 192
Query: 196 GINFSMFEESDLVKVGDVDVYPLAVKADASSDNHDGSNETETSSKPNSQITQAVFEKEKG 255
GI+ + F++S+L K D DV+PLAVKA+A+ S T N QITQ V+ KEKG
Sbjct: 193 GIDLTAFKDSELFKEVDTDVFPLAVKAEATPAEEGKSGST------NVQITQVVYTKEKG 246
Query: 256 EFRVKVVKQILSVNGMRYELQEIYGIGNSVESDVDDNEQGKECVICLSEPRDTIVHPCRH 315
E +++VVKQIL VN RYEL EIYGI E+ VD +++GKECV+CLSEPRDT V PCRH
Sbjct: 247 EIKIEVVKQILWVNKRRYELLEIYGI----ENTVDGSDEGKECVVCLSEPRDTTVLPCRH 302
Query: 316 MCMCSGCAKVLRFQTNRCPICRQPVERLLEI 346
MCMCSGCAK LRFQTN CP+CRQPVE LLEI
Sbjct: 303 MCMCSGCAKALRFQTNLCPVCRQPVEMLLEI 333
>UniRef100_Q9M8K4 Putative RING zinc finger protein [Arabidopsis thaliana]
Length = 546
Score = 280 bits (715), Expect = 6e-74
Identities = 159/366 (43%), Positives = 206/366 (55%), Gaps = 40/366 (10%)
Query: 10 HNRRRHATASRRTHPPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYY--PP 67
+NRRR R H PPPP P+ ++ P P P PP HY Y+Y Y+ PP
Sbjct: 195 NNRRRDNNNRRHLHHYPPPP--------PYYYLDPPPPPPPFPP-HY-DYNYSNYHLSPP 244
Query: 68 PIPMPH-------HHHPHPHPH------------PHMDPAWVSRYYPCGPVVNQPAPFVE 108
P P H+H HP P P M PA+ Y P P P P++E
Sbjct: 245 LPPQPQINSCSYGHYHYHPQPPQYFTTAQPNWWGPMMRPAY---YCPPQPQTQPPKPYLE 301
Query: 109 HQKAVTIRNDVNIKKETIVISPDEENPGFFLVSFTFDAAVSGSITIFFFAKEDEGCILTP 168
Q A +RNDVN+ ++T+ + D+ PG LVSF FDA GS TI FFAKE+ C + P
Sbjct: 302 QQNAKKVRNDVNVHRDTVRLEVDDLVPGHHLVSFVFDALFDGSFTITFFAKEEPNCTIIP 361
Query: 169 TKETHLAPVTVQFQQGLGQKFRQQAGTGINFSMFEESDLVKVGDVDVYPLAVKADASSDN 228
+P FQ+G GQKF Q +GTG + S F DL K + DVYPL + A+
Sbjct: 362 QFPEVYSPTRFHFQKGPGQKFLQPSGTGTDLSFFVLDDLSKPLEEDVYPLVISAETII-- 419
Query: 229 HDGSNETETSSKPNSQITQAVFEKEK-GEFRVKVVKQILSVNGMRYELQEIYG-IGNSVE 286
N S + Q+TQAV EK+ G F+VKVVKQIL + G+RYEL+E+YG
Sbjct: 420 --SPNSISEQSSVHKQVTQAVLEKDNDGSFKVKVVKQILWIEGVRYELRELYGSTTQGAA 477
Query: 287 SDVDDNEQGKECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQTNRCPICRQPVERLLEI 346
S +D++ G ECVIC++E +DT V PCRH+CMCS CAK LR Q+N+CPICRQP+E LLEI
Sbjct: 478 SGLDESGSGTECVICMTEAKDTAVLPCRHLCMCSDCAKELRLQSNKCPICRQPIEELLEI 537
Query: 347 KVGTEE 352
K+ + +
Sbjct: 538 KMNSSD 543
>UniRef100_Q8LA32 Putative RING zinc finger protein [Arabidopsis thaliana]
Length = 359
Score = 280 bits (715), Expect = 6e-74
Identities = 159/366 (43%), Positives = 206/366 (55%), Gaps = 40/366 (10%)
Query: 10 HNRRRHATASRRTHPPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYY--PP 67
+NRRR R H PPPP P+ ++ P P P PP HY Y+Y Y+ PP
Sbjct: 8 NNRRRDNNNRRHLHHYPPPP--------PYYYLDPPPPPPPFPP-HY-DYNYSNYHLSPP 57
Query: 68 PIPMPH-------HHHPHPHPH------------PHMDPAWVSRYYPCGPVVNQPAPFVE 108
P P H+H HP P P M PA+ Y P P P P++E
Sbjct: 58 LPPQPQINSCSYGHYHYHPQPPQYFTTAQPNWWGPMMRPAY---YCPPQPQTQPPKPYLE 114
Query: 109 HQKAVTIRNDVNIKKETIVISPDEENPGFFLVSFTFDAAVSGSITIFFFAKEDEGCILTP 168
Q A +RNDVN+ ++T+ + D+ PG LVSF FDA GS TI FFAKE+ C + P
Sbjct: 115 QQNAKKVRNDVNVHRDTVRLEVDDLVPGHHLVSFVFDALFDGSFTITFFAKEEPNCTIIP 174
Query: 169 TKETHLAPVTVQFQQGLGQKFRQQAGTGINFSMFEESDLVKVGDVDVYPLAVKADASSDN 228
+P FQ+G GQKF Q +GTG + S F DL K + DVYPL + A+
Sbjct: 175 QFPEVYSPTRFHFQKGPGQKFLQPSGTGTDLSFFVLDDLSKPLEEDVYPLVISAETII-- 232
Query: 229 HDGSNETETSSKPNSQITQAVFEKEK-GEFRVKVVKQILSVNGMRYELQEIYG-IGNSVE 286
N S + Q+TQAV EK+ G F+VKVVKQIL + G+RYEL+E+YG
Sbjct: 233 --SPNSISEQSSVHKQVTQAVLEKDNDGSFKVKVVKQILWIEGVRYELRELYGSTTQGAA 290
Query: 287 SDVDDNEQGKECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQTNRCPICRQPVERLLEI 346
S +D++ G ECVIC++E +DT V PCRH+CMCS CAK LR Q+N+CPICRQP+E LLEI
Sbjct: 291 SGLDESGSGTECVICMTEAKDTAVLPCRHLCMCSDCAKELRLQSNKCPICRQPIEELLEI 350
Query: 347 KVGTEE 352
K+ + +
Sbjct: 351 KMNSSD 356
>UniRef100_Q84ME1 At5g19080/T16G12_120 [Arabidopsis thaliana]
Length = 378
Score = 270 bits (689), Expect = 6e-71
Identities = 153/340 (45%), Positives = 194/340 (57%), Gaps = 32/340 (9%)
Query: 24 PPPP--PPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHP 81
PPPP PP + QP P Q Y PY YH YYP P + H
Sbjct: 57 PPPPAQPPSSSQPP--PSQISY---RPYGQ------NYHQNQYYPQQAPPYFTGYHHNGF 105
Query: 82 HPHMDPAWVSRYYPCGP----VVNQPAPFVEHQKAVTIRNDVNIKKETIVISPDEENPGF 137
+P M P + GP V+ PAP+VEHQ A ++NDVN+ K T+ + D+ NPG
Sbjct: 106 NPMMRPVYF------GPTPVAVMEPPAPYVEHQTAKKVKNDVNVNKATVRLVADDLNPGH 159
Query: 138 FLVSFTFDAAVSGSITIFFFAKEDEGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGTGI 197
+LVSF FDA GS TI FF +E+ C + P P+ V FQ+G GQKF Q GTGI
Sbjct: 160 YLVSFVFDALFDGSFTIIFFGEEESKCTIVPHLPEAFPPIKVPFQKGAGQKFLQAPGTGI 219
Query: 198 NFSMFEESDLVKVGDVDVYPLAVKADASSDNHDGSNETETSSKPNSQITQAVFEK-EKGE 256
+ F DL K +VYPL + A+ S E + QITQAV EK G
Sbjct: 220 DLGFFSLDDLSKPSPEEVYPLVISAETVISPSSVSEEPLV----HKQITQAVLEKTNDGS 275
Query: 257 FRVKVVKQILSVNGMRYELQEIYGIGNSVESDVD----DNEQGKECVICLSEPRDTIVHP 312
F+VKV+KQIL + G RYELQE+YGI NS+ ++ GKECVICL+EP+DT V P
Sbjct: 276 FKVKVMKQILWIEGERYELQELYGIDNSITQGTAASGLEDTGGKECVICLTEPKDTAVMP 335
Query: 313 CRHMCMCSGCAKVLRFQTNRCPICRQPVERLLEIKVGTEE 352
CRH+C+CS CA+ LRFQTN+CPICRQP+ L++IKV + +
Sbjct: 336 CRHLCLCSDCAEELRFQTNKCPICRQPIHELVKIKVESSD 375
>UniRef100_Q8L7V9 AT5g19080/T16G12_120 [Arabidopsis thaliana]
Length = 378
Score = 266 bits (679), Expect = 8e-70
Identities = 152/340 (44%), Positives = 193/340 (56%), Gaps = 32/340 (9%)
Query: 24 PPPP--PPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHP 81
PPPP PP + QP P Q Y PY YH YYP P + H
Sbjct: 57 PPPPAQPPSSSQPP--PSQISY---RPYGQ------NYHQNQYYPQQAPPYFTGYHHNGF 105
Query: 82 HPHMDPAWVSRYYPCGP----VVNQPAPFVEHQKAVTIRNDVNIKKETIVISPDEENPGF 137
+P M P + GP V+ PAP+VEHQ A ++NDVN+ K T+ + D+ NPG
Sbjct: 106 NPMMRPVYF------GPTPVAVMEPPAPYVEHQTAKKVKNDVNVNKATVRLVADDLNPGH 159
Query: 138 FLVSFTFDAAVSGSITIFFFAKEDEGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGTGI 197
+LVSF FDA GS TI FF +E+ C + P P+ V FQ+G GQKF Q GTGI
Sbjct: 160 YLVSFVFDALFDGSFTIIFFGEEESKCTIVPHLPEAFPPIKVPFQKGAGQKFLQAPGTGI 219
Query: 198 NFSMFEESDLVKVGDVDVYPLAVKADASSDNHDGSNETETSSKPNSQITQAVFEK-EKGE 256
+ F DL K +VYPL + A+ S E + QITQAV EK G
Sbjct: 220 DLGFFSLDDLSKPSPEEVYPLVISAETVISPSSVSEEPLV----HKQITQAVLEKTNDGS 275
Query: 257 FRVKVVKQILSVNGMRYELQEIYGIGNSVESDVD----DNEQGKECVICLSEPRDTIVHP 312
F+VKV+KQIL + G RYELQE+YGI NS+ ++ GKECVICL+EP+DT V P
Sbjct: 276 FKVKVMKQILWIEGERYELQELYGIDNSITQGTAASGLEDTGGKECVICLTEPKDTAVMP 335
Query: 313 CRHMCMCSGCAKVLRFQTNRCPICRQPVERLLEIKVGTEE 352
CRH+ +CS CA+ LRFQTN+CPICRQP+ L++IKV + +
Sbjct: 336 CRHLSLCSDCAEELRFQTNKCPICRQPIHELVKIKVESSD 375
>UniRef100_Q94E82 Mahogunin, ring finger 1-like protein [Oryza sativa]
Length = 313
Score = 254 bits (648), Expect = 3e-66
Identities = 143/310 (46%), Positives = 199/310 (64%), Gaps = 28/310 (9%)
Query: 45 GAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHPHPHMDPAWVSRYYPCGPVVNQPA 104
GA+P P PP H Y + PPP ++H +P P P P V PA
Sbjct: 9 GASPRPPPP-HLEAYRHG--VPPP-----YYHSYPRPPPGA---------AAPPPVPVPA 51
Query: 105 PFVEHQKAVTIRNDVNIKKETIVISPDEENPG--FFLVSFTFDAAVSGSITIFFFAKEDE 162
VE +AV + VN+K +T+ + PD+++ L++F+FDA GSIT+ FFA+ED+
Sbjct: 52 -HVERHRAVAVSVGVNVKGDTLRLVPDDDDDDGRSLLLAFSFDADGPGSITVCFFAQEDK 110
Query: 163 GCILTPTKETHLAPVTVQFQQGLGQKFRQQAGTGINFSMFEESDLVKVGDVDVYPLAVKA 222
C L KE L PVTV F++G GQ+F+Q +G+GI+ S FEES+L VG+ V+P+A K
Sbjct: 111 -CALKTAKENLLQPVTVPFKEGRGQEFKQPSGSGIDVSRFEESELTNVGEGGVFPVAFKV 169
Query: 223 D---ASSDNHDGSNETETSSKPNSQITQAVF-EKEKGEFRVKVVKQILSVNGMRYELQEI 278
+ + +G++ETE S + A+F +K+ E+ V VV+QIL VNG+RY LQEI
Sbjct: 170 QMDVSGNQESEGAHETEQSKY---LVKYAIFVKKDNAEYGVHVVQQILWVNGIRYVLQEI 226
Query: 279 YGIGNSVESDVDDNEQGKECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQTNRCPICRQ 338
YGIGN+ + + +++ GKECV+CLSEPRDT V PCRHMC+C CA+VL++QTN+CPICRQ
Sbjct: 227 YGIGNTADKNAHEDDSGKECVVCLSEPRDTAVLPCRHMCLCRECAQVLKYQTNKCPICRQ 286
Query: 339 PVERLLEIKV 348
PVE L EI+V
Sbjct: 287 PVEGLREIEV 296
>UniRef100_UPI0000248DFA UPI0000248DFA UniRef100 entry
Length = 541
Score = 151 bits (382), Expect = 2e-35
Identities = 103/277 (37%), Positives = 152/277 (54%), Gaps = 43/277 (15%)
Query: 98 PVV---NQPAPFVEHQKAVTIRNDVNIKKETIVI---SPDEENPG--------FFLVSFT 143
PVV N P P H+ T+R+ +NI+K+T+ + S D + PG + + FT
Sbjct: 39 PVVFPYNAPPP---HEPVKTLRSLINIRKDTLRLVRCSEDMKLPGQEVGKSHSCYNIEFT 95
Query: 144 FDAAVSGSITIFFFAKED--EGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGTGINFSM 201
FDA +ITI++ A E+ G + +++ L TV F++G+ Q+F + +N S
Sbjct: 96 FDADTQVAITIYYQAIEEFHNGVPIYLPQDSSLQSETVHFKRGVCQQFCLPSHY-VNLSE 154
Query: 202 FEESDLVKVGDVDVYPLAVKADASS-DNHDGSNETETSSKPNSQITQAVFEKEK-GEFRV 259
+ E +L+ D D+YP+ V+A D H G +S + A FEK G + V
Sbjct: 155 WAEDELLFDMDKDIYPMVVQAVVDEGDEHLG----------HSHVLLATFEKHMDGSYCV 204
Query: 260 KVVKQILSVNGMRYELQEIYGIGN---------SVESDVDDNEQGKECVICLSEPRDTIV 310
K +KQ V+G+ Y LQEIYGI N S E ++ DN ECV+CLS+ RDT++
Sbjct: 205 KPLKQKQVVDGVSYLLQEIYGIENKYNSQESKASAEDEISDNSA--ECVVCLSDVRDTLI 262
Query: 311 HPCRHMCMCSGCAKVLRFQTNRCPICRQPVERLLEIK 347
PCRH+C+C+ CA LR+Q N CPICR P LL+I+
Sbjct: 263 LPCRHLCLCNACADTLRYQANCCPICRLPFRALLQIR 299
>UniRef100_UPI00003AAE96 UPI00003AAE96 UniRef100 entry
Length = 514
Score = 149 bits (376), Expect = 1e-34
Identities = 107/300 (35%), Positives = 160/300 (52%), Gaps = 40/300 (13%)
Query: 77 PHPHPH---PHMD-------PAWVSRYYPCGPVVNQPAPFVEHQKAVTIRNDVNIKKETI 126
PHP + +MD P S Y P V PAP H+ T+R+ VNI+K+++
Sbjct: 18 PHPEGYLFGENMDLNFLGNRPVQASSYILSFPYVT-PAP---HEPVKTLRSLVNIRKDSL 73
Query: 127 --------VISPDEENPG---FFLVSFTFDAAVSGSITIFFFAKED--EGCILTPTKETH 173
V SP EEN + + FTFDA +ITI+ A E+ G + TK
Sbjct: 74 RLVRYKDDVDSPTEENGKQKVLYSLEFTFDADARVAITIYCQATEEFVGGMAVYSTKNPS 133
Query: 174 LAPVTVQFQQGLGQKFRQQAGTGINFSMFEESDLVKVGDVDVYPLAVKADASSDNHDGSN 233
L TV +++G+ Q+F + I+FS +++ +L D V+P+ ++A +G
Sbjct: 134 LQSETVHYKRGVSQQFSLPSFK-IDFSDWKDDELNFDLDRGVFPVVIRAVVD----EGDV 188
Query: 234 ETETSSKPNSQITQAVFEKE-KGEFRVKVVKQILSVNGMRYELQEIYGIGN-----SVES 287
E + ++ + A FEK G F VK +KQ V+ + Y LQEIYGI N + S
Sbjct: 189 VVEVTG--HAHVLLAAFEKHVDGSFSVKPLKQKQIVDRVSYLLQEIYGIENKNNQETKPS 246
Query: 288 DVDDNEQGKECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQTNRCPICRQPVERLLEIK 347
D ++++ ECV+CLS+ RDT++ PCRH+C+C+ CA LR+Q N CPICR P LL+I+
Sbjct: 247 DDENSDNSNECVVCLSDLRDTLILPCRHLCLCNSCADTLRYQANNCPICRLPFRALLQIR 306
>UniRef100_Q5XIQ4 Hypothetical protein [Rattus norvegicus]
Length = 533
Score = 142 bits (359), Expect = 1e-32
Identities = 105/321 (32%), Positives = 162/321 (49%), Gaps = 36/321 (11%)
Query: 51 NPPMHYPQYHYPGYYPPPIPMPHHHHPHPHPHPH-----MDPAWVSRYYPCGPVVNQPAP 105
N YP Y+ M PHP + MD ++ P V PAP
Sbjct: 20 NSAYRYPPKSAGNYFASHFFMGGEKFDTPHPEGYLFGENMDLNFLGSRPVQFPYVT-PAP 78
Query: 106 FVEHQKAVTIRNDVNIKKETIVI----------SPDEENPG-FFLVSFTFDAAVSGSITI 154
H+ T+R+ VNI+K+++ + + D E P + + FTFDA +ITI
Sbjct: 79 ---HEPVKTLRSLVNIRKDSLRLVRYKDDTDSPTEDGEKPRVLYSLEFTFDADARVAITI 135
Query: 155 FFFAKED--EGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGTGINFSMFEESDLVKVGD 212
+ A E+ G + K L TV +++G+ Q+F + I+FS +++ +L D
Sbjct: 136 YCQAVEEFVNGMTVYSCKNPSLQSETVHYKRGVSQQFSLPSFK-IDFSEWKDDELNFDLD 194
Query: 213 VDVYPLAVKADASSDNHDGSNETETSSKPNSQITQAVFEKE-KGEFRVKVVKQILSVNGM 271
V+P+ ++A D + E + ++ + A FEK G F VK +KQ V+ +
Sbjct: 195 RGVFPVVIQAVV-----DEGDVVEVTG--HAHVLLAAFEKHVDGSFSVKPLKQKQIVDRV 247
Query: 272 RYELQEIYGIGN-----SVESDVDDNEQGKECVICLSEPRDTIVHPCRHMCMCSGCAKVL 326
Y LQEIYGI N + SD ++++ ECV+CLS+ RDT++ PCRH+C+C+ CA L
Sbjct: 248 SYLLQEIYGIENKNNQETKPSDDENSDNSSECVVCLSDLRDTLILPCRHLCLCTSCADTL 307
Query: 327 RFQTNRCPICRQPVERLLEIK 347
R+Q N CPICR P LL+I+
Sbjct: 308 RYQANNCPICRLPFRALLQIR 328
>UniRef100_Q9D074 Mus musculus 10 days embryo whole body cDNA, RIKEN full-length
enriched library, clone:2610042J20 product:hypothetical
Crystallin/RING finger containing protein, full insert
sequence [Mus musculus]
Length = 532
Score = 141 bits (356), Expect = 2e-32
Identities = 105/317 (33%), Positives = 162/317 (50%), Gaps = 40/317 (12%)
Query: 59 YHYP----GYYPPPIPMPHHHHPHPHPHPH-----MDPAWVSRYYPCGPVVNQPAPFVEH 109
Y YP Y+ M PHP + MD ++ P V PAP H
Sbjct: 23 YRYPPKSGNYFASHFFMGGEKFDTPHPEGYLFGENMDLNFLGSRPVQFPYVT-PAP---H 78
Query: 110 QKAVTIRNDVNIKKETIVI----------SPDEENPG-FFLVSFTFDAAVSGSITIFFFA 158
+ T+R+ VNI+K+++ + + D E P + + FTFDA +ITI+ A
Sbjct: 79 EPVKTLRSLVNIRKDSLRLVRYKEDADSPTEDGEKPRVLYSLEFTFDADARVAITIYCQA 138
Query: 159 KED--EGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGTGINFSMFEESDLVKVGDVDVY 216
E+ G + K L TV +++G+ Q+F + I+FS +++ +L D V+
Sbjct: 139 VEELVNGVAVYSCKNPSLQSETVHYKRGVSQQFSLPSFK-IDFSEWKDDELNFDLDRGVF 197
Query: 217 PLAVKADASSDNHDGSNETETSSKPNSQITQAVFEKE-KGEFRVKVVKQILSVNGMRYEL 275
P+ ++A D + E + ++ + A FEK G F VK +KQ V+ + Y L
Sbjct: 198 PVVIQAVV-----DEGDVVEVTG--HAHVLLAAFEKHVDGSFSVKPLKQKQIVDRVSYLL 250
Query: 276 QEIYGIGN-----SVESDVDDNEQGKECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQT 330
QEIYGI N + SD ++++ ECV+CLS+ RDT++ PCRH+C+C+ CA LR+Q
Sbjct: 251 QEIYGIENKNNQETKPSDDENSDNSSECVVCLSDLRDTLILPCRHLCLCTSCADTLRYQA 310
Query: 331 NRCPICRQPVERLLEIK 347
N CPICR P LL+I+
Sbjct: 311 NNCPICRLPFRALLQIR 327
>UniRef100_Q86W76 Mahogunin, ring finger 1 [Homo sapiens]
Length = 576
Score = 141 bits (356), Expect = 2e-32
Identities = 105/317 (33%), Positives = 162/317 (50%), Gaps = 40/317 (12%)
Query: 59 YHYP----GYYPPPIPMPHHHHPHPHPHPH-----MDPAWVSRYYPCGPVVNQPAPFVEH 109
Y YP Y+ M PHP + MD ++ P V PAP H
Sbjct: 23 YRYPPKSGNYFASHFFMGGEKFDTPHPEGYLFGENMDLNFLGSRPVQFPYVT-PAP---H 78
Query: 110 QKAVTIRNDVNIKKETIVI--------SPDEENPG---FFLVSFTFDAAVSGSITIFFFA 158
+ T+R+ VNI+K+++ + SP E+ + + FTFDA +ITI+ A
Sbjct: 79 EPVKTLRSLVNIRKDSLRLVRYKDDADSPTEDGDKPRVLYSLEFTFDADARVAITIYCQA 138
Query: 159 KED--EGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGTGINFSMFEESDLVKVGDVDVY 216
E+ G + K L TV +++G+ Q+F + I+FS +++ +L D V+
Sbjct: 139 SEEFLNGRAVYSPKSPSLQSETVHYKRGVSQQFSLPSFK-IDFSEWKDDELNFDLDRGVF 197
Query: 217 PLAVKADASSDNHDGSNETETSSKPNSQITQAVFEKEK-GEFRVKVVKQILSVNGMRYEL 275
P+ ++A D + E + ++ + A FEK G F VK +KQ V+ + Y L
Sbjct: 198 PVVIQAVV-----DEGDVVEVTG--HAHVLLAAFEKHMDGSFSVKPLKQKQIVDRVSYLL 250
Query: 276 QEIYGIGN-----SVESDVDDNEQGKECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQT 330
QEIYGI N + SD ++++ ECV+CLS+ RDT++ PCRH+C+C+ CA LR+Q
Sbjct: 251 QEIYGIENKNNQETKPSDDENSDNSNECVVCLSDLRDTLILPCRHLCLCTSCADTLRYQA 310
Query: 331 NRCPICRQPVERLLEIK 347
N CPICR P LL+I+
Sbjct: 311 NNCPICRLPFRALLQIR 327
>UniRef100_O60291 KIAA0544 protein [Homo sapiens]
Length = 583
Score = 141 bits (356), Expect = 2e-32
Identities = 105/317 (33%), Positives = 162/317 (50%), Gaps = 40/317 (12%)
Query: 59 YHYP----GYYPPPIPMPHHHHPHPHPHPH-----MDPAWVSRYYPCGPVVNQPAPFVEH 109
Y YP Y+ M PHP + MD ++ P V PAP H
Sbjct: 54 YRYPPKSGNYFASHFFMGGEKFDTPHPEGYLFGENMDLNFLGSRPVQFPYVT-PAP---H 109
Query: 110 QKAVTIRNDVNIKKETIVI--------SPDEENPG---FFLVSFTFDAAVSGSITIFFFA 158
+ T+R+ VNI+K+++ + SP E+ + + FTFDA +ITI+ A
Sbjct: 110 EPVKTLRSLVNIRKDSLRLVRYKDDADSPTEDGDKPRVLYSLEFTFDADARVAITIYCQA 169
Query: 159 KED--EGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGTGINFSMFEESDLVKVGDVDVY 216
E+ G + K L TV +++G+ Q+F + I+FS +++ +L D V+
Sbjct: 170 SEEFLNGRAVYSPKSPSLQSETVHYKRGVSQQFSLPSFK-IDFSEWKDDELNFDLDRGVF 228
Query: 217 PLAVKADASSDNHDGSNETETSSKPNSQITQAVFEKEK-GEFRVKVVKQILSVNGMRYEL 275
P+ ++A D + E + ++ + A FEK G F VK +KQ V+ + Y L
Sbjct: 229 PVVIQAVV-----DEGDVVEVTG--HAHVLLAAFEKHMDGSFSVKPLKQKQIVDRVSYLL 281
Query: 276 QEIYGIGN-----SVESDVDDNEQGKECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQT 330
QEIYGI N + SD ++++ ECV+CLS+ RDT++ PCRH+C+C+ CA LR+Q
Sbjct: 282 QEIYGIENKNNQETKPSDDENSDNSNECVVCLSDLRDTLILPCRHLCLCTSCADTLRYQA 341
Query: 331 NRCPICRQPVERLLEIK 347
N CPICR P LL+I+
Sbjct: 342 NNCPICRLPFRALLQIR 358
>UniRef100_UPI000036AEB7 UPI000036AEB7 UniRef100 entry
Length = 470
Score = 139 bits (351), Expect = 9e-32
Identities = 102/273 (37%), Positives = 142/273 (51%), Gaps = 36/273 (13%)
Query: 98 PVVNQPAPFVEHQKAVTIRNDVNIKKETI--------VISPDEENPG---FFLVSFTFDA 146
PVV A + T+R+ VNI+K+T+ V SP EE + V FTFD
Sbjct: 29 PVVFPYAAPPPQEPVKTLRSLVNIRKDTLRLVKCAEEVKSPGEEASKAKVHYNVEFTFDT 88
Query: 147 AVSGSITIFFFAKED--EGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGTGINFSMFEE 204
+ITI++ A E+ G K+ L TVQ+++G+ Q+F + T ++ S + E
Sbjct: 89 DARVAITIYYQATEEFQNGIASYIPKDNSLQSETVQYKRGVCQQFCLPSHT-VDPSEWAE 147
Query: 205 SDLVKVGDVDVYPLAVKADASS-DNHDGSNETETSSKPNSQITQAVFEKEK-GEFRVKVV 262
+L D +VYPL V A D + G + + FEK G F VK +
Sbjct: 148 EELGFDLDREVYPLVVHAVVDEGDEYFG----------HCHVLLGTFEKHTDGTFCVKPL 197
Query: 263 KQILSVNGMRYELQEIYGIGNS--------VESDVDDNEQGKECVICLSEPRDTIVHPCR 314
KQ V+G+ Y LQEIYGI N E +V DN ECV+CLS+ RDT++ PCR
Sbjct: 198 KQKQVVDGVSYLLQEIYGIENKYNTQDSKVAEDEVSDNSA--ECVVCLSDVRDTLILPCR 255
Query: 315 HMCMCSGCAKVLRFQTNRCPICRQPVERLLEIK 347
H+C+C+ CA LR+Q N CPICR P LL+I+
Sbjct: 256 HLCLCNTCADTLRYQANNCPICRLPFRALLQIR 288
>UniRef100_Q96PX1 KIAA1917 protein [Homo sapiens]
Length = 702
Score = 139 bits (351), Expect = 9e-32
Identities = 102/273 (37%), Positives = 142/273 (51%), Gaps = 36/273 (13%)
Query: 98 PVVNQPAPFVEHQKAVTIRNDVNIKKETI--------VISPDEENPG---FFLVSFTFDA 146
PVV A + T+R+ VNI+K+T+ V SP EE + V FTFD
Sbjct: 90 PVVFPYAAPPPQEPVKTLRSLVNIRKDTLRLVKCAEEVKSPGEEASKAKVHYNVEFTFDT 149
Query: 147 AVSGSITIFFFAKED--EGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGTGINFSMFEE 204
+ITI++ A E+ G K+ L TVQ+++G+ Q+F + T ++ S + E
Sbjct: 150 DARVAITIYYQATEEFQNGIASYIPKDNSLQSETVQYKRGVCQQFCLPSHT-VDPSEWAE 208
Query: 205 SDLVKVGDVDVYPLAVKADASS-DNHDGSNETETSSKPNSQITQAVFEKEK-GEFRVKVV 262
+L D +VYPL V A D + G + + FEK G F VK +
Sbjct: 209 EELGFDLDREVYPLVVHAVVDEGDEYFG----------HCHVLLGTFEKHTDGTFCVKPL 258
Query: 263 KQILSVNGMRYELQEIYGIGNS--------VESDVDDNEQGKECVICLSEPRDTIVHPCR 314
KQ V+G+ Y LQEIYGI N E +V DN ECV+CLS+ RDT++ PCR
Sbjct: 259 KQKQVVDGVSYLLQEIYGIENKYNTQDSKVAEDEVSDNSA--ECVVCLSDVRDTLILPCR 316
Query: 315 HMCMCSGCAKVLRFQTNRCPICRQPVERLLEIK 347
H+C+C+ CA LR+Q N CPICR P LL+I+
Sbjct: 317 HLCLCNTCADTLRYQANNCPICRLPFRALLQIR 349
>UniRef100_Q8NB72 Hypothetical protein FLJ34148 [Homo sapiens]
Length = 619
Score = 139 bits (351), Expect = 9e-32
Identities = 102/273 (37%), Positives = 142/273 (51%), Gaps = 36/273 (13%)
Query: 98 PVVNQPAPFVEHQKAVTIRNDVNIKKETI--------VISPDEENPG---FFLVSFTFDA 146
PVV A + T+R+ VNI+K+T+ V SP EE + V FTFD
Sbjct: 29 PVVFPYAAPPPQEPVKTLRSLVNIRKDTLRLVKCAEEVKSPGEEASKAKVHYNVEFTFDT 88
Query: 147 AVSGSITIFFFAKED--EGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGTGINFSMFEE 204
+ITI++ A E+ G K+ L TVQ+++G+ Q+F + T ++ S + E
Sbjct: 89 DARVAITIYYQATEEFQNGIASYIPKDNSLQSETVQYKRGVCQQFCLPSHT-VDPSEWAE 147
Query: 205 SDLVKVGDVDVYPLAVKADASS-DNHDGSNETETSSKPNSQITQAVFEKEK-GEFRVKVV 262
+L D +VYPL V A D + G + + FEK G F VK +
Sbjct: 148 EELGFDLDREVYPLVVHAVVDEGDEYFG----------HCHVLLGTFEKHTDGTFCVKPL 197
Query: 263 KQILSVNGMRYELQEIYGIGNS--------VESDVDDNEQGKECVICLSEPRDTIVHPCR 314
KQ V+G+ Y LQEIYGI N E +V DN ECV+CLS+ RDT++ PCR
Sbjct: 198 KQKQVVDGVSYLLQEIYGIENKYNTQDSKVAEDEVSDNSA--ECVVCLSDVRDTLILPCR 255
Query: 315 HMCMCSGCAKVLRFQTNRCPICRQPVERLLEIK 347
H+C+C+ CA LR+Q N CPICR P LL+I+
Sbjct: 256 HLCLCNTCADTLRYQANNCPICRLPFRALLQIR 288
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.136 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 672,794,437
Number of Sequences: 2790947
Number of extensions: 33560683
Number of successful extensions: 311971
Number of sequences better than 10.0: 5857
Number of HSP's better than 10.0 without gapping: 1434
Number of HSP's successfully gapped in prelim test: 4703
Number of HSP's that attempted gapping in prelim test: 226023
Number of HSP's gapped (non-prelim): 32132
length of query: 352
length of database: 848,049,833
effective HSP length: 128
effective length of query: 224
effective length of database: 490,808,617
effective search space: 109941130208
effective search space used: 109941130208
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC148995.10


