

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148970.7 - phase: 0
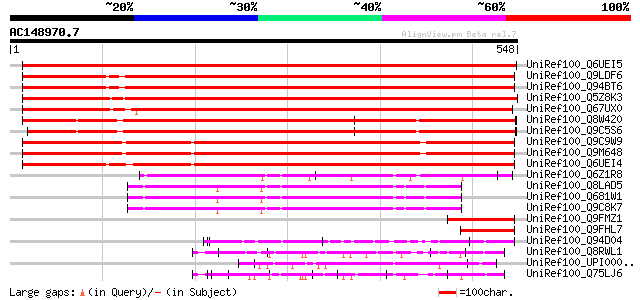
(548 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6UEI5 ZEITLUPE [Mesembryanthemum crystallinum] 976 0.0
UniRef100_Q9LDF6 Clock-associated PAS protein ZTL [Arabidopsis t... 972 0.0
UniRef100_Q94BT6 AT5g57360/MSF19_2 [Arabidopsis thaliana] 962 0.0
UniRef100_Q5Z8K3 Putative ZEITLUPE [Oryza sativa] 922 0.0
UniRef100_Q67UX0 Putative ZEITLUPE [Oryza sativa] 879 0.0
UniRef100_Q8W420 LOV kelch protein 2 [Arabidopsis thaliana] 846 0.0
UniRef100_Q9C5S6 Adagio 2 [Arabidopsis thaliana] 833 0.0
UniRef100_Q9C9W9 Hypothetical protein T23K23.10 [Arabidopsis tha... 729 0.0
UniRef100_Q9M648 FKF1 [Arabidopsis thaliana] 726 0.0
UniRef100_Q6UEI4 FKF1 [Mesembryanthemum crystallinum] 716 0.0
UniRef100_Q6Z1R8 Putative F-box protein [Oryza sativa] 159 3e-37
UniRef100_Q8LAD5 F-box protein ZEITLUPE/FKF/LKP/ADAGIO family [A... 129 2e-28
UniRef100_Q681W1 Hypothetical protein At1g51550 [Arabidopsis tha... 126 2e-27
UniRef100_Q9C8K7 Hypothetical protein F5D21.4 [Arabidopsis thali... 126 2e-27
UniRef100_Q9FMZ1 Arabidopsis thaliana genomic DNA, chromosome 5,... 120 1e-25
UniRef100_Q9FHL7 Arabidopsis thaliana genomic DNA, chromosome 5,... 104 8e-21
UniRef100_Q94D04 P0682B08.13 protein [Oryza sativa] 90 2e-16
UniRef100_Q8RWL1 Hypothetical protein At2g36360 [Arabidopsis tha... 90 2e-16
UniRef100_UPI00003C2253 UPI00003C2253 UniRef100 entry 89 3e-16
UniRef100_Q75LJ6 Putative transcription factor [Oryza sativa] 89 3e-16
>UniRef100_Q6UEI5 ZEITLUPE [Mesembryanthemum crystallinum]
Length = 615
Score = 976 bits (2523), Expect = 0.0
Identities = 459/534 (85%), Positives = 501/534 (92%)
Query: 14 RFLQCRGPFAKRRHPLVDSSVISEIRKCIDEGVEFQGELLNFRKDGSPLMNRLRLTPIYG 73
RFLQCRGPFAKRRHPLVDS V++EIR+C+D+GVEFQGELLNFRKDGSPLMNRLRLTPIYG
Sbjct: 76 RFLQCRGPFAKRRHPLVDSVVVAEIRRCLDDGVEFQGELLNFRKDGSPLMNRLRLTPIYG 135
Query: 74 EDDEITHVIGIQLFTEANIDLGPLPGSTIKESLKSSGRFHSVLSSLQPPPLGDRNVSRGI 133
+DD ITH+IGIQL +EAN+DLGPLP +KES KSS ++ S LS +R++ +
Sbjct: 136 DDDTITHIIGIQLLSEANVDLGPLPSFPVKESTKSSDQYRSGLSLFPSFQSRERSICLEV 195
Query: 134 CGIFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGSETTRVL 193
CG+ Q+SDEVL+LKI +RLTPRDIA+V SVC RLYE+TRNEDLWRMVCQNAWG+ETTRVL
Sbjct: 196 CGLLQISDEVLALKIFSRLTPRDIAAVGSVCRRLYELTRNEDLWRMVCQNAWGTETTRVL 255
Query: 194 ETVPGAKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVN 253
ETVPGAKRLGWGRLARELTTLEAAAWRKLTVGG VEPSRCNFSACAVGNRVVLFGGEGVN
Sbjct: 256 ETVPGAKRLGWGRLARELTTLEAAAWRKLTVGGAVEPSRCNFSACAVGNRVVLFGGEGVN 315
Query: 254 MQPMNDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVF 313
MQPMNDTFVLDLN++NPEWQHV+VSSPPPGRWGHTLSC+NGS LVVFGGCGTQGLLNDVF
Sbjct: 316 MQPMNDTFVLDLNASNPEWQHVKVSSPPPGRWGHTLSCMNGSNLVVFGGCGTQGLLNDVF 375
Query: 314 VLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSME 373
VLDLDA PTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLD++M+
Sbjct: 376 VLDLDAKQPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDIAMD 435
Query: 374 NPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCW 433
PVWREIPVTW+PPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDV+TMDLSED+PCW
Sbjct: 436 KPVWREIPVTWSPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVYTMDLSEDDPCW 495
Query: 434 RCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEK 493
RCVTGS MPGAGNP G+APPPRLDHVAVSLPGGRIL+FGGSVAGLHSASQLY+LDPT+EK
Sbjct: 496 RCVTGSAMPGAGNPGGVAPPPRLDHVAVSLPGGRILVFGGSVAGLHSASQLYLLDPTEEK 555
Query: 494 PTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSLANSV 547
PTWRILNVPGRPP FAWGHSTCVVGGTRAIVLGGQTGE+WML+D+HELSLA++V
Sbjct: 556 PTWRILNVPGRPPSFAWGHSTCVVGGTRAIVLGGQTGEDWMLTDIHELSLASTV 609
>UniRef100_Q9LDF6 Clock-associated PAS protein ZTL [Arabidopsis thaliana]
Length = 609
Score = 972 bits (2513), Expect = 0.0
Identities = 463/532 (87%), Positives = 499/532 (93%), Gaps = 8/532 (1%)
Query: 14 RFLQCRGPFAKRRHPLVDSSVISEIRKCIDEGVEFQGELLNFRKDGSPLMNRLRLTPIYG 73
RFLQCRGPFAKRRHPLVDS V+SEIRKCIDEG+EFQGELLNFRKDGSPLMNRLRLTPIYG
Sbjct: 83 RFLQCRGPFAKRRHPLVDSMVVSEIRKCIDEGIEFQGELLNFRKDGSPLMNRLRLTPIYG 142
Query: 74 EDDEITHVIGIQLFTEANIDLGPLPGSTIKESLKSSGRFHSVLSSLQPPPLGDRNVSRGI 133
+DD ITH+IGIQ F E +IDLGP+ GS+ KE KS +S L++ G+RNVSRG+
Sbjct: 143 DDDTITHIIGIQFFIETDIDLGPVLGSSTKE--KSIDGIYSALAA------GERNVSRGM 194
Query: 134 CGIFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGSETTRVL 193
CG+FQLSDEV+S+KIL+RLTPRD+ASVSSVC RLY +T+NEDLWR VCQNAWGSETTRVL
Sbjct: 195 CGLFQLSDEVVSMKILSRLTPRDVASVSSVCRRLYVLTKNEDLWRRVCQNAWGSETTRVL 254
Query: 194 ETVPGAKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVN 253
ETVPGAKRLGWGRLARELTTLEAAAWRKL+VGG VEPSRCNFSACAVGNRVVLFGGEGVN
Sbjct: 255 ETVPGAKRLGWGRLARELTTLEAAAWRKLSVGGSVEPSRCNFSACAVGNRVVLFGGEGVN 314
Query: 254 MQPMNDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVF 313
MQPMNDTFVLDLNS+ PEWQHV+VSSPPPGRWGHTL+CVNGS LVVFGGCG QGLLNDVF
Sbjct: 315 MQPMNDTFVLDLNSDYPEWQHVKVSSPPPGRWGHTLTCVNGSNLVVFGGCGQQGLLNDVF 374
Query: 314 VLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSME 373
VL+LDA PPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLD+S+E
Sbjct: 375 VLNLDAKPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDLSIE 434
Query: 374 NPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCW 433
PVWREIP WTPPSRLGHTLSVYGGRKILMFGGLAKSGPL+FRSSDVFTMDLSE+EPCW
Sbjct: 435 KPVWREIPAAWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLKFRSSDVFTMDLSEEEPCW 494
Query: 434 RCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEK 493
RCVTGSGMPGAGNP G+APPPRLDHVAV+LPGGRILIFGGSVAGLHSASQLY+LDPT++K
Sbjct: 495 RCVTGSGMPGAGNPGGVAPPPRLDHVAVNLPGGRILIFGGSVAGLHSASQLYLLDPTEDK 554
Query: 494 PTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSLAN 545
PTWRILN+PGRPPRFAWGH TCVVGGTRAIVLGGQTGEEWMLS+LHELSLA+
Sbjct: 555 PTWRILNIPGRPPRFAWGHGTCVVGGTRAIVLGGQTGEEWMLSELHELSLAS 606
>UniRef100_Q94BT6 AT5g57360/MSF19_2 [Arabidopsis thaliana]
Length = 609
Score = 962 bits (2488), Expect = 0.0
Identities = 459/532 (86%), Positives = 496/532 (92%), Gaps = 8/532 (1%)
Query: 14 RFLQCRGPFAKRRHPLVDSSVISEIRKCIDEGVEFQGELLNFRKDGSPLMNRLRLTPIYG 73
RFLQCRGPFAKRRHPLVDS V+SEIRKCIDEG+EFQGELLNFRKDGSPLMNRLRLTPIYG
Sbjct: 83 RFLQCRGPFAKRRHPLVDSMVVSEIRKCIDEGIEFQGELLNFRKDGSPLMNRLRLTPIYG 142
Query: 74 EDDEITHVIGIQLFTEANIDLGPLPGSTIKESLKSSGRFHSVLSSLQPPPLGDRNVSRGI 133
+DD ITH+IGIQ F E +IDLGP+ GS+ KE KS +S L++ G+RNVSRG+
Sbjct: 143 DDDTITHIIGIQFFIETDIDLGPVLGSSTKE--KSIDGIYSALAA------GERNVSRGM 194
Query: 134 CGIFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGSETTRVL 193
CG+FQLSDEV+S+KIL+RLTPRD+ASVSSVC RLY +T+NEDLWR VCQNAWGSETTRVL
Sbjct: 195 CGLFQLSDEVVSMKILSRLTPRDVASVSSVCRRLYVLTKNEDLWRRVCQNAWGSETTRVL 254
Query: 194 ETVPGAKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVN 253
ETVPGAKRLGWGRLARELTTLEAAAWRKL+VGG VEPSRCNFSACAVGNRVVLFGGEGVN
Sbjct: 255 ETVPGAKRLGWGRLARELTTLEAAAWRKLSVGGSVEPSRCNFSACAVGNRVVLFGGEGVN 314
Query: 254 MQPMNDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVF 313
MQPMNDTFVLDLNS+ PEWQHV+VSSPPPGRWGHTL+CVNGS LVVFGGCG QGLLNDVF
Sbjct: 315 MQPMNDTFVLDLNSDYPEWQHVKVSSPPPGRWGHTLTCVNGSNLVVFGGCGQQGLLNDVF 374
Query: 314 VLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSME 373
VL+LDA PPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLD+S+E
Sbjct: 375 VLNLDAKPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDLSIE 434
Query: 374 NPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCW 433
PVWREIP WTPPSRLGHTLSVYGGR+ L GGLAKSGPL+FRSSDVFTMDLSE+EPCW
Sbjct: 435 KPVWREIPAAWTPPSRLGHTLSVYGGRRNLDVGGLAKSGPLKFRSSDVFTMDLSEEEPCW 494
Query: 434 RCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEK 493
RCVTGSGMPGAGNP G+APPPRLDHVAV+LPGGRILIFGGSVAGLHSASQLY+LDPT++K
Sbjct: 495 RCVTGSGMPGAGNPGGVAPPPRLDHVAVNLPGGRILIFGGSVAGLHSASQLYLLDPTEDK 554
Query: 494 PTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSLAN 545
PTWRILN+PGRPPRFAWGH TCVVGGTRAIVLGGQTGEEWMLS+LHELSLA+
Sbjct: 555 PTWRILNIPGRPPRFAWGHGTCVVGGTRAIVLGGQTGEEWMLSELHELSLAS 606
>UniRef100_Q5Z8K3 Putative ZEITLUPE [Oryza sativa]
Length = 630
Score = 922 bits (2382), Expect = 0.0
Identities = 436/537 (81%), Positives = 482/537 (89%), Gaps = 4/537 (0%)
Query: 14 RFLQCRGPFAKRRHPLVDSSVISEIRKCIDEGVEFQGELLNFRKDGSPLMNRLRLTPIYG 73
RFLQCRGPFA+RRHPLVD+ V+SEIRKCID G EF+G+LLNFRKDGSPLMN+L LTPIYG
Sbjct: 96 RFLQCRGPFAQRRHPLVDAMVVSEIRKCIDNGTEFRGDLLNFRKDGSPLMNKLHLTPIYG 155
Query: 74 EDDEITHVIGIQLFTEANIDLGPLPGSTIKESLKSSGRF--HSVLSSLQPPPLGDRNVSR 131
+D+ ITH +GIQ FT AN+DLGPLPGS KE ++S+ RF + + P G N R
Sbjct: 156 DDETITHYMGIQFFTNANVDLGPLPGSLTKEPVRST-RFTPDNFFRPISTGP-GQSNFCR 213
Query: 132 GICGIFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGSETTR 191
+FQL+DEVL IL+RL+PRDIASVSSVC RLY +TRNEDLWRMVCQNAWGSETTR
Sbjct: 214 EYSSLFQLTDEVLCQSILSRLSPRDIASVSSVCRRLYLLTRNEDLWRMVCQNAWGSETTR 273
Query: 192 VLETVPGAKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEG 251
LETVP AKRLGWGRLARELTTLEA AWRKLTVGG VEPSRCNFSACAVGNRVVLFGGEG
Sbjct: 274 ALETVPAAKRLGWGRLARELTTLEAVAWRKLTVGGAVEPSRCNFSACAVGNRVVLFGGEG 333
Query: 252 VNMQPMNDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLND 311
VNMQPMNDTFVLDLN++NPEW+HV VSS PPGRWGHTLSC+NGS LVVFGGCG QGLLND
Sbjct: 334 VNMQPMNDTFVLDLNASNPEWRHVNVSSAPPGRWGHTLSCLNGSLLVVFGGCGRQGLLND 393
Query: 312 VFVLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMS 371
VF LDLDA PTWREI G+APP+PRSWHSSCTLDGTKL+VSGGCADSGVLLSDT+LLD++
Sbjct: 394 VFTLDLDAKQPTWREIPGVAPPVPRSWHSSCTLDGTKLVVSGGCADSGVLLSDTYLLDVT 453
Query: 372 MENPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEP 431
M+ PVWRE+P +WTPPSRLGH++SVYGGRKILMFGGLAKSGPLR RSSDVFTMDLSE+EP
Sbjct: 454 MDKPVWREVPASWTPPSRLGHSMSVYGGRKILMFGGLAKSGPLRLRSSDVFTMDLSEEEP 513
Query: 432 CWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTD 491
CWRC+TGSGMPGAGNP G PPPRLDHVAVSLPGGR+LIFGGSVAGLHSASQLY+LDPT+
Sbjct: 514 CWRCLTGSGMPGAGNPAGAGPPPRLDHVAVSLPGGRVLIFGGSVAGLHSASQLYLLDPTE 573
Query: 492 EKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSLANSVI 548
EKPTWRILNVPGRPPRFAWGHSTCVVGGT+AIVLGGQTGEEWML+++HELSLA+S +
Sbjct: 574 EKPTWRILNVPGRPPRFAWGHSTCVVGGTKAIVLGGQTGEEWMLTEIHELSLASSTV 630
>UniRef100_Q67UX0 Putative ZEITLUPE [Oryza sativa]
Length = 635
Score = 879 bits (2271), Expect = 0.0
Identities = 411/534 (76%), Positives = 473/534 (87%), Gaps = 11/534 (2%)
Query: 14 RFLQCRGPFAKRRHPLVDSSVISEIRKCIDEGVEFQGELLNFRKDGSPLMNRLRLTPIYG 73
RFLQCRGPFAKRRHPLVD++V+++IR+C++EG FQG+LLNFRKDGSP M +L+LTPIYG
Sbjct: 92 RFLQCRGPFAKRRHPLVDTTVVTDIRRCLEEGTVFQGDLLNFRKDGSPFMAKLQLTPIYG 151
Query: 74 EDDEITHVIGIQLFTEANIDLGPLPGSTIKESLKSSGRFHSVLSSLQPPPLGDRNVSRGI 133
+D+ ITH +G+Q F ++N+DLGPL ST KE ++S+ + ++++P P+G +G
Sbjct: 152 DDETITHYMGMQFFNDSNVDLGPLSVSTTKEIVRST--LITPDNTIRPSPMG-----KGF 204
Query: 134 CG----IFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGSET 189
C +F LSDEVL KIL+RL+PRDIASV+SVC RLY +TRN+DLWRMVCQNAWGSE
Sbjct: 205 CSEHSDLFLLSDEVLCQKILSRLSPRDIASVNSVCKRLYHLTRNDDLWRMVCQNAWGSEA 264
Query: 190 TRVLETVPGAKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGG 249
T+VLETV G + L WGRLARELTTLEA WRKLTVGG VEPSRCNFSACA GNRVVLFGG
Sbjct: 265 TQVLETVAGTRSLAWGRLARELTTLEAVTWRKLTVGGAVEPSRCNFSACAAGNRVVLFGG 324
Query: 250 EGVNMQPMNDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLL 309
EGVNMQPMNDTFVLDLN++ PEW+H+ V S PPGRWGHTLSC+NGSRLV+FGGCG QGLL
Sbjct: 325 EGVNMQPMNDTFVLDLNASKPEWRHINVRSAPPGRWGHTLSCLNGSRLVLFGGCGRQGLL 384
Query: 310 NDVFVLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLD 369
NDVF+LDLDA PTWREI GLAPP+PRSWHSSCTLDGTKL+VSGGCADSGVLLSDT+LLD
Sbjct: 385 NDVFMLDLDAQQPTWREIPGLAPPVPRSWHSSCTLDGTKLVVSGGCADSGVLLSDTYLLD 444
Query: 370 MSMENPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSED 429
++ME PVWREIP +WTPP RLGH+LSVY GRKILMFGGLAKSGPLR RS+DVFT+DLSE+
Sbjct: 445 VTMERPVWREIPASWTPPCRLGHSLSVYDGRKILMFGGLAKSGPLRLRSNDVFTLDLSEN 504
Query: 430 EPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDP 489
+PCWRC+TGSGMPGA NP G+ PPPRLDHVAVSLPGGRILIFGGSVAGLHSAS+LY+LDP
Sbjct: 505 KPCWRCITGSGMPGASNPAGVGPPPRLDHVAVSLPGGRILIFGGSVAGLHSASKLYLLDP 564
Query: 490 TDEKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSL 543
T+EKPTWRILNVPGRPPRFAWGHSTCVVGGT+AIVLGGQTGEEW L++LHELSL
Sbjct: 565 TEEKPTWRILNVPGRPPRFAWGHSTCVVGGTKAIVLGGQTGEEWTLTELHELSL 618
>UniRef100_Q8W420 LOV kelch protein 2 [Arabidopsis thaliana]
Length = 611
Score = 846 bits (2186), Expect = 0.0
Identities = 405/534 (75%), Positives = 464/534 (86%), Gaps = 11/534 (2%)
Query: 14 RFLQCRGPFAKRRHPLVDSSVISEIRKCIDEGVEFQGELLNFRKDGSPLMNRLRLTPIYG 73
RFLQCRGPF KRRHP+VDS++++++R+C++ G+EFQGELLNFRKDGSPLMN+LRL PI
Sbjct: 83 RFLQCRGPFTKRRHPMVDSTIVAKMRQCLENGIEFQGELLNFRKDGSPLMNKLRLVPIR- 141
Query: 74 EDDEITHVIGIQLFTEANIDLGPLPGSTIKESLKSSGRFHSVLSSLQPPPLGDRNVSRGI 133
E+DEITH IG+ LFT+A IDLGP P + KE + S F S L P+G+RNVSRG+
Sbjct: 142 EEDEITHFIGVLLFTDAKIDLGPSPDLSAKEIPRISRSFTSAL------PIGERNVSRGL 195
Query: 134 CGIFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGSETTRVL 193
CGIF+LSDEV+++KIL++LTP DIASV VC RL E+T+N+D+WRMVCQN WG+E TRVL
Sbjct: 196 CGIFELSDEVIAIKILSQLTPGDIASVGCVCRRLNELTKNDDVWRMVCQNTWGTEATRVL 255
Query: 194 ETVPGAKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVN 253
E+VPGAKR+GW RLARE TT EA AWRK +VGG VEPSRCNFSACAVGNR+V+FGGEGVN
Sbjct: 256 ESVPGAKRIGWVRLAREFTTHEATAWRKFSVGGTVEPSRCNFSACAVGNRIVIFGGEGVN 315
Query: 254 MQPMNDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVF 313
MQPMNDTFVLDL S++PEW+ V VSSPPPGRWGHTLSCVNGSRLVVFGG G+ GLLNDVF
Sbjct: 316 MQPMNDTFVLDLGSSSPEWKSVLVSSPPPGRWGHTLSCVNGSRLVVFGGYGSHGLLNDVF 375
Query: 314 VLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSME 373
+LDLDA PP+WRE+SGLAPP+PRSWHSSCTLDGTKLIVSGGCADSG LLSDTFLLD+SM+
Sbjct: 376 LLDLDADPPSWREVSGLAPPIPRSWHSSCTLDGTKLIVSGGCADSGALLSDTFLLDLSMD 435
Query: 374 NPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCW 433
P WREIPV WTPPSRLGHTL+VYG RKILMFGGLAK+G LRFRS+DV+TMDLSEDEP W
Sbjct: 436 IPAWREIPVPWTPPSRLGHTLTVYGDRKILMFGGLAKNGTLRFRSNDVYTMDLSEDEPSW 495
Query: 434 RCVTGSGMPGAGNPEGI-APPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDE 492
R V G G+ P G+ APPPRLDHVA+SLPGGRILIFGGSVAGL SASQLY+LDP +E
Sbjct: 496 RPVIGY---GSSLPGGMAAPPPRLDHVAISLPGGRILIFGGSVAGLDSASQLYLLDPNEE 552
Query: 493 KPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSLANS 546
KP WRILNV G PPRFAWGH+TCVVGGTR +VLGGQTGEEWML++ HEL LA S
Sbjct: 553 KPAWRILNVQGGPPRFAWGHTTCVVGGTRLVVLGGQTGEEWMLNEAHELLLATS 606
Score = 71.2 bits (173), Expect = 7e-11
Identities = 55/177 (31%), Positives = 81/177 (45%), Gaps = 19/177 (10%)
Query: 373 ENPVWREIPVTWT-PPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEP 431
E WR+ V T PSR + G R I++FGG G +D F +DL P
Sbjct: 277 EATAWRKFSVGGTVEPSRCNFSACAVGNR-IVIFGG---EGVNMQPMNDTFVLDLGSSSP 332
Query: 432 CWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSA-SQLYILDPT 490
W+ V S PP R H + G R+++FGG G H + +++LD
Sbjct: 333 EWKSVLVSS----------PPPGRWGHTLSCVNGSRLVVFGGY--GSHGLLNDVFLLDLD 380
Query: 491 DEKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSLANSV 547
+ P+WR ++ P +W HS+C + GT+ IV GG +LSD L L+ +
Sbjct: 381 ADPPSWREVSGLAPPIPRSW-HSSCTLDGTKLIVSGGCADSGALLSDTFLLDLSMDI 436
>UniRef100_Q9C5S6 Adagio 2 [Arabidopsis thaliana]
Length = 597
Score = 833 bits (2152), Expect = 0.0
Identities = 399/528 (75%), Positives = 458/528 (86%), Gaps = 11/528 (2%)
Query: 20 GPFAKRRHPLVDSSVISEIRKCIDEGVEFQGELLNFRKDGSPLMNRLRLTPIYGEDDEIT 79
GPF KRRHP+VDS++++++R+C++ G+EFQGELLNFRKDGSPLMN+LRL PI E+DEIT
Sbjct: 75 GPFTKRRHPMVDSTIVAKMRQCLENGIEFQGELLNFRKDGSPLMNKLRLVPIR-EEDEIT 133
Query: 80 HVIGIQLFTEANIDLGPLPGSTIKESLKSSGRFHSVLSSLQPPPLGDRNVSRGICGIFQL 139
H IG+ LFT+A IDLGP P + KE + S F S L P+G+RNVSRG+CGIF+L
Sbjct: 134 HFIGVLLFTDAKIDLGPSPDLSAKEIPRISRSFTSAL------PIGERNVSRGLCGIFEL 187
Query: 140 SDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGSETTRVLETVPGA 199
SDEV+++KIL++LTP DIASV VC RL E+T+N+D+WRMVCQN WG+E TRVLE+VPGA
Sbjct: 188 SDEVIAIKILSQLTPGDIASVGCVCRRLNELTKNDDVWRMVCQNTWGTEATRVLESVPGA 247
Query: 200 KRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMND 259
KR+GW RLARE TT EA AWRK +VGG VEPSRCNFSACAVGNR+V+FGGEGVNMQPMND
Sbjct: 248 KRIGWVRLAREFTTHEATAWRKFSVGGTVEPSRCNFSACAVGNRIVIFGGEGVNMQPMND 307
Query: 260 TFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDA 319
TFVLDL S++PEW+ V VSSPPPGRWGHTLSCVNGSRLVVFGG G+ GLLNDVF+LDLDA
Sbjct: 308 TFVLDLGSSSPEWKSVLVSSPPPGRWGHTLSCVNGSRLVVFGGYGSHGLLNDVFLLDLDA 367
Query: 320 TPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWRE 379
PP+WRE+SGLAPP+PRSWHSSCTLDGTKLIVSGGCADSG LLSDTFLLD+SM+ P WRE
Sbjct: 368 DPPSWREVSGLAPPIPRSWHSSCTLDGTKLIVSGGCADSGALLSDTFLLDLSMDIPAWRE 427
Query: 380 IPVTWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGS 439
IPV WTPPSRLGHTL+VYG RKILMFGGLAK+G LRFRS+DV+TMDLSEDEP WR V G
Sbjct: 428 IPVPWTPPSRLGHTLTVYGDRKILMFGGLAKNGTLRFRSNDVYTMDLSEDEPSWRPVIGY 487
Query: 440 GMPGAGNPEGI-APPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWRI 498
G+ P G+ APPPRLDHVA+SLPGGRILIFGGSVAGL SASQLY+LDP +EKP WRI
Sbjct: 488 ---GSSLPGGMAAPPPRLDHVAISLPGGRILIFGGSVAGLDSASQLYLLDPNEEKPAWRI 544
Query: 499 LNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSLANS 546
LNV G PPRFAWGH+TCVVGGTR +VLGGQTGEEWML++ HEL LA S
Sbjct: 545 LNVQGGPPRFAWGHTTCVVGGTRLVVLGGQTGEEWMLNEAHELLLATS 592
Score = 71.2 bits (173), Expect = 7e-11
Identities = 55/177 (31%), Positives = 81/177 (45%), Gaps = 19/177 (10%)
Query: 373 ENPVWREIPVTWT-PPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEP 431
E WR+ V T PSR + G R I++FGG G +D F +DL P
Sbjct: 263 EATAWRKFSVGGTVEPSRCNFSACAVGNR-IVIFGG---EGVNMQPMNDTFVLDLGSSSP 318
Query: 432 CWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSA-SQLYILDPT 490
W+ V S PP R H + G R+++FGG G H + +++LD
Sbjct: 319 EWKSVLVSS----------PPPGRWGHTLSCVNGSRLVVFGGY--GSHGLLNDVFLLDLD 366
Query: 491 DEKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSLANSV 547
+ P+WR ++ P +W HS+C + GT+ IV GG +LSD L L+ +
Sbjct: 367 ADPPSWREVSGLAPPIPRSW-HSSCTLDGTKLIVSGGCADSGALLSDTFLLDLSMDI 422
>UniRef100_Q9C9W9 Hypothetical protein T23K23.10 [Arabidopsis thaliana]
Length = 619
Score = 729 bits (1883), Expect = 0.0
Identities = 347/532 (65%), Positives = 422/532 (79%), Gaps = 10/532 (1%)
Query: 14 RFLQCRGPFAKRRHPLVDSSVISEIRKCIDEGVEFQGELLNFRKDGSPLMNRLRLTPIYG 73
RFLQ R P A+RRHPLVD V+SEIR+C++EG+EFQGELLNFRKDG+PL+NRLRL PI
Sbjct: 92 RFLQYRDPRAQRRHPLVDPVVVSEIRRCLEEGIEFQGELLNFRKDGTPLVNRLRLAPIRD 151
Query: 74 EDDEITHVIGIQLFTEANIDLGPLPGSTIKESLKSSGRFHSVLSSLQPPPLGDRNVSRGI 133
+D ITHVIGIQ+F+E IDL + K + + S P +
Sbjct: 152 DDGTITHVIGIQVFSETTIDLDRVSYPVFKHKQQLDQTSECLFPSGSPR---FKEHHEDF 208
Query: 134 CGIFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGSETTRVL 193
CGI QLSDEVL+ IL+RLTPRD+AS+ S C RL ++T+NE + +MVCQNAWG E T L
Sbjct: 209 CGILQLSDEVLAHNILSRLTPRDVASIGSACRRLRQLTKNESVRKMVCQNAWGKEITGTL 268
Query: 194 ETVPGAKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVN 253
E + K+L WGRLARELTTLEA WRK TVGG V+PSRCNFSACAVGNR+VLFGGEGVN
Sbjct: 269 EIM--TKKLRWGRLARELTTLEAVCWRKFTVGGIVQPSRCNFSACAVGNRLVLFGGEGVN 326
Query: 254 MQPMNDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVF 313
MQP++DTFVL+L++ PEWQ V+V+S PPGRWGHTLSC+NGS LVVFGGCG QGLLNDVF
Sbjct: 327 MQPLDDTFVLNLDAECPEWQRVRVTSSPPGRWGHTLSCLNGSWLVVFGGCGRQGLLNDVF 386
Query: 314 VLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSME 373
VLDLDA PTW+E++G PPLPRSWHSSCT++G+KL+VSGGC D+GVLLSDTFLLD++ +
Sbjct: 387 VLDLDAKHPTWKEVAGGTPPLPRSWHSSCTIEGSKLVVSGGCTDAGVLLSDTFLLDLTTD 446
Query: 374 NPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCW 433
P W+EIP +W PPSRLGH+LSV+G KILMFGGLA SG L+ RS + +T+DL ++EP W
Sbjct: 447 KPTWKEIPTSWAPPSRLGHSLSVFGRTKILMFGGLANSGHLKLRSGEAYTIDLEDEEPRW 506
Query: 434 RCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEK 493
R + S PG + PPPRLDHVAVS+P GR++IFGGS+AGLHS SQL+++DP +EK
Sbjct: 507 RELECSAFPGV-----VVPPPRLDHVAVSMPCGRVIIFGGSIAGLHSPSQLFLIDPAEEK 561
Query: 494 PTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSLAN 545
P+WRILNVPG+PP+ AWGHSTCVVGGTR +VLGG TGEEW+L++LHEL LA+
Sbjct: 562 PSWRILNVPGKPPKLAWGHSTCVVGGTRVLVLGGHTGEEWILNELHELCLAS 613
>UniRef100_Q9M648 FKF1 [Arabidopsis thaliana]
Length = 619
Score = 726 bits (1874), Expect = 0.0
Identities = 345/532 (64%), Positives = 421/532 (78%), Gaps = 10/532 (1%)
Query: 14 RFLQCRGPFAKRRHPLVDSSVISEIRKCIDEGVEFQGELLNFRKDGSPLMNRLRLTPIYG 73
RFLQ R P A+RRHPLVD V+SEIR+C++EG+EFQGELLNFRKDG+PL+NRLRL PI
Sbjct: 92 RFLQYRDPRAQRRHPLVDPVVVSEIRRCLEEGIEFQGELLNFRKDGTPLVNRLRLAPIRD 151
Query: 74 EDDEITHVIGIQLFTEANIDLGPLPGSTIKESLKSSGRFHSVLSSLQPPPLGDRNVSRGI 133
+D ITHVIGIQ+F+E IDL + K + + S P +
Sbjct: 152 DDGTITHVIGIQVFSETTIDLDRVSYPVFKHKQQLDQTSECLFPSGSPR---FKEHHEDF 208
Query: 134 CGIFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGSETTRVL 193
CGI QLSDEVL+ IL+RLTPRD+AS+ S C RL ++T+NE + +MVCQNAWG E T L
Sbjct: 209 CGILQLSDEVLAHNILSRLTPRDVASIGSACRRLRQLTKNESVRKMVCQNAWGKEITGTL 268
Query: 194 ETVPGAKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVN 253
E + K+L WGRLARELTTLEA WRK TVGG V+PSRCNFSACAVGNR+VLFGGEGVN
Sbjct: 269 EIM--TKKLRWGRLARELTTLEAVCWRKFTVGGIVQPSRCNFSACAVGNRLVLFGGEGVN 326
Query: 254 MQPMNDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVF 313
MQP++DTFVL+L++ PEWQ V+V+S PPGRWGHTLSC+NGS LVVFGGCG QGLLNDVF
Sbjct: 327 MQPLDDTFVLNLDAECPEWQRVRVTSSPPGRWGHTLSCLNGSWLVVFGGCGRQGLLNDVF 386
Query: 314 VLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSME 373
VLDLDA PTW+E++G PPLPRSWHSSCT++G+KL+VSGGC D+GVLLSDTFLLD++ +
Sbjct: 387 VLDLDAKHPTWKEVAGGTPPLPRSWHSSCTIEGSKLVVSGGCTDAGVLLSDTFLLDLTTD 446
Query: 374 NPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCW 433
P W+EIP +W PPSRLGH+LSV+G KILMFGGLA G L+ RS + +T+DL ++EP W
Sbjct: 447 KPTWKEIPTSWAPPSRLGHSLSVFGRTKILMFGGLANIGHLKLRSGEAYTIDLEDEEPRW 506
Query: 434 RCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEK 493
R + S PG + PPPRLDHVAVS+P GR++IFGGS+AGLHS SQL+++DP +EK
Sbjct: 507 RELECSAFPGV-----VVPPPRLDHVAVSMPCGRVIIFGGSIAGLHSPSQLFLIDPAEEK 561
Query: 494 PTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSLAN 545
P+WRILNVPG+PP+ AWGH+TCVVGGTR +VLGG TGEEW+L++LHEL LA+
Sbjct: 562 PSWRILNVPGKPPKLAWGHNTCVVGGTRVLVLGGHTGEEWILNELHELCLAS 613
>UniRef100_Q6UEI4 FKF1 [Mesembryanthemum crystallinum]
Length = 634
Score = 716 bits (1847), Expect = 0.0
Identities = 340/533 (63%), Positives = 416/533 (77%), Gaps = 11/533 (2%)
Query: 14 RFLQCRGPFAKRRHPLVDSSVISEIRKCIDEGVEFQGELLNFRKDGSPLMNRLRLTPIYG 73
RFLQ P A+RRHPLVD ++SE+R+C++E V+F+GELLNF+KDG+PL+NRLRL PI+
Sbjct: 106 RFLQYMDPHAQRRHPLVDPVMVSEMRRCLEEAVQFEGELLNFKKDGTPLVNRLRLQPIHA 165
Query: 74 EDDEITHVIGIQLFTEANIDLGPLPGSTIKESLKSSGRFHSVLSSLQPPPLGDRNVSRGI 133
+D ITHVIGIQ+F+EA ++L + KE K +F GD
Sbjct: 166 DDGTITHVIGIQIFSEAKVNLNHVSYPVYKE--KCDHQFDCADDDHPTKDGGD------F 217
Query: 134 CGIFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGSETTRVL 193
CGI QLSDEVL+ IL+RLTPRD+AS+ SVC R+Y++T+NE + +MVCQN WG E L
Sbjct: 218 CGILQLSDEVLAHNILSRLTPRDVASIGSVCRRIYQLTKNELVRKMVCQNLWGREVIGNL 277
Query: 194 ETVPGAKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVN 253
E + K+LGW RLARELTTLEA WRK TVGG VEPSRCNFSACA GNR+VLFGGEGV+
Sbjct: 278 ELM--TKKLGWVRLARELTTLEAVRWRKFTVGGAVEPSRCNFSACAAGNRLVLFGGEGVD 335
Query: 254 MQPMNDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVF 313
MQPM+DTFVL+L++ +PEW+ V V S PPGRWGHTLSC+NGS LVVFGGCG QGLLNDVF
Sbjct: 336 MQPMDDTFVLNLDAEHPEWRRVSVKSSPPGRWGHTLSCLNGSWLVVFGGCGKQGLLNDVF 395
Query: 314 VLDLDATPPTWREI-SGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSM 372
V+DLDA PTW EI G PP+PRSWHSSCT+DG+KL+VSGGC+DSGVLLSDT LLD+
Sbjct: 396 VIDLDAKQPTWTEIPGGSTPPVPRSWHSSCTIDGSKLVVSGGCSDSGVLLSDTHLLDLMT 455
Query: 373 ENPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPC 432
P WREIP +W PPSRLGH+L+VYG I MFGGL KSG LR RS + +T+D+ D P
Sbjct: 456 NTPAWREIPTSWAPPSRLGHSLTVYGKTNIFMFGGLVKSGQLRLRSGEAYTIDIGNDNPR 515
Query: 433 WRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDE 492
WR + G G G+ PPPRLDHVAVS+P GRI+IFGGS+AGLHS+SQ++++DP++E
Sbjct: 516 WRQLECGAHAGGGTQGGVVPPPRLDHVAVSMPCGRIIIFGGSIAGLHSSSQIFLIDPSEE 575
Query: 493 KPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSLAN 545
KP+WRI+NVPG+PP+FAWGHSTCVVGGTR +VLGG TGEEW+L++LHEL LA+
Sbjct: 576 KPSWRIINVPGQPPKFAWGHSTCVVGGTRVLVLGGHTGEEWILNELHELCLAS 628
>UniRef100_Q6Z1R8 Putative F-box protein [Oryza sativa]
Length = 448
Score = 159 bits (401), Expect = 3e-37
Identities = 123/418 (29%), Positives = 194/418 (45%), Gaps = 42/418 (10%)
Query: 141 DEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGSETTRVL--ETVPG 198
D+VLS IL L + S ++ C + ++ LW +C+ WG+ T L G
Sbjct: 13 DQVLS--ILHLLPAESVLSFAAACRAFHAWASSDALWEALCRRDWGARATAALAERRRRG 70
Query: 199 AKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMN 258
+ W R+ E+ L A + R++ V G R + S V +VLFGG + ++
Sbjct: 71 GGGVPWRRIYAEVALLGALSARRVPVKGASPRPRASHSLNLVAGWLVLFGGGCEGGRHLD 130
Query: 259 DTFVLDLNSNNPE-------WQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGL-LN 310
DT+V + + WQ + S P GR+GH+ S V G LV+FGG QG LN
Sbjct: 131 DTWVAYVGNGAGNRSSAVFSWQQLD-SGTPSGRFGHSCSIV-GDALVLFGGINDQGQRLN 188
Query: 311 DVFVLDLDATPP-----TWREIS-GLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSD 364
D ++ + +WR + G P PR H++C +D +++ GG SG L D
Sbjct: 189 DTWIGQIICEESRRMKISWRLLEVGPHAPYPRGAHAACCVDDKFIVIHGGIGQSGSRLGD 248
Query: 365 TFLLDMS--MENPVWREIPVTW-TPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDV 421
T+LLD+S + + +W +I T P SR GHTL+ GG ++++FGG + +DV
Sbjct: 249 TWLLDLSNGLRSGIWHQIEDTEPLPLSRSGHTLTWIGGSRMVLFGGRGSEFDV---LNDV 305
Query: 422 FTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSA 481
+ +D++E P W+ + + G P PR+ H A + GG+IL++GG +
Sbjct: 306 WLLDINERYPKWKELKYD----LSSVLGEMPFPRVGHSATLVLGGKILVYGGEDSQRRRK 361
Query: 482 SQLYILD-----------PTDEKPTWRILNVPGRPPRFAWGHSTCV-VGGTRAIVLGG 527
+ LD K W+ L + G+ P + H CV G + GG
Sbjct: 362 DDFWTLDLPALLQFESGSKKMTKRMWKKLRIDGQCPNYRSFHGACVDTSGCHVYIFGG 419
Score = 53.9 bits (128), Expect = 1e-05
Identities = 58/227 (25%), Positives = 96/227 (41%), Gaps = 29/227 (12%)
Query: 331 APPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLL-------DMSMENPVWREIPVT 383
A P PR+ HS + G ++ GGC + G L DT++ + S W+++ +
Sbjct: 99 ASPRPRASHSLNLVAGWLVLFGGGC-EGGRHLDDTWVAYVGNGAGNRSSAVFSWQQLD-S 156
Query: 384 WTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEP-----CWRCVTG 438
TP R GH+ S+ G +L FGG+ G R +D + + +E WR +
Sbjct: 157 GTPSGRFGHSCSIVGDALVL-FGGINDQGQ---RLNDTWIGQIICEESRRMKISWRLLEV 212
Query: 439 SGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTD--EKPTW 496
AP PR H A + I+I GG ++LD ++ W
Sbjct: 213 GPH---------APYPRGAHAACCVDDKFIVIHGGIGQSGSRLGDTWLLDLSNGLRSGIW 263
Query: 497 RILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSL 543
+ P GH+ +GG+R ++ GG+ E +L+D+ L +
Sbjct: 264 HQIEDTEPLPLSRSGHTLTWIGGSRMVLFGGRGSEFDVLNDVWLLDI 310
>UniRef100_Q8LAD5 F-box protein ZEITLUPE/FKF/LKP/ADAGIO family [Arabidopsis thaliana]
Length = 478
Score = 129 bits (324), Expect = 2e-28
Identities = 106/379 (27%), Positives = 173/379 (44%), Gaps = 27/379 (7%)
Query: 128 NVSRGICGIFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGS 187
+ SR I L D+ L L IL L I S S C R + ++ LW +C+ WG
Sbjct: 13 STSRSSSPIINLPDDHL-LTILLLLPVDSILSFSMTCKRYKSLACSDSLWEALCEREWGP 71
Query: 188 ETTRVLETVPGAKRLGWGRLARELTTLEAAAWRKLT-------VGGGVEPSRCNFSACAV 240
+ L+ W + + + +++ K++ R + S V
Sbjct: 72 TSVDALKLSSLRDGFSWMLMFQRVYKMDSVCCHKISDPDDDDEESSSFPIPRASHSLNFV 131
Query: 241 GNRVVLFGGEGVNMQPMNDTFVLDLNSNNP---EWQHVQVSSPPPGRWGHTLSCVNGSRL 297
+ +VLFGG + ++DT+ ++ +N +W+ V+ S P GR+GHT + G L
Sbjct: 132 NDHLVLFGGGCQGGRHLDDTWTSYVDKSNQSILKWKKVK-SGTPSGRFGHTCIVI-GEYL 189
Query: 298 VVFGGCGTQG-LLNDVFVLDLDATPP-TWR--EISGLAPPLP--RSWHSSCTLDGTKLIV 351
++FGG +G LND ++ + +W+ + L P P R HS+C + K++V
Sbjct: 190 LLFGGINDRGERLNDTWIGQVFCHEGLSWKLLNVGSLQRPRPPPRGAHSACCIAEKKMVV 249
Query: 352 SGGCADSGVLLSDTFLLDMS--MENPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGLA 409
GG +GV L DT++L++S + W + PP R GHTL+ ++++FGG
Sbjct: 250 HGGIGQNGVRLGDTWILELSEDFSSGTWHMVESPQLPPPRSGHTLTCIRENQVVLFGGRG 309
Query: 410 KSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRIL 469
+ DV+ +D+ EPC PE A PR+ H A + GGRIL
Sbjct: 310 LGYDV---LDDVWILDI--QEPCEEKWIQIFYDFQDVPE-YASLPRVGHSATLVLGGRIL 363
Query: 470 IFGGSVAGLHSASQLYILD 488
I+GG + H ++LD
Sbjct: 364 IYGGEDSYRHRKDDFWVLD 382
>UniRef100_Q681W1 Hypothetical protein At1g51550 [Arabidopsis thaliana]
Length = 476
Score = 126 bits (316), Expect = 2e-27
Identities = 105/379 (27%), Positives = 173/379 (44%), Gaps = 27/379 (7%)
Query: 128 NVSRGICGIFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGS 187
+ S+ I L D+ L L IL L I S S C R + ++ LW +C+ WG
Sbjct: 11 STSQSSSPIINLPDDHL-LTILLLLPVDSILSFSMTCKRYKSLACSDSLWEALCEREWGP 69
Query: 188 ETTRVLETVPGAKRLGWGRLARELTTLEAAAWRKLT-------VGGGVEPSRCNFSACAV 240
+ L+ W + + + +++ K++ R + S V
Sbjct: 70 TSVDALKLSSLRDGFSWMLMFQRVYKMDSVCCHKISDPDDDDEESSSFPIPRASHSLNFV 129
Query: 241 GNRVVLFGGEGVNMQPMNDTFVLDLNSNNP---EWQHVQVSSPPPGRWGHTLSCVNGSRL 297
+ +VLFGG + ++DT+ ++ +N +W+ V+ S P GR+GHT + G L
Sbjct: 130 NDHLVLFGGGCQGGRHLDDTWTSYVDKSNQSILKWKKVK-SGTPSGRFGHTCIVI-GEYL 187
Query: 298 VVFGGCGTQG-LLNDVFVLDLDATPP-TWR--EISGLAPPLP--RSWHSSCTLDGTKLIV 351
++FGG +G LND ++ + +W+ + L P P R HS+C + K++V
Sbjct: 188 LLFGGINDRGERLNDTWIGQVFCHEGLSWKLLNVGSLQRPRPPPRGAHSACCIAEKKMVV 247
Query: 352 SGGCADSGVLLSDTFLLDMS--MENPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGLA 409
GG +GV L DT++L++S + W + PP R GHTL+ ++++FGG
Sbjct: 248 HGGIGLNGVRLGDTWILELSEDFSSGTWHMVESPQLPPPRSGHTLTCIRENQVVLFGGRG 307
Query: 410 KSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRIL 469
+ DV+ +D+ EPC PE A PR+ H A + GGRIL
Sbjct: 308 LGYDV---LDDVWILDI--QEPCEEKWIQIFYDFQDVPE-YASLPRVGHSATLVLGGRIL 361
Query: 470 IFGGSVAGLHSASQLYILD 488
I+GG + H ++LD
Sbjct: 362 IYGGEDSYRHRKDDFWVLD 380
>UniRef100_Q9C8K7 Hypothetical protein F5D21.4 [Arabidopsis thaliana]
Length = 478
Score = 126 bits (316), Expect = 2e-27
Identities = 105/379 (27%), Positives = 173/379 (44%), Gaps = 27/379 (7%)
Query: 128 NVSRGICGIFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGS 187
+ S+ I L D+ L L IL L I S S C R + ++ LW +C+ WG
Sbjct: 13 STSQSSSPIINLPDDHL-LTILLLLPVDSILSFSMTCKRYKSLACSDSLWEALCEREWGP 71
Query: 188 ETTRVLETVPGAKRLGWGRLARELTTLEAAAWRKLT-------VGGGVEPSRCNFSACAV 240
+ L+ W + + + +++ K++ R + S V
Sbjct: 72 TSVDALKLSSLRDGFSWMLMFQRVYKMDSVCCHKISDPDDDDEESSSFPIPRASHSLNFV 131
Query: 241 GNRVVLFGGEGVNMQPMNDTFVLDLNSNNP---EWQHVQVSSPPPGRWGHTLSCVNGSRL 297
+ +VLFGG + ++DT+ ++ +N +W+ V+ S P GR+GHT + G L
Sbjct: 132 NDHLVLFGGGCQGGRHLDDTWTSYVDKSNQSILKWKKVK-SGTPSGRFGHTCIVI-GEYL 189
Query: 298 VVFGGCGTQG-LLNDVFVLDLDATPP-TWR--EISGLAPPLP--RSWHSSCTLDGTKLIV 351
++FGG +G LND ++ + +W+ + L P P R HS+C + K++V
Sbjct: 190 LLFGGINDRGERLNDTWIGQVFCHEGLSWKLLNVGSLQRPRPPPRGAHSACCIAEKKMVV 249
Query: 352 SGGCADSGVLLSDTFLLDMS--MENPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGLA 409
GG +GV L DT++L++S + W + PP R GHTL+ ++++FGG
Sbjct: 250 HGGIGLNGVRLGDTWILELSEDFSSGTWHMVESPQLPPPRSGHTLTCIRENQVVLFGGRG 309
Query: 410 KSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRIL 469
+ DV+ +D+ EPC PE A PR+ H A + GGRIL
Sbjct: 310 LGYDV---LDDVWILDI--QEPCEEKWIQIFYDFQDVPE-YASLPRVGHSATLVLGGRIL 363
Query: 470 IFGGSVAGLHSASQLYILD 488
I+GG + H ++LD
Sbjct: 364 IYGGEDSYRHRKDDFWVLD 382
>UniRef100_Q9FMZ1 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MJB21
[Arabidopsis thaliana]
Length = 95
Score = 120 bits (300), Expect = 1e-25
Identities = 51/72 (70%), Positives = 63/72 (86%)
Query: 474 SVAGLHSASQLYILDPTDEKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEW 533
SV GLHS S L+++DP+ EKP+WRILNVPG+PP+ AWGHSTCVVGGTR +VLGG GEEW
Sbjct: 18 SVEGLHSPSLLFLIDPSKEKPSWRILNVPGKPPKLAWGHSTCVVGGTRVLVLGGHNGEEW 77
Query: 534 MLSDLHELSLAN 545
+L++LHEL LA+
Sbjct: 78 ILNELHELCLAS 89
Score = 41.2 bits (95), Expect = 0.079
Identities = 21/59 (35%), Positives = 34/59 (57%), Gaps = 2/59 (3%)
Query: 261 FVLDLNSNNPEWQHVQVSSPPPG-RWGHTLSCVNGSRLVVFGG-CGTQGLLNDVFVLDL 317
F++D + P W+ + V PP WGH+ V G+R++V GG G + +LN++ L L
Sbjct: 29 FLIDPSKEKPSWRILNVPGKPPKLAWGHSTCVVGGTRVLVLGGHNGEEWILNELHELCL 87
Score = 39.3 bits (90), Expect = 0.30
Identities = 19/43 (44%), Positives = 25/43 (57%), Gaps = 1/43 (2%)
Query: 366 FLLDMSMENPVWREIPVTWTPPS-RLGHTLSVYGGRKILMFGG 407
FL+D S E P WR + V PP GH+ V GG ++L+ GG
Sbjct: 29 FLIDPSKEKPSWRILNVPGKPPKLAWGHSTCVVGGTRVLVLGG 71
Score = 35.4 bits (80), Expect = 4.3
Identities = 15/44 (34%), Positives = 27/44 (61%), Gaps = 1/44 (2%)
Query: 312 VFVLDLDATPPTWREISGLAPPLPRSW-HSSCTLDGTKLIVSGG 354
+F++D P+WR ++ P +W HS+C + GT+++V GG
Sbjct: 28 LFLIDPSKEKPSWRILNVPGKPPKLAWGHSTCVVGGTRVLVLGG 71
>UniRef100_Q9FHL7 Arabidopsis thaliana genomic DNA, chromosome 5, TAC clone:K19M13
[Arabidopsis thaliana]
Length = 84
Score = 104 bits (259), Expect = 8e-21
Identities = 43/58 (74%), Positives = 52/58 (89%)
Query: 488 DPTDEKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSLAN 545
DP+ EKP+WRILNVPG+PP+ AWGHSTCVVGGTR +VLGG GEEW+L++LHEL LA+
Sbjct: 21 DPSKEKPSWRILNVPGKPPKLAWGHSTCVVGGTRVLVLGGHNGEEWILNELHELCLAS 78
Score = 38.1 bits (87), Expect = 0.67
Identities = 21/58 (36%), Positives = 32/58 (54%), Gaps = 2/58 (3%)
Query: 262 VLDLNSNNPEWQHVQVSSPPPG-RWGHTLSCVNGSRLVVFGG-CGTQGLLNDVFVLDL 317
V D + P W+ + V PP WGH+ V G+R++V GG G + +LN++ L L
Sbjct: 19 VEDPSKEKPSWRILNVPGKPPKLAWGHSTCVVGGTRVLVLGGHNGEEWILNELHELCL 76
Score = 34.7 bits (78), Expect = 7.4
Identities = 17/40 (42%), Positives = 22/40 (54%), Gaps = 1/40 (2%)
Query: 369 DMSMENPVWREIPVTWTPPSRL-GHTLSVYGGRKILMFGG 407
D S E P WR + V PP GH+ V GG ++L+ GG
Sbjct: 21 DPSKEKPSWRILNVPGKPPKLAWGHSTCVVGGTRVLVLGG 60
>UniRef100_Q94D04 P0682B08.13 protein [Oryza sativa]
Length = 624
Score = 89.7 bits (221), Expect = 2e-16
Identities = 92/290 (31%), Positives = 128/290 (43%), Gaps = 31/290 (10%)
Query: 214 LEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQ 273
LE AW L G +R + A VG+R+++FGG + +ND VLDL + EW
Sbjct: 54 LETMAWSSLATTGARPGTRDSHGAALVGHRMMVFGGTN-GSKKVNDLHVLDLRTK--EWT 110
Query: 274 HVQVSSPPPG-RWGHTLSCVNG-SRLVVFGGC--GTQGLLNDVFVLDLDATPPTWREISG 329
PP R HT++ G RLVVFGG G LNDV VLD+ + E+ G
Sbjct: 111 KPPCKGTPPSPRESHTVTACGGCDRLVVFGGSGEGEGNYLNDVHVLDVATMTWSSPEVKG 170
Query: 330 LAPPLPRSWHSSCTLDGTKLIVSGG-CADSGVLLSDTFLLDMSMENPVWREIPVTWTPPS 388
P PR H + + G++L+V GG C D + +LD M+ W V P
Sbjct: 171 DVVPAPRDSHGAVAV-GSRLVVYGGDCGDR--YHGEVDVLD--MDAMAWSRFAVKGASPG 225
Query: 389 -RLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNP 447
R GH +V G K+ + GG+ + SD + +D++ W + G P
Sbjct: 226 VRAGHA-AVGVGSKVYVIGGVGD----KQYYSDAWILDVANRS--WTQLEICGQ----QP 274
Query: 448 EGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWR 497
+G R H AV L I I+GG ++L IL E P R
Sbjct: 275 QG-----RFSHSAVVL-NTDIAIYGGCGEDERPLNELLILQLGSEHPNGR 318
Score = 83.2 bits (204), Expect = 2e-14
Identities = 93/331 (28%), Positives = 138/331 (41%), Gaps = 27/331 (8%)
Query: 217 AAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQ 276
A W V G P R SAC V +FGG ++ + VL LN W +
Sbjct: 7 AMWLYPKVVGFNPPERWGHSACFFEGVVYVFGG---CCGGLHFSDVLTLNLETMAWSSLA 63
Query: 277 VSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISGLAPPLPR 336
+ PG + + G R++VFGG +ND+ VLDL T G PP PR
Sbjct: 64 TTGARPGTRDSHGAALVGHRMMVFGGTNGSKKVNDLHVLDLRTKEWTKPPCKG-TPPSPR 122
Query: 337 SWHSSCTLDG-TKLIVSGGCAD-SGVLLSDTFLLDMSMENPVWREIPVTWTPPSRLGHTL 394
H+ G +L+V GG + G L+D +LD++ E+ P R H
Sbjct: 123 ESHTVTACGGCDRLVVFGGSGEGEGNYLNDVHVLDVATMTWSSPEVKGDVVPAPRDSHG- 181
Query: 395 SVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPP 454
+V G +++++GG G DV MD W +G +P
Sbjct: 182 AVAVGSRLVVYGG--DCGDRYHGEVDVLDMDAM----AWSRFA---------VKGASPGV 226
Query: 455 RLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWRILNVPGRPPRFAWGHST 514
R H AV + G ++ + GG V S +ILD + +W L + G+ P+ + HS
Sbjct: 227 RAGHAAVGV-GSKVYVIGG-VGDKQYYSDAWILDVANR--SWTQLEICGQQPQGRFSHS- 281
Query: 515 CVVGGTRAIVLGGQTGEEWMLSDLHELSLAN 545
VV T + GG +E L++L L L +
Sbjct: 282 AVVLNTDIAIYGGCGEDERPLNELLILQLGS 312
Score = 58.9 bits (141), Expect = 4e-07
Identities = 36/131 (27%), Positives = 66/131 (49%), Gaps = 6/131 (4%)
Query: 210 ELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNN 269
++ ++A AW + V G R +A VG++V + GG G + Q +D ++LD+ N
Sbjct: 205 DVLDMDAMAWSRFAVKGASPGVRAGHAAVGVGSKVYVIGGVG-DKQYYSDAWILDV--AN 261
Query: 270 PEWQHVQV-SSPPPGRWGHTLSCVNGSRLVVFGGCG-TQGLLNDVFVLDLDATPPTWREI 327
W +++ P GR+ H+ +N + + ++GGCG + LN++ +L L + P R
Sbjct: 262 RSWTQLEICGQQPQGRFSHSAVVLN-TDIAIYGGCGEDERPLNELLILQLGSEHPNGRYN 320
Query: 328 SGLAPPLPRSW 338
+ L W
Sbjct: 321 ISMCKVLSNHW 331
>UniRef100_Q8RWL1 Hypothetical protein At2g36360 [Arabidopsis thaliana]
Length = 496
Score = 89.7 bits (221), Expect = 2e-16
Identities = 81/268 (30%), Positives = 115/268 (42%), Gaps = 35/268 (13%)
Query: 279 SPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLD-----ATPPTWREISGLAPP 333
+PP R GHT V S +VVFGG + L+D+ V D++ T E G P
Sbjct: 15 TPPQARSGHTAVNVGKSMVVVFGGLVDKKFLSDIIVYDIENKLWFEPECTGSESEGQVGP 74
Query: 334 LPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTW---TPPSRL 390
PR++H + T+D I G G L D ++LD +W+ +T P R
Sbjct: 75 TPRAFHVAITIDCHMFIFGG--RSGGKRLGDFWVLD----TDIWQWSELTSFGDLPTPRD 128
Query: 391 GHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGI 450
+ G +KI++ GG L SDV+ MD E V+GS
Sbjct: 129 FAAAAAIGSQKIVLCGGWDGKKWL----SDVYVMDTMSLEWLELSVSGS----------- 173
Query: 451 APPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLY----ILDPTDEKPTWRILNVPGRPP 506
PPPR H A ++ R+L+FGG G L+ ++D E P W L +PG+ P
Sbjct: 174 LPPPRCGHTA-TMVEKRLLVFGGRGGGGPIMGDLWALKGLIDEERETPGWTQLKLPGQAP 232
Query: 507 RFAWGHSTCVVGGTRAIVLGGQTGEEWM 534
GH T GG ++ GG W+
Sbjct: 233 SSRCGH-TVTSGGHYLLLFGGHGTGGWL 259
Score = 76.3 bits (186), Expect = 2e-12
Identities = 85/275 (30%), Positives = 128/275 (45%), Gaps = 39/275 (14%)
Query: 198 GAKRLG--WGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQ 255
G KRLG W + + W +LT G + R +A A+G++ ++ G +
Sbjct: 97 GGKRLGDFW------VLDTDIWQWSELTSFGDLPTPRDFAAAAAIGSQKIVLCGGWDGKK 150
Query: 256 PMNDTFVLDLNSNNPEWQHVQVS-SPPPGRWGHTLSCVNGSRLVVFGGCGTQG-LLNDVF 313
++D +V+D S EW + VS S PP R GHT + V RL+VFGG G G ++ D++
Sbjct: 151 WLSDVYVMDTMS--LEWLELSVSGSLPPPRCGHTATMVE-KRLLVFGGRGGGGPIMGDLW 207
Query: 314 VL----DLDATPPTWREIS--GLAPPLPRSWHSSCTLDGTKLIVSGGCADSG------VL 361
L D + P W ++ G AP R H + T G L++ GG G V
Sbjct: 208 ALKGLIDEERETPGWTQLKLPGQAPS-SRCGH-TVTSGGHYLLLFGGHGTGGWLSRYDVY 265
Query: 362 LSDTFLLDMSMENPVWREIPV-TWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSD 420
+DT +LD W+ +P+ PP R HT++ G R +L+ GG G L F D
Sbjct: 266 YNDTIILDRVTAQ--WKRLPIGNEPPPPRAYHTMTCIGARHLLI-GGF--DGKLTF--GD 318
Query: 421 VFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPR 455
++ + + ED+P S +P NP I R
Sbjct: 319 LWWL-VPEDDP---IAKRSSVPQVVNPPEIKESER 349
Score = 72.8 bits (177), Expect = 2e-11
Identities = 80/280 (28%), Positives = 128/280 (45%), Gaps = 35/280 (12%)
Query: 226 GGVEPS-RCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVSS----P 280
G V P+ R A + + +FGG + + D +VLD + WQ +++S P
Sbjct: 70 GQVGPTPRAFHVAITIDCHMFIFGGRSGGKR-LGDFWVLDTDI----WQWSELTSFGDLP 124
Query: 281 PPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISGLAPPLPRSWHS 340
P + + + ++V+ GG + L+DV+V+D + +SG PP PR H+
Sbjct: 125 TPRDFA-AAAAIGSQKIVLCGGWDGKKWLSDVYVMDTMSLEWLELSVSGSLPP-PRCGHT 182
Query: 341 SCTLDGTKLIVSGGCADSGVLLSDTF----LLDMSMENPVWREIPVTW-TPPSRLGHTLS 395
+ T+ +L+V GG G ++ D + L+D E P W ++ + P SR GHT++
Sbjct: 183 A-TMVEKRLLVFGGRGGGGPIMGDLWALKGLIDEERETPGWTQLKLPGQAPSSRCGHTVT 241
Query: 396 VYGGRKILMFGGLAKSGPLRFRSSDVF---TMDLSEDEPCWRCVTGSGMPGAGNPEGIAP 452
GG +L+FGG G L DV+ T+ L W+ +P P P
Sbjct: 242 S-GGHYLLLFGGHGTGGWLS--RYDVYYNDTIILDRVTAQWK-----RLPIGNEP----P 289
Query: 453 PPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDE 492
PPR H + G R L+ GG G + L+ L P D+
Sbjct: 290 PPRAYHTMTCI-GARHLLIGG-FDGKLTFGDLWWLVPEDD 327
>UniRef100_UPI00003C2253 UPI00003C2253 UniRef100 entry
Length = 1203
Score = 89.0 bits (219), Expect = 3e-16
Identities = 90/307 (29%), Positives = 136/307 (43%), Gaps = 35/307 (11%)
Query: 248 GGEGVNMQPMN-DTFVLDLNSN----NPEWQHVQV----SSPPPGRWGHTLSCVNGSRLV 298
GG N P N + LDL++N +P WQ + ++ PP + HT+S +N S L+
Sbjct: 74 GGYTYNSAPNNGQMYTLDLSTNFSLASPPWQTISTQIDSAAAPPVAF-HTISPLNNSALL 132
Query: 299 VFGGCG-----TQGLLNDVFVLDLDATPPTWREISGLAPPLPRSWH---------SSCTL 344
+FGG G Q + +++ LD P T R S P+P SW+ S
Sbjct: 133 LFGGDGAPIVPVQTGNDSAYIVHLDG-PATNRTAS--YQPVPTSWNQPMRRVHHTSESNA 189
Query: 345 DGTKLIVSGGCAD-SGVLLSDTFLLDMSMENPVWREIPVTWTPP--SRLGHTLSVYGGRK 401
GT I+ G AD SG++ + + +D + + + PP S +G T ++
Sbjct: 190 RGTVWILGGEKADGSGLIFDEQWNIDTTTISSSSPSFQLVSEPPSGSLVGSTSTMLSDGT 249
Query: 402 ILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAV 461
+L+ GGL SG L+ +++ S + + P PR H+AV
Sbjct: 250 LLLIGGLDVSGQLQ-SMQNLYAYSAKSRSWSQTATQSSTNATFSARQNLFPSPRRGHIAV 308
Query: 462 SLPGGRILIFGGSVAGLHSA-SQLYILDPTDEKPTWRILNVPGRP-PRFAWGHSTCVVGG 519
SLP R+ I GG+ A L + S +ILD + P W ++ G P PRF GHS G
Sbjct: 309 SLPNQRVFIQGGASADLSTVYSDAWILDWSVNPPVWTEVDSTGGPGPRF--GHSAVAYGR 366
Query: 520 TRAIVLG 526
I G
Sbjct: 367 QIVISFG 373
Score = 65.9 bits (159), Expect = 3e-09
Identities = 66/251 (26%), Positives = 107/251 (42%), Gaps = 31/251 (12%)
Query: 265 LNSNNPEWQHVQVSSPPPGRW-GHTLSCVNGSRLVVFGGCGTQGLLNDVFVL-DLDATPP 322
++S++P +Q VS PP G G T + ++ L++ GG G L + L A
Sbjct: 219 ISSSSPSFQ--LVSEPPSGSLVGSTSTMLSDGTLLLIGGLDVSGQLQSMQNLYAYSAKSR 276
Query: 323 TWREISGLAP------------PLPRSWHSSCTLDGTKLIVSGGC-ADSGVLLSDTFLLD 369
+W + + + P PR H + +L ++ + GG AD + SD ++LD
Sbjct: 277 SWSQTATQSSTNATFSARQNLFPSPRRGHIAVSLPNQRVFIQGGASADLSTVYSDAWILD 336
Query: 370 MSMENPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGL-AKSGPLRFRSSDVFTMDLSE 428
S+ PVW E+ T P R GH+ YG + ++ FG + + P D T+ S
Sbjct: 337 WSVNPPVWTEVDSTGGPGPRFGHSAVAYGRQIVISFGWAGSNAAPTPVIVFDASTLTSSA 396
Query: 429 DEPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSL----PGGRILIFGGSVAGLHSASQL 484
GS G+ + AP P++ + + P G G S G S+S
Sbjct: 397 SS------KGSWSGGSWSATTYAPDPQMTTASTTASSESPSGS--PNGSSANGSSSSSGG 448
Query: 485 YILDP-TDEKP 494
+ P +D KP
Sbjct: 449 DLSSPASDSKP 459
Score = 42.7 bits (99), Expect = 0.027
Identities = 29/119 (24%), Positives = 50/119 (41%), Gaps = 4/119 (3%)
Query: 199 AKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPM- 257
AK W + A + +T + R+ P R + + RV + GG ++ +
Sbjct: 273 AKSRSWSQTATQSSTNATFSARQNLFPS---PRRGHIAVSLPNQRVFIQGGASADLSTVY 329
Query: 258 NDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLD 316
+D ++LD + N P W V + P R+GH+ ++ FG G+ V V D
Sbjct: 330 SDAWILDWSVNPPVWTEVDSTGGPGPRFGHSAVAYGRQIVISFGWAGSNAAPTPVIVFD 388
>UniRef100_Q75LJ6 Putative transcription factor [Oryza sativa]
Length = 476
Score = 89.0 bits (219), Expect = 3e-16
Identities = 83/264 (31%), Positives = 113/264 (42%), Gaps = 31/264 (11%)
Query: 281 PPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLD----ATPP-TWREISGLAPPLP 335
P R GHT + S++VVFGG + L+D+ V D++ TP G A P P
Sbjct: 25 PAPRSGHTAVSIGKSKVVVFGGFADKRFLSDIAVYDVENRIWYTPECNGSGSDGQAGPSP 84
Query: 336 RSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTWTPPS-RLGHTL 394
R++H + +D I G G L D ++LD + W E+ PS R
Sbjct: 85 RAFHVAIVIDCNMFIFGG--RSGGKRLGDFWMLDTDIWQ--WSELTGFGDLPSPREFAAA 140
Query: 395 SVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPP 454
S G RKI+M+GG L SDV+ MD E VTGS PPP
Sbjct: 141 SAIGNRKIVMYGGWDGKKWL----SDVYIMDTMSLEWTELSVTGS-----------VPPP 185
Query: 455 RLDHVAVSLPGGRILIFGGSVAGLHSASQLYIL----DPTDEKPTWRILNVPGRPPRFAW 510
R H A + R+L+FGG L+ L + +E P W L +PG+ P
Sbjct: 186 RCGHSATMIE-KRLLVFGGRGGAGPIMGDLWALKGVTEEDNETPGWTQLKLPGQSPSPRC 244
Query: 511 GHSTCVVGGTRAIVLGGQTGEEWM 534
GHS GG ++ GG W+
Sbjct: 245 GHSV-TSGGPYLLLFGGHGTGGWL 267
Score = 73.2 bits (178), Expect = 2e-11
Identities = 67/249 (26%), Positives = 118/249 (46%), Gaps = 33/249 (13%)
Query: 237 ACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVSS----PPPGRWGHTLSCV 292
A + + +FGG + + D ++LD + WQ +++ P P + S +
Sbjct: 90 AIVIDCNMFIFGGRSGGKR-LGDFWMLDTDI----WQWSELTGFGDLPSPREFA-AASAI 143
Query: 293 NGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTKLIVS 352
++V++GG + L+DV+++D + T ++G PP PR HS+ ++ +L+V
Sbjct: 144 GNRKIVMYGGWDGKKWLSDVYIMDTMSLEWTELSVTGSVPP-PRCGHSATMIE-KRLLVF 201
Query: 353 GGCADSGVLLSDTFLL----DMSMENPVWREIPVTWTPPS-RLGHTLSVYGGRKILMFGG 407
GG +G ++ D + L + E P W ++ + PS R GH+++ GG +L+FGG
Sbjct: 202 GGRGGAGPIMGDLWALKGVTEEDNETPGWTQLKLPGQSPSPRCGHSVTS-GGPYLLLFGG 260
Query: 408 LAKSGPLRFRSSDVFTMD---LSEDEPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLP 464
G L DV+ + L W+ + S P PPPR H +++
Sbjct: 261 HGTGGWLS--RYDVYYNECIILDRVSVQWKLLATSNEP---------PPPRAYH-SMTCI 308
Query: 465 GGRILIFGG 473
G R L+FGG
Sbjct: 309 GSRFLLFGG 317
Score = 68.9 bits (167), Expect = 4e-10
Identities = 50/148 (33%), Positives = 80/148 (53%), Gaps = 15/148 (10%)
Query: 219 WRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVL----DLNSNNPEWQH 274
W +L+V G V P RC SA + R+++FGG G M D + L + ++ P W
Sbjct: 173 WTELSVTGSVPPPRCGHSATMIEKRLLVFGGRGGAGPIMGDLWALKGVTEEDNETPGWTQ 232
Query: 275 VQV--SSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLN--DVF---VLDLDATPPTWREI 327
+++ SP P R GH+++ G L++FGG GT G L+ DV+ + LD W+ +
Sbjct: 233 LKLPGQSPSP-RCGHSVTS-GGPYLLLFGGHGTGGWLSRYDVYYNECIILDRVSVQWKLL 290
Query: 328 -SGLAPPLPRSWHSSCTLDGTKLIVSGG 354
+ PP PR++H S T G++ ++ GG
Sbjct: 291 ATSNEPPPPRAYH-SMTCIGSRFLLFGG 317
Score = 68.6 bits (166), Expect = 5e-10
Identities = 69/228 (30%), Positives = 112/228 (48%), Gaps = 33/228 (14%)
Query: 198 GAKRLG--WGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQ 255
G KRLG W + + W +LT G + R +A A+GNR ++ G +
Sbjct: 105 GGKRLGDFW------MLDTDIWQWSELTGFGDLPSPREFAAASAIGNRKIVMYGGWDGKK 158
Query: 256 PMNDTFVLDLNSNNPEWQHVQVS-SPPPGRWGHTLSCVNGSRLVVFGGCGTQG-LLNDVF 313
++D +++D S EW + V+ S PP R GH+ + + RL+VFGG G G ++ D++
Sbjct: 159 WLSDVYIMDTMS--LEWTELSVTGSVPPPRCGHSATMIE-KRLLVFGGRGGAGPIMGDLW 215
Query: 314 VL----DLDATPPTWREIS--GLAPPLPRSWHSSCTLDGTKLIVSGGCADSG------VL 361
L + D P W ++ G +P PR H S T G L++ GG G V
Sbjct: 216 ALKGVTEEDNETPGWTQLKLPGQSPS-PRCGH-SVTSGGPYLLLFGGHGTGGWLSRYDVY 273
Query: 362 LSDTFLLD-MSMENPVWREIPVT-WTPPSRLGHTLSVYGGRKILMFGG 407
++ +LD +S++ W+ + + PP R H+++ G R L+FGG
Sbjct: 274 YNECIILDRVSVQ---WKLLATSNEPPPPRAYHSMTCIGSR-FLLFGG 317
Score = 50.1 bits (118), Expect = 2e-04
Identities = 35/120 (29%), Positives = 58/120 (48%), Gaps = 10/120 (8%)
Query: 215 EAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPM------NDTFVLDLNSN 268
E W +L + G RC S + G ++LFGG G N+ +LD S
Sbjct: 226 ETPGWTQLKLPGQSPSPRCGHSVTSGGPYLLLFGGHGTGGWLSRYDVYYNECIILDRVS- 284
Query: 269 NPEWQHVQVSS-PPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREI 327
+W+ + S+ PPP R H+++C+ GSR ++FGG + D++ L + P R++
Sbjct: 285 -VQWKLLATSNEPPPPRAYHSMTCI-GSRFLLFGGFDGKNTFGDLWWLVPEGDPIAKRDL 342
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.139 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,026,222,105
Number of Sequences: 2790947
Number of extensions: 46670936
Number of successful extensions: 101654
Number of sequences better than 10.0: 686
Number of HSP's better than 10.0 without gapping: 254
Number of HSP's successfully gapped in prelim test: 438
Number of HSP's that attempted gapping in prelim test: 98228
Number of HSP's gapped (non-prelim): 2016
length of query: 548
length of database: 848,049,833
effective HSP length: 132
effective length of query: 416
effective length of database: 479,644,829
effective search space: 199532248864
effective search space used: 199532248864
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC148970.7


