

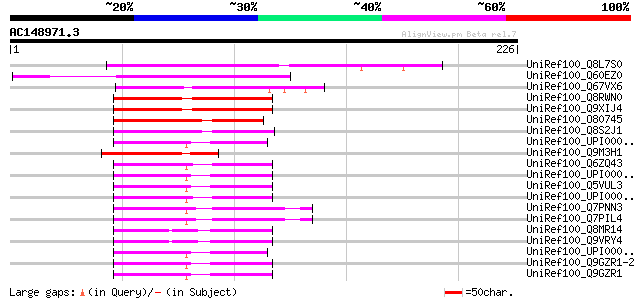
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148971.3 - phase: 0
(226 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8L7S0 At1g09730/F21M12_12 [Arabidopsis thaliana] 103 4e-21
UniRef100_Q60EZ0 Hypothetical protein OJ1430_B02.6 [Oryza sativa] 95 1e-18
UniRef100_Q67VX6 Ulp1 protease-like [Oryza sativa] 67 3e-10
UniRef100_Q8RWN0 Similar to protein-tyrosine phosphatase 2 [Arab... 67 5e-10
UniRef100_Q9XIJ4 T10O24.20 [Arabidopsis thaliana] 65 1e-09
UniRef100_O80745 T13D8.11 protein [Arabidopsis thaliana] 60 3e-08
UniRef100_Q8S2J1 OSJNBb0021A09.14 protein [Oryza sativa] 60 6e-08
UniRef100_UPI000032EE05 UPI000032EE05 UniRef100 entry 51 2e-05
UniRef100_Q9M3H1 Hypothetical protein T29H11_250 [Arabidopsis th... 51 2e-05
UniRef100_Q6ZQ43 MKIAA0797 protein [Mus musculus] 50 3e-05
UniRef100_UPI000036D86F UPI000036D86F UniRef100 entry 50 4e-05
UniRef100_Q5VUL3 OTTHUMP00000039934 [Homo sapiens] 50 4e-05
UniRef100_UPI00001D0038 UPI00001D0038 UniRef100 entry 50 6e-05
UniRef100_Q7PNN3 ENSANGP00000004517 [Anopheles gambiae str. PEST] 50 6e-05
UniRef100_Q7PIL4 ENSANGP00000023666 [Anopheles gambiae str. PEST] 50 6e-05
UniRef100_Q8MR14 LD44235p [Drosophila melanogaster] 49 8e-05
UniRef100_Q9VRY4 CG10107-PA, isoform A [Drosophila melanogaster] 49 8e-05
UniRef100_UPI00003630DE UPI00003630DE UniRef100 entry 49 1e-04
UniRef100_Q9GZR1-2 Splice isoform 2 of Q9GZR1 [Homo sapiens] 49 1e-04
UniRef100_Q9GZR1 Sentrin-specific protease 6 [Homo sapiens] 49 1e-04
>UniRef100_Q8L7S0 At1g09730/F21M12_12 [Arabidopsis thaliana]
Length = 378
Score = 103 bits (256), Expect = 4e-21
Identities = 59/153 (38%), Positives = 87/153 (56%), Gaps = 7/153 (4%)
Query: 44 FFFQLPQQDNFYDCGLFLLYFVERFLEEAPIKFNPFKITKFSKFLNSNWFPSDEASLRRS 103
F FQLPQQ+N +DCGLFLL+++E FL EAP+ F+PFKI S FL NWFP EASL+R+
Sbjct: 11 FLFQLPQQENSFDCGLFLLHYLELFLAEAPLNFSPFKIYNASNFLYLNWFPPAEASLKRT 70
Query: 104 HIQNLIYDIFENGSLKAPPIDCRGKGPLSELPGVIEHKVEADSSGASCYPGI-WHGNLSN 162
IQ LI+++ EN S + + E P + + + C P I +G+++
Sbjct: 71 LIQKLIFELLENRSREV----SNEQNQSCESPVAVNDDMGIEVLSERCSPLIDCNGDMTQ 126
Query: 163 GSTETDIQFRPV--SPVRAASCSRDPGIVFKDL 193
+ I+ + S +R + D G+V +DL
Sbjct: 127 TQDDQGIEMTLLERSSMRHIQAANDSGMVLRDL 159
>UniRef100_Q60EZ0 Hypothetical protein OJ1430_B02.6 [Oryza sativa]
Length = 991
Score = 95.1 bits (235), Expect = 1e-18
Identities = 53/124 (42%), Positives = 67/124 (53%), Gaps = 29/124 (23%)
Query: 2 ADDFSSKFLQLRFISLEVSSLQMNLTNSVELFIDFCFSLPPFFFFQLPQQDNFYDCGLFL 61
A D S KFL LRFISLE LPQQDN +DCGLFL
Sbjct: 455 ASDCSDKFLNLRFISLE-----------------------------LPQQDNSFDCGLFL 485
Query: 62 LYFVERFLEEAPIKFNPFKITKFSKFLNSNWFPSDEASLRRSHIQNLIYDIFENGSLKAP 121
L++VE FL + P FNP KI F+ +L+ +WFP EASL+RS I+ LI+ + + S P
Sbjct: 486 LHYVELFLMDTPRSFNPLKIDSFANYLSDDWFPPAEASLKRSLIRKLIHKLLKEPSQDFP 545
Query: 122 PIDC 125
+ C
Sbjct: 546 KLVC 549
>UniRef100_Q67VX6 Ulp1 protease-like [Oryza sativa]
Length = 522
Score = 67.0 bits (162), Expect = 3e-10
Identities = 39/102 (38%), Positives = 55/102 (53%), Gaps = 12/102 (11%)
Query: 48 LPQQDNFYDCGLFLLYFVERFLEEAPIKFNPFKITKFSKFLNSNWFPSDEASLRRSHIQN 107
+PQQDN YDCG+F+LY++ RF+EEAP + N S WF +EAS R +Q
Sbjct: 414 VPQQDNEYDCGVFVLYYMRRFIEEAPERLNN---KDSSNMFGEGWFQREEASALRKEMQA 470
Query: 108 LIYDIFE----NGSLKAP--PIDCRGKGP---LSELPGVIEH 140
L+ +FE N ++ P P+ + P LS P V +H
Sbjct: 471 LLLRLFEEAKDNNHMRDPTTPVSATAEHPVEVLSTEPAVPDH 512
>UniRef100_Q8RWN0 Similar to protein-tyrosine phosphatase 2 [Arabidopsis thaliana]
Length = 571
Score = 66.6 bits (161), Expect = 5e-10
Identities = 31/71 (43%), Positives = 45/71 (62%), Gaps = 4/71 (5%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFNPFKITKFSKFLNSNWFPSDEASLRRSHIQ 106
Q+PQQ N +DCGLFLL+F+ RF+EEAP + + K ++ WF +EAS R I
Sbjct: 502 QVPQQKNDFDCGLFLLFFIRRFIEEAPQRLT----LQDLKMIHKKWFKPEEASALRIKIW 557
Query: 107 NLIYDIFENGS 117
N++ D+F G+
Sbjct: 558 NILVDLFRKGN 568
>UniRef100_Q9XIJ4 T10O24.20 [Arabidopsis thaliana]
Length = 582
Score = 65.5 bits (158), Expect = 1e-09
Identities = 30/71 (42%), Positives = 45/71 (63%), Gaps = 4/71 (5%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFNPFKITKFSKFLNSNWFPSDEASLRRSHIQ 106
++PQQ N +DCGLFLL+F+ RF+EEAP + + K ++ WF +EAS R I
Sbjct: 513 EVPQQKNDFDCGLFLLFFIRRFIEEAPQRLT----LQDLKMIHKKWFKPEEASALRIKIW 568
Query: 107 NLIYDIFENGS 117
N++ D+F G+
Sbjct: 569 NILVDLFRKGN 579
>UniRef100_O80745 T13D8.11 protein [Arabidopsis thaliana]
Length = 547
Score = 60.5 bits (145), Expect = 3e-08
Identities = 29/67 (43%), Positives = 42/67 (62%), Gaps = 4/67 (5%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFNPFKITKFSKFLNSNWFPSDEASLRRSHIQ 106
Q+PQQ N +DCG F+L+F++RF+EEAP + + F K WF DEAS R I+
Sbjct: 478 QVPQQKNDFDCGPFVLFFIKRFIEEAPQRLKRKDLGMFDK----KWFRPDEASALRIKIR 533
Query: 107 NLIYDIF 113
N + ++F
Sbjct: 534 NTLIELF 540
>UniRef100_Q8S2J1 OSJNBb0021A09.14 protein [Oryza sativa]
Length = 506
Score = 59.7 bits (143), Expect = 6e-08
Identities = 27/72 (37%), Positives = 43/72 (59%), Gaps = 4/72 (5%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFNPFKITKFSKFLNSNWFPSDEASLRRSHIQ 106
Q+P Q N YDCG+F+L+++ERF++EAP + + F + WF E S R I+
Sbjct: 408 QVPSQRNKYDCGIFMLHYIERFIQEAPERLTRENLCMFGR----KWFDPKETSGLRDRIR 463
Query: 107 NLIYDIFENGSL 118
L++D FE+ +
Sbjct: 464 ALMFDAFESARM 475
>UniRef100_UPI000032EE05 UPI000032EE05 UniRef100 entry
Length = 160
Score = 51.2 bits (121), Expect = 2e-05
Identities = 24/70 (34%), Positives = 40/70 (56%), Gaps = 9/70 (12%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFN-PFKITKFSKFLNSNWFPSDEASLRRSHI 105
Q+PQQDN DCGL+LL + + FL+ + F P ++ NWFP + +R I
Sbjct: 98 QVPQQDNSSDCGLYLLQYAQSFLQNPVVHFELPLRL--------GNWFPRQQVRQKREEI 149
Query: 106 QNLIYDIFEN 115
++LI ++ ++
Sbjct: 150 RSLILELHQS 159
>UniRef100_Q9M3H1 Hypothetical protein T29H11_250 [Arabidopsis thaliana]
Length = 169
Score = 51.2 bits (121), Expect = 2e-05
Identities = 22/52 (42%), Positives = 32/52 (61%), Gaps = 3/52 (5%)
Query: 42 PFFFFQLPQQDNFYDCGLFLLYFVERFLEEAPIKFNPFKITKFSKFLNSNWF 93
PF+ +PQQ N +CG F+LY++ RF+E+AP FN + FL +WF
Sbjct: 102 PFYVPMVPQQTNDVECGSFVLYYIHRFIEDAPENFN---VEDMPYFLKEDWF 150
>UniRef100_Q6ZQ43 MKIAA0797 protein [Mus musculus]
Length = 1174
Score = 50.4 bits (119), Expect = 3e-05
Identities = 25/72 (34%), Positives = 39/72 (53%), Gaps = 9/72 (12%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFN-PFKITKFSKFLNSNWFPSDEASLRRSHI 105
++PQQ+NF DCG+++L +VE F E + F P + NWFP +R I
Sbjct: 1081 KVPQQNNFSDCGVYVLQYVESFFENPVLNFELPMNLV--------NWFPPPRMKTKREEI 1132
Query: 106 QNLIYDIFENGS 117
+N+I + E+ S
Sbjct: 1133 RNIILKLQESQS 1144
>UniRef100_UPI000036D86F UPI000036D86F UniRef100 entry
Length = 1107
Score = 50.1 bits (118), Expect = 4e-05
Identities = 25/72 (34%), Positives = 40/72 (54%), Gaps = 9/72 (12%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFN-PFKITKFSKFLNSNWFPSDEASLRRSHI 105
++PQQ+NF DCG+++L +VE F E + F P + +NWFP +R I
Sbjct: 1015 KVPQQNNFSDCGVYVLQYVESFFENPILSFELPMNL--------ANWFPPPRMRTKREEI 1066
Query: 106 QNLIYDIFENGS 117
+N+I + E+ S
Sbjct: 1067 RNIILKLQEDQS 1078
>UniRef100_Q5VUL3 OTTHUMP00000039934 [Homo sapiens]
Length = 1107
Score = 50.1 bits (118), Expect = 4e-05
Identities = 25/72 (34%), Positives = 40/72 (54%), Gaps = 9/72 (12%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFN-PFKITKFSKFLNSNWFPSDEASLRRSHI 105
++PQQ+NF DCG+++L +VE F E + F P + +NWFP +R I
Sbjct: 1015 KVPQQNNFSDCGVYVLQYVESFFENPILSFELPMNL--------ANWFPPPRMRTKREEI 1066
Query: 106 QNLIYDIFENGS 117
+N+I + E+ S
Sbjct: 1067 RNIILKLQEDQS 1078
>UniRef100_UPI00001D0038 UPI00001D0038 UniRef100 entry
Length = 1530
Score = 49.7 bits (117), Expect = 6e-05
Identities = 25/72 (34%), Positives = 38/72 (52%), Gaps = 9/72 (12%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFN-PFKITKFSKFLNSNWFPSDEASLRRSHI 105
++PQQ+NF DCG+++L +VE F E + F P + NWFP +R I
Sbjct: 1438 KVPQQNNFSDCGVYVLQYVESFFENPILNFELPMNLV--------NWFPPPRMKTKREEI 1489
Query: 106 QNLIYDIFENGS 117
+N+I + E S
Sbjct: 1490 RNIILKLQEEQS 1501
>UniRef100_Q7PNN3 ENSANGP00000004517 [Anopheles gambiae str. PEST]
Length = 279
Score = 49.7 bits (117), Expect = 6e-05
Identities = 29/90 (32%), Positives = 48/90 (53%), Gaps = 14/90 (15%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFN-PFKITKFSKFLNSNWFPSDEASLRRSHI 105
++PQQ+N+ DCGL+LL +VE F + + ++ P K + +WF + + +R I
Sbjct: 198 KVPQQNNYTDCGLYLLQYVEHFFLDPILDYHLPIKQLQ-------DWFETITVTKKREDI 250
Query: 106 QNLIYDIFENGSLKAPPIDCRGKGPLSELP 135
NLI ++ + APP+ P ELP
Sbjct: 251 SNLIKELIDKHDPSAPPL------PSIELP 274
>UniRef100_Q7PIL4 ENSANGP00000023666 [Anopheles gambiae str. PEST]
Length = 916
Score = 49.7 bits (117), Expect = 6e-05
Identities = 29/90 (32%), Positives = 48/90 (53%), Gaps = 14/90 (15%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFN-PFKITKFSKFLNSNWFPSDEASLRRSHI 105
++PQQ+N+ DCGL+LL +VE F + + ++ P K + +WF + + +R I
Sbjct: 811 KVPQQNNYTDCGLYLLQYVEHFFLDPILDYHLPIKQLQ-------DWFETITVTKKREDI 863
Query: 106 QNLIYDIFENGSLKAPPIDCRGKGPLSELP 135
NLI ++ + APP+ P ELP
Sbjct: 864 SNLIKELIDKHDPSAPPL------PSIELP 887
>UniRef100_Q8MR14 LD44235p [Drosophila melanogaster]
Length = 711
Score = 49.3 bits (116), Expect = 8e-05
Identities = 27/71 (38%), Positives = 41/71 (57%), Gaps = 6/71 (8%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFNPFKITKFSKFLNSNWFPSDEASLRRSHIQ 106
++PQQ+NF DCGL+LL +VE+F E PI+ I + +NWF + +R I
Sbjct: 492 KVPQQNNFTDCGLYLLQYVEQFFGE-PIRDYRLPIKQL-----TNWFDFLTVTKKREDIA 545
Query: 107 NLIYDIFENGS 117
NLI + + G+
Sbjct: 546 NLIQQLMDEGN 556
>UniRef100_Q9VRY4 CG10107-PA, isoform A [Drosophila melanogaster]
Length = 1833
Score = 49.3 bits (116), Expect = 8e-05
Identities = 27/71 (38%), Positives = 41/71 (57%), Gaps = 6/71 (8%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFNPFKITKFSKFLNSNWFPSDEASLRRSHIQ 106
++PQQ+NF DCGL+LL +VE+F E PI+ I + +NWF + +R I
Sbjct: 1614 KVPQQNNFTDCGLYLLQYVEQFFGE-PIRDYRLPIKQL-----TNWFDFLTVTKKREDIA 1667
Query: 107 NLIYDIFENGS 117
NLI + + G+
Sbjct: 1668 NLIQQLMDEGN 1678
>UniRef100_UPI00003630DE UPI00003630DE UniRef100 entry
Length = 115
Score = 48.9 bits (115), Expect = 1e-04
Identities = 24/70 (34%), Positives = 39/70 (55%), Gaps = 9/70 (12%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFN-PFKITKFSKFLNSNWFPSDEASLRRSHI 105
++PQQDN DCGL+LL + E FL+ + F P ++ NWFP + +R I
Sbjct: 53 RVPQQDNSSDCGLYLLQYAESFLQNPVVHFELPVRL--------DNWFPRQQVRQKREEI 104
Query: 106 QNLIYDIFEN 115
++LI + ++
Sbjct: 105 RSLIMKMHQS 114
>UniRef100_Q9GZR1-2 Splice isoform 2 of Q9GZR1 [Homo sapiens]
Length = 1105
Score = 48.9 bits (115), Expect = 1e-04
Identities = 24/72 (33%), Positives = 40/72 (55%), Gaps = 9/72 (12%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFN-PFKITKFSKFLNSNWFPSDEASLRRSHI 105
++PQQ+NF DCG+++L +VE F + + F P + +NWFP +R I
Sbjct: 1013 KVPQQNNFSDCGVYVLQYVESFFQNPILSFELPMNL--------ANWFPPPRMRTKREEI 1064
Query: 106 QNLIYDIFENGS 117
+N+I + E+ S
Sbjct: 1065 RNIILKLQEDQS 1076
>UniRef100_Q9GZR1 Sentrin-specific protease 6 [Homo sapiens]
Length = 1112
Score = 48.9 bits (115), Expect = 1e-04
Identities = 24/72 (33%), Positives = 40/72 (55%), Gaps = 9/72 (12%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFN-PFKITKFSKFLNSNWFPSDEASLRRSHI 105
++PQQ+NF DCG+++L +VE F + + F P + +NWFP +R I
Sbjct: 1020 KVPQQNNFSDCGVYVLQYVESFFQNPILSFELPMNL--------ANWFPPPRMRTKREEI 1071
Query: 106 QNLIYDIFENGS 117
+N+I + E+ S
Sbjct: 1072 RNIILKLQEDQS 1083
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.324 0.141 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 392,210,904
Number of Sequences: 2790947
Number of extensions: 16841587
Number of successful extensions: 38373
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 72
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 38271
Number of HSP's gapped (non-prelim): 85
length of query: 226
length of database: 848,049,833
effective HSP length: 123
effective length of query: 103
effective length of database: 504,763,352
effective search space: 51990625256
effective search space used: 51990625256
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 72 (32.3 bits)
Medicago: description of AC148971.3


