

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148970.5 + phase: 0
(402 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
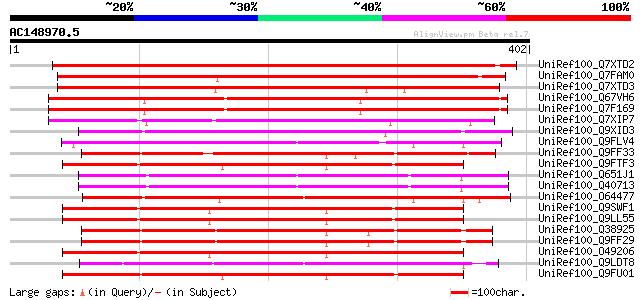
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XTD2 OSJNBb0022F16.13 protein [Oryza sativa] 375 e-102
UniRef100_Q7FAM0 OSJNBa0071I13.6 protein [Oryza sativa] 356 7e-97
UniRef100_Q7XTD3 OSJNBb0022F16.12 protein [Oryza sativa] 350 4e-95
UniRef100_Q67VH6 S-receptor kinase PK3-like [Oryza sativa] 330 3e-89
UniRef100_Q7F169 S-receptor kinase PK3-like protein [Oryza sativa] 326 8e-88
UniRef100_Q7XIP7 Receptor-like kinase TAK33-like protein [Oryza ... 307 3e-82
UniRef100_Q9XID3 F23M19.5 protein [Arabidopsis thaliana] 258 3e-67
UniRef100_Q9FLV4 Receptor-like protein kinase [Arabidopsis thali... 257 5e-67
UniRef100_Q9FF33 Receptor serine/threonine kinase-like protein [... 255 1e-66
UniRef100_Q9FTF3 Putative receptor serine/threonine kinase PR5K ... 254 2e-66
UniRef100_Q651J1 Putative receptor-like protein kinase [Oryza sa... 254 3e-66
UniRef100_Q40713 Protein kinase [Oryza sativa] 254 3e-66
UniRef100_O64477 Putative receptor-like protein kinase [Arabidop... 253 7e-66
UniRef100_Q9SWF1 Receptor kinase [Oryza sativa] 252 1e-65
UniRef100_Q9LL55 Receptor-like kinase [Oryza sativa] 252 1e-65
UniRef100_Q38925 Receptor serine/threonine kinase PR5K [Arabidop... 251 3e-65
UniRef100_Q9FF29 Receptor serine/threonine kinase [Arabidopsis t... 250 6e-65
UniRef100_O49206 Receptor-like protein kinase [Oryza sativa] 250 6e-65
UniRef100_Q9LDT8 Putative S-receptor kinase [Oryza sativa] 249 7e-65
UniRef100_Q9FU01 Putative rust resistance kinase Lr10 [Oryza sat... 249 1e-64
>UniRef100_Q7XTD2 OSJNBb0022F16.13 protein [Oryza sativa]
Length = 419
Score = 375 bits (962), Expect = e-102
Identities = 191/361 (52%), Positives = 241/361 (65%), Gaps = 5/361 (1%)
Query: 34 DSKHEFATMERIFSNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVA 93
D + E +++R +I REKP RFTPE L E T Y+ LG+G FGVV++G G VA
Sbjct: 59 DVRVEMGSVDRFLDDILREKPARFTPENLREFTGDYAERLGAGGFGVVYRGRFPGGVQVA 118
Query: 94 VKVLN-CLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYI 152
VK+L+ LD EEQF AEV T GRTYHINLV+LYGFCF +ALVYEY+ENGSLD+ +
Sbjct: 119 VKILHRTLDRRAEEQFMAEVATAGRTYHINLVRLYGFCFDATTKALVYEYLENGSLDRVL 178
Query: 153 FGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFG 212
F + +F LH I +GTA+G+ YLHEEC+HRIIHYDIKP NVLL PK+ADFG
Sbjct: 179 FDAAAAAALEFDTLHGIVVGTARGVRYLHEECQHRIIHYDIKPGNVLLAGDYAPKVADFG 238
Query: 213 LAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSS 272
LAKL SR++ T RGT GYAAPE+W P PVT+KCDVYSFG+L+FEI+GRRR+ D+
Sbjct: 239 LAKLCSRDNTHLTMTGARGTPGYAAPELWLPLPVTHKCDVYSFGMLVFEILGRRRNLDTQ 298
Query: 273 Y-SESQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLM 331
+ESQ+W+PRW W+ F+ ++A I KD E AERM KVALWC+QY P RP M
Sbjct: 299 RPAESQEWYPRWAWQRFDQGRFGEVMAASGIRSKDGEKAERMCKVALWCIQYQPEARPSM 358
Query: 332 STVVKMLEGEIDISSPPFPFHNLVPAKENSTQEGSTADSDTTTSSWRTESSRESGFKTKH 391
S+VV+MLEGE I+ P PF + S+ T TS+ +S +S T+H
Sbjct: 359 SSVVRMLEGEEQIARPVNPFAYMATMDAISSSSSGGGGVSTATSA---SASGDSAQSTRH 415
Query: 392 N 392
+
Sbjct: 416 D 416
>UniRef100_Q7FAM0 OSJNBa0071I13.6 protein [Oryza sativa]
Length = 430
Score = 356 bits (913), Expect = 7e-97
Identities = 179/352 (50%), Positives = 242/352 (67%), Gaps = 7/352 (1%)
Query: 38 EFATMERIFSNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVL 97
E +E+ + I EKP+RFT E+L T YS+ LGSG FGVV++GEL G VAVKVL
Sbjct: 65 EIGPVEKFLNEILSEKPMRFTSEQLAACTGNYSSELGSGGFGVVYRGELPKGLQVAVKVL 124
Query: 98 NC-LDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSK 156
++ ++E F AE+ TIGRTYH++LV+LYGFCF D +ALVYE++ENGSL+KY++G
Sbjct: 125 KVSMNKKVQEAFMAEIGTIGRTYHVHLVRLYGFCFDADTKALVYEFLENGSLEKYLYGGG 184
Query: 157 NRN---DFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGL 213
+ +++ LH IA+GTAKGI YLHEEC+ RI+HYDIKP N+LL PK+ADFGL
Sbjct: 185 GEDRGKKLEWRTLHDIAVGTAKGIRYLHEECQQRIVHYDIKPANILLTADFTPKVADFGL 244
Query: 214 AKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFD-SS 272
A+L RE+ T RGT GYAAPE+W P T KCDVYSFG++LFE++GRRR++D ++
Sbjct: 245 ARLGERENTHMSLTGGRGTPGYAAPELWMALPATEKCDVYSFGMVLFEVLGRRRNYDLAA 304
Query: 273 YSESQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMS 332
+ESQ+WFP+W W+ +E ++ +++ I E+D AE M KVALWCVQ+ P+ RP MS
Sbjct: 305 QAESQEWFPKWVWDRYEQGDMECVVSAAGIGEEDRAKAEMMCKVALWCVQFQPSARPKMS 364
Query: 333 TVVKMLEGEIDISSPPFPFHNLVPAKENSTQEGSTADSDTTTSSWRTESSRE 384
+VV+MLEGE+ I P PFH ++ S+ T+ S +S T S E
Sbjct: 365 SVVRMLEGEMAIVPPVNPFHYVMSGGSGSST--LTSSSTNLSSGGTTTGSSE 414
>UniRef100_Q7XTD3 OSJNBb0022F16.12 protein [Oryza sativa]
Length = 431
Score = 350 bits (898), Expect = 4e-95
Identities = 186/357 (52%), Positives = 233/357 (65%), Gaps = 15/357 (4%)
Query: 38 EFATMERIFSNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVL 97
E +M + E+PVRF+ +L T+ Y+ +GSG FGVV++G +G VAVKVL
Sbjct: 73 EMGSMSHFIEGLQNERPVRFSARQLRAFTKSYAHKVGSGGFGVVYRGVFPSGAPVAVKVL 132
Query: 98 NC-LDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSK 156
N L EEQF AEV TIGRTYHINLV+LYGFCF D +ALVYEY+E GSLD+Y+F S
Sbjct: 133 NSTLGKRAEEQFMAEVGTIGRTYHINLVRLYGFCFDADVKALVYEYMEKGSLDRYLFDSS 192
Query: 157 NR---NDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGL 213
F+KLH+IA+GTAK + YLHEEC RIIHYDIKPENVLL + PK++DFGL
Sbjct: 193 PSPAAERIGFEKLHEIAVGTAKAVRYLHEECAQRIIHYDIKPENVLLGAGMAPKVSDFGL 252
Query: 214 AKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSY 273
AKL RE T RGT GYAAPE+W P PVT+KCDVYS+G+LLFE++GRRR+ +
Sbjct: 253 AKLCDREDTHLTITGARGTPGYAAPELWMPLPVTHKCDVYSYGMLLFEMLGRRRNLELGA 312
Query: 274 SE------SQQWFPRWTWEMFENNELVVMLALCEIEE----KDSEIAERMLKVALWCVQY 323
SQ+W+PRW W FE E +LA ++ E AER+ VALWCVQY
Sbjct: 313 GAGAHGHGSQEWYPRWVWHRFEAGETEAVLARATAAAAGGGREREKAERVCMVALWCVQY 372
Query: 324 SPNDRPLMSTVVKMLEGEIDISSPPFPFHNLVP-AKENSTQEGSTADSDTTTSSWRT 379
P DRP M VV+MLEGE I++P PF +L P + S+ +TA +++ SS RT
Sbjct: 373 RPEDRPSMGNVVRMLEGEDHIAAPRNPFAHLAPYSAAGSSPTTTTATTESDGSSART 429
>UniRef100_Q67VH6 S-receptor kinase PK3-like [Oryza sativa]
Length = 444
Score = 330 bits (847), Expect = 3e-89
Identities = 183/364 (50%), Positives = 233/364 (63%), Gaps = 12/364 (3%)
Query: 31 VQTDSKHEFATMERIFSNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGE 90
V DS+ AT+ER I EKP+RFT ++L T YS LG+G FG V+KG L NG
Sbjct: 72 VVPDSQIRDATVERFLKEIAGEKPIRFTAQQLAGFTNNYSARLGAGGFGTVYKGMLPNGL 131
Query: 91 NVAVKVLNCLDMG-----MEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVEN 145
VAVK L+ G +EQF AEV ++GR +HINLV+L+GFCF D RALVYEY++N
Sbjct: 132 TVAVKRLHVGGHGDGWSTSQEQFMAEVGSVGRIHHINLVRLFGFCFDADVRALVYEYMDN 191
Query: 146 GSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLE 205
G+LD Y+F ++ IA+G A+G+ YLHEEC+H+I+HYDIKP NVLLD L
Sbjct: 192 GALDAYLFDRSRAVAVATRRA--IAVGVARGLRYLHEECQHKIVHYDIKPGNVLLDGGLT 249
Query: 206 PKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGR 265
PK+ADFGLA+L SR + RGT GYAAPEMW VT KCDVYSFG+LLFEIV R
Sbjct: 250 PKVADFGLARLASRGDTHVSVSGMRGTPGYAAPEMWMQAGVTEKCDVYSFGVLLFEIVRR 309
Query: 266 RRHFD--SSYSESQQWFPRWTWEMFENNELVVMLALCE-IEEKDSEIAERMLKVALWCVQ 322
RR+ D + QQWFP W E L + C+ +++++ E ERM KVA WCVQ
Sbjct: 310 RRNLDDGGAPGSQQQWFPMLAWSKHEAGHLAEAIEGCDAMDKQERETVERMCKVAFWCVQ 369
Query: 323 YSPNDRPLMSTVVKMLEGEIDISSPPF-PFHNLVPAKENSTQEGSTADSDTTTSSWRTES 381
P RP MS VV+MLEGE+DI +PP PF +LV + + + ST DS + +S R+
Sbjct: 370 QQPEARPPMSAVVRMLEGEVDIDAPPVNPFQHLVASPAAALRWTSTTDSAESDNSLRS-G 428
Query: 382 SRES 385
SR+S
Sbjct: 429 SRQS 432
>UniRef100_Q7F169 S-receptor kinase PK3-like protein [Oryza sativa]
Length = 444
Score = 326 bits (835), Expect = 8e-88
Identities = 181/364 (49%), Positives = 230/364 (62%), Gaps = 12/364 (3%)
Query: 31 VQTDSKHEFATMERIFSNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGE 90
V DS+ AT+ER I EKP+RFT ++L T YS LG+G FG V+KG L NG
Sbjct: 72 VVPDSQIRDATVERFLKEIAGEKPIRFTAQQLAGFTNNYSARLGAGGFGTVYKGMLPNGL 131
Query: 91 NVAVKVLNCLDMG-----MEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVEN 145
VAVK L+ G +EQF AEV ++GR +HINLV+L+GFCF D RALVYEY++N
Sbjct: 132 TVAVKRLHVGGHGDGWSTSQEQFMAEVGSVGRIHHINLVRLFGFCFDADVRALVYEYMDN 191
Query: 146 GSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLE 205
G+LD Y+F ++ IA+G A+G+ YLHEEC+H+I+HYDIKP NVLLD L
Sbjct: 192 GALDAYLFDRSRAVPVATRRA--IAVGVARGLRYLHEECQHKIVHYDIKPGNVLLDGGLT 249
Query: 206 PKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGR 265
PK+ADFGLA+L SR + RGT GYAAPEMW VT KCDVYSFG+ LFEIV R
Sbjct: 250 PKVADFGLARLASRGDTHVSVSGMRGTPGYAAPEMWMQAGVTEKCDVYSFGVHLFEIVRR 309
Query: 266 RRHFD--SSYSESQQWFPRWTWEMFENNELVVMLALCE-IEEKDSEIAERMLKVALWCVQ 322
RR+ D QWFP W E L + C+ +++++ E ERM KVA WCVQ
Sbjct: 310 RRNLDDGGEPGSQHQWFPMLAWSKHEAGHLAEAIEGCDAMDKQERETVERMCKVAFWCVQ 369
Query: 323 YSPNDRPLMSTVVKMLEGEIDISSPPF-PFHNLVPAKENSTQEGSTADSDTTTSSWRTES 381
P RP MS VV+MLEGE+DI +PP PF +LV + + + ST DS + +S R+
Sbjct: 370 QQPEARPPMSAVVRMLEGEVDIDAPPVNPFQHLVASPAAALRWTSTTDSAESDNSLRS-G 428
Query: 382 SRES 385
SR+S
Sbjct: 429 SRQS 432
>UniRef100_Q7XIP7 Receptor-like kinase TAK33-like protein [Oryza sativa]
Length = 447
Score = 307 bits (787), Expect = 3e-82
Identities = 180/377 (47%), Positives = 221/377 (57%), Gaps = 37/377 (9%)
Query: 31 VQTDSKHEFATMERIFSNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGE 90
V D+ AT+ER + EKP+RFTP +L T YS LG+G FG V+ G L NG
Sbjct: 61 VVPDAAMRSATVERFLWEMAHEKPIRFTPRQLAGFTRGYSARLGAGVFGTVYGGALPNGL 120
Query: 91 NVAVKVLNCLDMGM-----EEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVEN 145
VAVKVL GM EEQF AEV TIGRT+HINLV+L+GFC+ RALVYEY+ N
Sbjct: 121 AVAVKVLRG---GMDRRRSEEQFMAEVGTIGRTHHINLVRLFGFCYDAAVRALVYEYMGN 177
Query: 146 GSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLE 205
G+LD Y+F D IAIG A+G+ YLHEEC+H+I+HYDIKP NVLLD +
Sbjct: 178 GALDAYLFDLSR--DVGVPARRAIAIGVARGLRYLHEECEHKIVHYDIKPGNVLLDGGMT 235
Query: 206 PKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGR 265
PK+ADFGLA+L +R + RGT GYAAPE VT KCDVYSFG+LL +IVGR
Sbjct: 236 PKVADFGLARLVNRGDTHVSVSGMRGTPGYAAPETLMQSGVTEKCDVYSFGMLLLKIVGR 295
Query: 266 RRHFDSSYSESQQWFPRWTWEMFENNEL-------------------------VVMLALC 300
RR+FD + ESQQW+P W +E EL VV +A
Sbjct: 296 RRNFDEAAPESQQWWPMEAWARYERGELMMVDDAAAAINHPSDEICSGSDGEAVVTVAEA 355
Query: 301 EIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFPFHNLV--PAK 358
+ E + E RM +VA WCVQ P RP M VVKMLEGE+D++ P PF +L+ PA
Sbjct: 356 DDERRCKEAVVRMYQVAFWCVQQRPEARPPMGAVVKMLEGEMDVAPPVNPFLHLMAAPAP 415
Query: 359 ENSTQEGSTADSDTTTS 375
+ +TA S S
Sbjct: 416 VPNPWATTTASSGNAVS 432
>UniRef100_Q9XID3 F23M19.5 protein [Arabidopsis thaliana]
Length = 829
Score = 258 bits (658), Expect = 3e-67
Identities = 142/338 (42%), Positives = 202/338 (59%), Gaps = 5/338 (1%)
Query: 54 PVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVI 113
PV+FT ++L T+ + LG+G FG V++G L+N VAVK L ++ G E+QF+ EV
Sbjct: 471 PVQFTYKELQRCTKSFKEKLGAGGFGTVYRGVLTNRTVVAVKQLEGIEQG-EKQFRMEVA 529
Query: 114 TIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGT 173
TI T+H+NLV+L GFC R LVYE++ NGSLD ++F + + ++ IA+GT
Sbjct: 530 TISSTHHLNLVRLIGFCSQGRHRLLVYEFMRNGSLDNFLFTTDSAKFLTWEYRFNIALGT 589
Query: 174 AKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTR 233
AKGI YLHEEC+ I+H DIKPEN+L+D K++DFGLAKL + + N + RGTR
Sbjct: 590 AKGITYLHEECRDCIVHCDIKPENILVDDNFAAKVSDFGLAKLLNPKDNRYNMSSVRGTR 649
Query: 234 GYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFE--NN 291
GY APE P+T K DVYS+G++L E+V +R+FD S + + F W +E FE N
Sbjct: 650 GYLAPEWLANLPITSKSDVYSYGMVLLELVSGKRNFDVSEKTNHKKFSIWAYEEFEKGNT 709
Query: 292 ELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFPF 351
+ ++ L E + D E RM+K + WC+Q P RP M VV+MLEG +I +P P
Sbjct: 710 KAILDTRLSEDQTVDMEQVMRMVKTSFWCIQEQPLQRPTMGKVVQMLEGITEIKNPLCP- 768
Query: 352 HNLVPAKENSTQEGSTADSDTTTSSWRTESSRESGFKT 389
+ S ST+ + +S T SS S ++
Sbjct: 769 -KTISEVSFSGNSMSTSHASMFVASGPTRSSSFSATRS 805
>UniRef100_Q9FLV4 Receptor-like protein kinase [Arabidopsis thaliana]
Length = 872
Score = 257 bits (656), Expect = 5e-67
Identities = 154/363 (42%), Positives = 213/363 (58%), Gaps = 28/363 (7%)
Query: 41 TMERIFSN--INREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLN 98
T++R N I + PV FT L T +S +LGSG FG V+KG ++ VAVK L+
Sbjct: 502 TLKRAAKNSLILCDSPVSFTYRDLQNCTNNFSQLLGSGGFGTVYKGTVAGETLVAVKRLD 561
Query: 99 CLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGS-KN 157
E +F EV TIG +H+NLV+L G+C R LVYEY+ NGSLDK+IF S +
Sbjct: 562 RALSHGEREFITEVNTIGSMHHMNLVRLCGYCSEDSHRLLVYEYMINGSLDKWIFSSEQT 621
Query: 158 RNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLR 217
N D++ +IA+ TA+GIAY HE+C++RIIH DIKPEN+LLD PK++DFGLAK+
Sbjct: 622 ANLLDWRTRFEIAVATAQGIAYFHEQCRNRIIHCDIKPENILLDDNFCPKVSDFGLAKMM 681
Query: 218 SRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQ 277
RE + + T RGTRGY APE P+T K DVYS+G+LL EIVG RR+ D SY
Sbjct: 682 GREHS-HVVTMIRGTRGYLAPEWVSNRPITVKADVYSYGMLLLEIVGGRRNLDMSYDAED 740
Query: 278 QWFPRWTWEMFENNELVVMLALCEIEEKDSEIAE-----RMLKVALWCVQYSPNDRPLMS 332
++P W ++ EL +L ++++ +AE + LKVA WC+Q + RP M
Sbjct: 741 FFYPGWAYK-----ELTNGTSLKAVDKRLQGVAEEEEVVKALKVAFWCIQDEVSMRPSMG 795
Query: 333 TVVKMLEGEID-ISSPPFP-------------FHNLVPAKENSTQEGSTADSDTTTSSWR 378
VVK+LEG D I+ PP P + + + N+ T ++ TT+ S+R
Sbjct: 796 EVVKLLEGTSDEINLPPMPQTILELIEEGLEDVYRAMRREFNNQLSSLTVNTITTSQSYR 855
Query: 379 TES 381
+ S
Sbjct: 856 SSS 858
>UniRef100_Q9FF33 Receptor serine/threonine kinase-like protein [Arabidopsis
thaliana]
Length = 611
Score = 255 bits (652), Expect = 1e-66
Identities = 136/327 (41%), Positives = 211/327 (63%), Gaps = 16/327 (4%)
Query: 56 RFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITI 115
R++ ++ +IT+ ++ ++G G FG V+KG L +G VAVK+L + G E F EV +I
Sbjct: 293 RYSYRQIKKITKSFTEVVGRGGFGTVYKGNLRDGRKVAVKILKDSN-GNCEDFINEVASI 351
Query: 116 GRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAK 175
+T H+N+V L GFCF + KRA+VYE++ENGSLD+ ++ D L+ IA+G A+
Sbjct: 352 SQTSHVNIVSLLGFCFEKSKRAIVYEFLENGSLDQ-------SSNLDVSTLYGIALGVAR 404
Query: 176 GIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGY 235
GI YLH CK RI+H+DIKP+NVLLD L+PK+ADFGLAKL ++ +I RGT GY
Sbjct: 405 GIEYLHFGCKKRIVHFDIKPQNVLLDENLKPKVADFGLAKLCEKQESILSLLDTRGTIGY 464
Query: 236 AAPEMWKPY--PVTYKCDVYSFGILLFEIVGRR---RHFDSSYSESQQWFPRWTWEMFEN 290
APE++ V++K DVYS+G+L+ E+ G R R ++ + S +FP W ++ EN
Sbjct: 465 IAPELFSRVYGNVSHKSDVYSYGMLVLEMTGARNKERVQNADSNNSSAYFPDWIFKDLEN 524
Query: 291 NELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID-ISSPPF 349
+ V +LA + ++ +IA++M+ V LWC+Q+ P+DRP M+ VV M+EG +D + PP
Sbjct: 525 GDYVKLLA-DGLTREEEDIAKKMILVGLWCIQFRPSDRPSMNKVVGMMEGNLDSLDPPPK 583
Query: 350 PFHNLVPAKENSTQEGSTADSDTTTSS 376
P ++ P + N+ + ++ D++ S
Sbjct: 584 PLLHM-PMQNNNAESSQPSEEDSSIYS 609
>UniRef100_Q9FTF3 Putative receptor serine/threonine kinase PR5K [Oryza sativa]
Length = 621
Score = 254 bits (650), Expect = 2e-66
Identities = 136/317 (42%), Positives = 200/317 (62%), Gaps = 10/317 (3%)
Query: 42 MERIFSNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLD 101
+E KP+R+T + +IT ++ LG G FG V+KGEL NG VAVK+L
Sbjct: 299 VEMFLKTYGTSKPMRYTFSDVKKITRRFKNKLGHGGFGSVYKGELPNGVPVAVKMLEN-S 357
Query: 102 MGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDF 161
+G E+F EV TIGR +H N+V+L GFC +RAL+YE++ N SL+KYIF + +
Sbjct: 358 LGEGEEFINEVATIGRIHHANIVRLLGFCSEGTRRALIYEFMPNESLEKYIFSNGSNISR 417
Query: 162 DF---QKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRS 218
+F +K+ IA+G A+G+ YLH+ C RI+H+DIKP N+LLD PKI+DFGLAKL +
Sbjct: 418 EFLVPKKMLDIALGIARGMEYLHQGCNQRILHFDIKPHNILLDYSFSPKISDFGLAKLCA 477
Query: 219 RESNIELNTHFRGTRGYAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFDSSY-SE 275
R+ +I T RGT GY APE++ ++YK DVYSFG+L+ E+V RR+ D + ++
Sbjct: 478 RDQSIVTLTAARGTMGYIAPELYSRSFGAISYKSDVYSFGMLVLEMVSGRRNTDPTVENQ 537
Query: 276 SQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVV 335
++ +FP W +E N + +V+ E + + E ++ VALWC+Q++P +RP M+ VV
Sbjct: 538 NEFYFPEWIYERVINGQELVL--NMETTQGEKETVRQLAIVALWCIQWNPTNRPSMTKVV 595
Query: 336 KMLEGEID-ISSPPFPF 351
ML G + + PP PF
Sbjct: 596 NMLTGRLQKLQVPPKPF 612
>UniRef100_Q651J1 Putative receptor-like protein kinase [Oryza sativa]
Length = 825
Score = 254 bits (649), Expect = 3e-66
Identities = 145/339 (42%), Positives = 203/339 (59%), Gaps = 9/339 (2%)
Query: 54 PVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVI 113
PVRFT +L + T + LG G FG V+ G L +G +AVK L + G +E F++EV
Sbjct: 488 PVRFTYRELQDATSNFCNKLGQGGFGSVYLGTLPDGSRIAVKKLEGIGQGKKE-FRSEVT 546
Query: 114 TIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDF-DFQKLHKIAIG 172
IG +HI+LVKL GFC R L YEY+ NGSLDK+IF SK + D+ IA+G
Sbjct: 547 IIGSIHHIHLVKLRGFCTEGPHRLLAYEYMANGSLDKWIFHSKEDDHLLDWDTRFNIALG 606
Query: 173 TAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGT 232
TAKG+AYLH++C +I+H DIKPENVLLD K++DFGLAKL +RE + + T RGT
Sbjct: 607 TAKGLAYLHQDCDSKIVHCDIKPENVLLDDNFIAKVSDFGLAKLMTREQS-HVFTTLRGT 665
Query: 233 RGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNE 292
RGY APE Y ++ K DVYS+G++L EI+G R+ +D S + FP + ++ E +
Sbjct: 666 RGYLAPEWLTNYAISEKSDVYSYGMVLLEIIGGRKSYDPSEISEKAHFPSFAFKKLEEGD 725
Query: 293 LV-VMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPP--- 348
L + A + +KD + E +KVALWC+Q RP MS VV+MLEG ++ PP
Sbjct: 726 LQDIFDAKLKYNDKDGRV-ETAIKVALWCIQDDFYQRPSMSKVVQMLEGVCEVLQPPVSS 784
Query: 349 -FPFHNLVPAKENSTQEGSTADSDTTTSSWRTESSRESG 386
+ A ++S++EG+++ S + R SG
Sbjct: 785 QIGYRLYANAFKSSSEEGTSSGMSDYNSDALLSAVRLSG 823
>UniRef100_Q40713 Protein kinase [Oryza sativa]
Length = 824
Score = 254 bits (649), Expect = 3e-66
Identities = 145/339 (42%), Positives = 203/339 (59%), Gaps = 9/339 (2%)
Query: 54 PVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVI 113
PVRFT +L + T + LG G FG V+ G L +G +AVK L + G +E F++EV
Sbjct: 487 PVRFTYRELQDATSNFCNKLGQGGFGSVYLGTLPDGSRIAVKKLEGIGQGKKE-FRSEVT 545
Query: 114 TIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDF-DFQKLHKIAIG 172
IG +HI+LVKL GFC R L YEY+ NGSLDK+IF SK + D+ IA+G
Sbjct: 546 IIGSIHHIHLVKLRGFCTEGPHRLLAYEYMANGSLDKWIFHSKEDDHLLDWDTRFNIALG 605
Query: 173 TAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGT 232
TAKG+AYLH++C +I+H DIKPENVLLD K++DFGLAKL +RE + + T RGT
Sbjct: 606 TAKGLAYLHQDCDSKIVHCDIKPENVLLDDNFIAKVSDFGLAKLMTREQS-HVFTTLRGT 664
Query: 233 RGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNE 292
RGY APE Y ++ K DVYS+G++L EI+G R+ +D S + FP + ++ E +
Sbjct: 665 RGYLAPEWLTNYAISEKSDVYSYGMVLLEIIGGRKSYDPSEISEKAHFPSFAFKKLEEGD 724
Query: 293 LV-VMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPP--- 348
L + A + +KD + E +KVALWC+Q RP MS VV+MLEG ++ PP
Sbjct: 725 LQDIFDAKLKYNDKDGRV-ETAIKVALWCIQDDFYQRPSMSKVVQMLEGVCEVLQPPVSS 783
Query: 349 -FPFHNLVPAKENSTQEGSTADSDTTTSSWRTESSRESG 386
+ A ++S++EG+++ S + R SG
Sbjct: 784 QIGYRLYANAFKSSSEEGTSSGMSDYNSDALLSAVRLSG 822
>UniRef100_O64477 Putative receptor-like protein kinase [Arabidopsis thaliana]
Length = 828
Score = 253 bits (646), Expect = 7e-66
Identities = 144/340 (42%), Positives = 208/340 (60%), Gaps = 10/340 (2%)
Query: 57 FTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIG 116
F+ +L T+ +S LG G FG VFKG L + ++AVK L + G E+QF+ EV+TIG
Sbjct: 483 FSYRELQNATKNFSDKLGGGGFGSVFKGALPDSSDIAVKRLEGISQG-EKQFRTEVVTIG 541
Query: 117 RTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDF--DFQKLHKIAIGTA 174
H+NLV+L GFC K+ LVY+Y+ NGSLD ++F ++ ++ +IA+GTA
Sbjct: 542 TIQHVNLVRLRGFCSEGSKKLLVYDYMPNGSLDSHLFLNQVEEKIVLGWKLRFQIALGTA 601
Query: 175 KGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRG 234
+G+AYLH+EC+ IIH DIKPEN+LLD + PK+ADFGLAKL R+ + L T RGTRG
Sbjct: 602 RGLAYLHDECRDCIIHCDIKPENILLDSQFCPKVADFGLAKLVGRDFSRVLTT-MRGTRG 660
Query: 235 YAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELV 294
Y APE +T K DVYS+G++LFE+V RR+ + S +E ++FP W + + +
Sbjct: 661 YLAPEWISGVAITAKADVYSYGMMLFELVSGRRNTEQSENEKVRFFPSWAATILTKDGDI 720
Query: 295 VMLALCEIEEKDSEIAE--RMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFP-- 350
L +E +I E R KVA WC+Q + RP MS VV++LEG ++++ PPFP
Sbjct: 721 RSLVDPRLEGDAVDIEEVTRACKVACWCIQDEESHRPAMSQVVQILEGVLEVNPPPFPRS 780
Query: 351 FHNLVPAKENST--QEGSTADSDTTTSSWRTESSRESGFK 388
LV + E+ E S++ S ++ + + SS S K
Sbjct: 781 IQALVVSDEDVVFFTESSSSSSHNSSQNHKHSSSSSSSKK 820
>UniRef100_Q9SWF1 Receptor kinase [Oryza sativa]
Length = 657
Score = 252 bits (643), Expect = 1e-65
Identities = 138/317 (43%), Positives = 200/317 (62%), Gaps = 10/317 (3%)
Query: 42 MERIFSNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLD 101
+E KP R+T ++ +I ++ LG GAFG V+KGEL NG VAVK+L
Sbjct: 335 VEMFLKTYGTSKPTRYTFSEVKKIARRFKDKLGHGAFGTVYKGELPNGVPVAVKMLEN-S 393
Query: 102 MGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIF--GSK-NR 158
+G ++F EV TIGR +H N+V+L GFC +RAL+YE + N SL+KYIF GS +R
Sbjct: 394 VGEGQEFINEVATIGRIHHANIVRLLGFCSEGTRRALIYELMPNESLEKYIFPHGSNISR 453
Query: 159 NDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRS 218
K+ IA+G A+G+ YLH+ C RI+H+DIKP N+LLD PKI+DFGLAKL +
Sbjct: 454 ELLVPDKMLDIALGIARGMEYLHQGCNQRILHFDIKPHNILLDYSFNPKISDFGLAKLCA 513
Query: 219 RESNIELNTHFRGTRGYAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFD-SSYSE 275
R+ +I T RGT GY APE++ ++YK DVYSFG+L+ E+V RR+ D + ++
Sbjct: 514 RDQSIVTLTAARGTMGYIAPELYSRNFGAISYKSDVYSFGMLVLEMVSGRRNTDPPAENQ 573
Query: 276 SQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVV 335
++ +FP W +E N + +V+ E + + E+ ++ VALWC+Q++P DRP M+ VV
Sbjct: 574 NEFYFPEWVFERVMNGQDLVL--TMETTQGEKEMVRQLAIVALWCIQWNPKDRPSMTKVV 631
Query: 336 KMLEGEI-DISSPPFPF 351
ML G + ++ PP PF
Sbjct: 632 NMLTGRLQNLQVPPKPF 648
>UniRef100_Q9LL55 Receptor-like kinase [Oryza sativa]
Length = 657
Score = 252 bits (643), Expect = 1e-65
Identities = 138/317 (43%), Positives = 200/317 (62%), Gaps = 10/317 (3%)
Query: 42 MERIFSNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLD 101
+E KP R+T ++ +I ++ LG GAFG V+KGEL NG VAVK+L
Sbjct: 335 VEMFLKTYGTSKPTRYTFSEVKKIARRFKDKLGHGAFGTVYKGELPNGVPVAVKMLEN-S 393
Query: 102 MGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIF--GSK-NR 158
+G ++F EV TIGR +H N+V+L GFC +RAL+YE + N SL+KYIF GS +R
Sbjct: 394 VGEGQEFINEVATIGRIHHANIVRLLGFCSEGTRRALIYELMPNESLEKYIFPHGSNISR 453
Query: 159 NDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRS 218
K+ IA+G A+G+ YLH+ C RI+H+DIKP N+LLD PKI+DFGLAKL +
Sbjct: 454 ELLVPDKMLDIALGIARGMEYLHQGCNQRILHFDIKPHNILLDYSFNPKISDFGLAKLCA 513
Query: 219 RESNIELNTHFRGTRGYAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFD-SSYSE 275
R+ +I T RGT GY APE++ ++YK DVYSFG+L+ E+V RR+ D + ++
Sbjct: 514 RDQSIVTLTAARGTMGYIAPELYSRNFGAISYKSDVYSFGMLVLEMVSGRRNTDPPAENQ 573
Query: 276 SQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVV 335
++ +FP W +E N + +V+ E + + E+ ++ VALWC+Q++P DRP M+ VV
Sbjct: 574 NEFYFPEWVFERVMNGQDLVL--TMETTQGEKEMVRQLAIVALWCIQWNPKDRPSMTKVV 631
Query: 336 KMLEGEI-DISSPPFPF 351
ML G + ++ PP PF
Sbjct: 632 NMLTGRLQNLQVPPKPF 648
>UniRef100_Q38925 Receptor serine/threonine kinase PR5K [Arabidopsis thaliana]
Length = 665
Score = 251 bits (641), Expect = 3e-65
Identities = 130/326 (39%), Positives = 212/326 (64%), Gaps = 13/326 (3%)
Query: 56 RFTPEKLDEITEKYSTILGSGAFGVVFKGELSN-GENVAVKVLNCLDMGMEEQFKAEVIT 114
R++ ++ ++T ++ +LG G FG V+KG+L++ G +VAVK+L + G E+F EV +
Sbjct: 320 RYSYTRVKKMTNSFAHVLGKGGFGTVYKGKLADSGRDVAVKILKVSE-GNGEEFINEVAS 378
Query: 115 IGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTA 174
+ RT H+N+V L GFC+ ++KRA++YE++ NGSLDKYI + + ++++L+ +A+G +
Sbjct: 379 MSRTSHVNIVSLLGFCYEKNKRAIIYEFMPNGSLDKYISANMSTK-MEWERLYDVAVGIS 437
Query: 175 KGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRG 234
+G+ YLH C RI+H+DIKP+N+L+D L PKI+DFGLAKL + +I H RGT G
Sbjct: 438 RGLEYLHNRCVTRIVHFDIKPQNILMDENLCPKISDFGLAKLCKNKESIISMLHMRGTFG 497
Query: 235 YAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFDSSYSES---QQWFPRWTWEMFE 289
Y APEM+ V++K DVYS+G+++ E++G + YSES +FP W ++ FE
Sbjct: 498 YIAPEMFSKNFGAVSHKSDVYSYGMVVLEMIGAKNIEKVEYSESNNGSMYFPEWVYKDFE 557
Query: 290 NNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID-ISSPP 348
E+ + I E++ + A++++ VALWC+Q +P+DRP M V +MLEG ++ + PP
Sbjct: 558 KGEITRIFG-NSITEEEEKFAKKLVLVALWCIQMNPSDRPPMIKVTEMLEGNLEALQVPP 616
Query: 349 FPFHNLVPAKENSTQEGSTADSDTTT 374
P L+ + E + + DT+T
Sbjct: 617 NP---LLFSPEETVPDTLEDSDDTST 639
>UniRef100_Q9FF29 Receptor serine/threonine kinase [Arabidopsis thaliana]
Length = 665
Score = 250 bits (638), Expect = 6e-65
Identities = 129/326 (39%), Positives = 213/326 (64%), Gaps = 13/326 (3%)
Query: 56 RFTPEKLDEITEKYSTILGSGAFGVVFKGELSN-GENVAVKVLNCLDMGMEEQFKAEVIT 114
R++ ++ ++T ++ +LG G FG V+KG+L++ G +VAVK+L + G E+F EV +
Sbjct: 320 RYSYTRVKKMTNSFAHVLGKGGFGTVYKGKLADSGRDVAVKILKVSE-GNGEEFINEVAS 378
Query: 115 IGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTA 174
+ RT H+N+V L GFC+ ++KRA++YE++ NGSLDKYI + + ++++L+ +A+G +
Sbjct: 379 MSRTSHVNIVSLLGFCYEKNKRAIIYEFMPNGSLDKYISANMSTK-MEWERLYDVAVGIS 437
Query: 175 KGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRG 234
+G+ YLH C RI+H+DIKP+N+L+D L PKI+DFGLAKL + +I H RGT G
Sbjct: 438 RGLEYLHNRCVTRIVHFDIKPQNILMDENLCPKISDFGLAKLCKNKESIISMLHMRGTFG 497
Query: 235 YAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFDSSYSES---QQWFPRWTWEMFE 289
Y APEM+ V++K DVYS+G+++ E++G + YS S +FP W ++ FE
Sbjct: 498 YIAPEMFSKNFGAVSHKSDVYSYGMVVLEMIGAKNIEKVEYSGSNNGSMYFPEWVYKDFE 557
Query: 290 NNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID-ISSPP 348
E+ + I +++ +IA++++ VALWC+Q +P+DRP M V++MLEG ++ + PP
Sbjct: 558 KGEITRIFG-DSITDEEEKIAKKLVLVALWCIQMNPSDRPPMIKVIEMLEGNLEALQVPP 616
Query: 349 FPFHNLVPAKENSTQEGSTADSDTTT 374
P L+ + E + + DT+T
Sbjct: 617 NP---LLFSPEETVPDTLEDSDDTST 639
>UniRef100_O49206 Receptor-like protein kinase [Oryza sativa]
Length = 413
Score = 250 bits (638), Expect = 6e-65
Identities = 138/317 (43%), Positives = 198/317 (61%), Gaps = 8/317 (2%)
Query: 42 MERIFSNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLD 101
+E KP R+T ++ +I ++ LG GAFG V+KGEL NG VAVK+L
Sbjct: 89 VEMFLKTYGTSKPTRYTFSEVKKIARRFKDKLGHGAFGTVYKGELPNGVPVAVKMLEN-S 147
Query: 102 MGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIF--GSK-NR 158
+G ++F EV TIGR +H N+V+L GFC +RAL+YE + N SL+KYIF GS +R
Sbjct: 148 VGEGQEFINEVATIGRIHHANIVRLLGFCSEGTRRALIYELMPNESLEKYIFPHGSNISR 207
Query: 159 NDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRS 218
K+ IA+G A+G+ YLH+ C RI+H+DIKP N+LLD PKI+DFGLAKL +
Sbjct: 208 ELLVPDKMLDIALGIARGMEYLHQGCNQRILHFDIKPHNILLDYSFNPKISDFGLAKLCA 267
Query: 219 RESNIELNTHFRGTRGYAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFDSSY-SE 275
R+ +I T RGT GY APE++ ++YK DVYSFG+L+ E+V RR+ D + S+
Sbjct: 268 RDQSIVTLTAARGTMGYIAPELYSRNFGAISYKSDVYSFGMLVLEMVSGRRNTDPTVESQ 327
Query: 276 SQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVV 335
++ +FP W +E N + +V+ EK+ ++ +ALWC+Q++P DRP M+ VV
Sbjct: 328 NEFYFPEWIYERVINGQDLVLTMETTQGEKEMVRQLAIVALALWCIQWNPKDRPSMTKVV 387
Query: 336 KMLEGEI-DISSPPFPF 351
ML G + ++ PP PF
Sbjct: 388 NMLTGRLQNLQVPPKPF 404
>UniRef100_Q9LDT8 Putative S-receptor kinase [Oryza sativa]
Length = 813
Score = 249 bits (637), Expect = 7e-65
Identities = 144/327 (44%), Positives = 192/327 (58%), Gaps = 16/327 (4%)
Query: 55 VRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGM-EEQFKAEVI 113
V ++ ++ + TE +S LG G FG VF+G L G V V N +G E+QF+AEV
Sbjct: 495 VVYSYAQIKKATENFSDKLGEGGFGSVFRGTLP-GSTTVVAVKNLKGLGYAEKQFRAEVQ 553
Query: 114 TIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGT 173
T+G H NLV+L GFC +++ LVYEY+ NGSLD +IF K+ + +Q ++IAIG
Sbjct: 554 TVGMIRHTNLVRLLGFCVKGNRKLLVYEYMPNGSLDAHIFSQKS-SPLSWQVRYQIAIGI 612
Query: 174 AKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTR 233
A+G+AYLHEEC+H IIH DIKPEN+LLD + PKIADFG+AKL RE N L T RGTR
Sbjct: 613 ARGLAYLHEECEHCIIHCDIKPENILLDEEFRPKIADFGMAKLLGREFNAALTT-IRGTR 671
Query: 234 GYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNEL 293
GY APE P+T K DVYSFGI+LFE++ R + S +++P + ++
Sbjct: 672 GYLAPEWLYGQPITKKADVYSFGIVLFEMISGIRSTVTMKFGSHRYYPSYAAAQMHEGDV 731
Query: 294 VVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFP--F 351
+ +L + E + +VA WC+Q DRP M VV+MLEG +D PP P F
Sbjct: 732 LCLLDSRLEGNANVEELDITCRVACWCIQDREGDRPSMGHVVRMLEGVVDTEMPPIPASF 791
Query: 352 HNLVPAKENSTQEGSTADSDTTTSSWR 378
NLV DSD +WR
Sbjct: 792 QNLVDGD----------DSDIYEENWR 808
>UniRef100_Q9FU01 Putative rust resistance kinase Lr10 [Oryza sativa]
Length = 635
Score = 249 bits (636), Expect = 1e-64
Identities = 132/317 (41%), Positives = 197/317 (61%), Gaps = 10/317 (3%)
Query: 42 MERIFSNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLD 101
+E KP R+T ++ +I ++ +G G FG V++GEL NG VAVK+L +
Sbjct: 313 VEMFLKTYGTSKPTRYTFSEVKKIARRFKVKVGQGGFGSVYRGELPNGVPVAVKMLENSE 372
Query: 102 MGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDF 161
G ++F EV TIGR +H N+V+L GFC +RAL+YEY+ N SL+KY+F +
Sbjct: 373 -GEGDEFINEVATIGRIHHANIVRLLGFCSEGTRRALIYEYMPNDSLEKYVFSHDSDTSQ 431
Query: 162 DF---QKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRS 218
+ K+ IAIG A+G+ YLH+ C RI+H+DIKP N+LLD PKI+DFGLAKL +
Sbjct: 432 EVLVPSKMLDIAIGIARGMEYLHQGCNQRILHFDIKPNNILLDYNFSPKISDFGLAKLCA 491
Query: 219 RESNIELNTHFRGTRGYAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFDSSY-SE 275
R+ +I T RGT GY APE++ ++YK DVYSFG+L+ E+V RR+ D S S+
Sbjct: 492 RDQSIVTLTAARGTMGYIAPELYSRNFGEISYKSDVYSFGMLVLEMVSGRRNSDPSVESQ 551
Query: 276 SQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVV 335
+ +FP W +E + + + + E+ +++ E ++ VALWC+Q++P +RP M+ VV
Sbjct: 552 NVVYFPEWIYEQVNSGQDLALGR--EMTQEEKETVRQLAIVALWCIQWNPKNRPSMTKVV 609
Query: 336 KMLEGEI-DISSPPFPF 351
ML G + ++ PP PF
Sbjct: 610 NMLTGRLQNLQVPPKPF 626
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.134 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 685,902,668
Number of Sequences: 2790947
Number of extensions: 29744524
Number of successful extensions: 123484
Number of sequences better than 10.0: 18081
Number of HSP's better than 10.0 without gapping: 7222
Number of HSP's successfully gapped in prelim test: 10859
Number of HSP's that attempted gapping in prelim test: 89755
Number of HSP's gapped (non-prelim): 20454
length of query: 402
length of database: 848,049,833
effective HSP length: 130
effective length of query: 272
effective length of database: 485,226,723
effective search space: 131981668656
effective search space used: 131981668656
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC148970.5


