

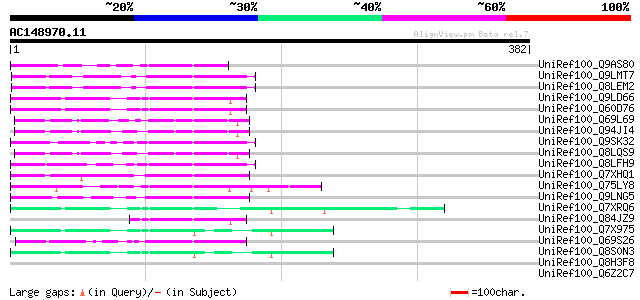
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148970.11 + phase: 0
(382 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9AS80 P0028E10.12 protein [Oryza sativa] 65 3e-09
UniRef100_Q9LMT7 F2H15.15 protein [Arabidopsis thaliana] 63 1e-08
UniRef100_Q8LEM2 Hypothetical protein [Arabidopsis thaliana] 63 1e-08
UniRef100_Q9LD66 Similar to maize transposon MuDR mudrA protein ... 62 2e-08
UniRef100_Q60D76 Putative polyprotein [Oryza sativa] 62 2e-08
UniRef100_Q69L69 Calcineurin-like phosphoesterase-like protein [... 59 3e-07
UniRef100_Q94JI4 P0638D12.5 protein [Oryza sativa] 57 9e-07
UniRef100_Q9SK32 Expressed protein [Arabidopsis thaliana] 57 9e-07
UniRef100_Q8LQS9 P0702H08.15 protein [Oryza sativa] 57 9e-07
UniRef100_Q8LFH9 Hypothetical protein [Arabidopsis thaliana] 57 9e-07
UniRef100_Q7XHQ1 Calcineurin-like phosphoesterase-like [Oryza sa... 56 2e-06
UniRef100_Q75LY8 Putative MuDR family transposase [Oryza sativa] 55 4e-06
UniRef100_Q9LNG5 F21D18.16 [Arabidopsis thaliana] 54 6e-06
UniRef100_Q7XRQ6 OSJNBb0096E05.18 protein [Oryza sativa] 54 6e-06
UniRef100_Q84JZ9 Mutator-like transposase [Oryza sativa] 52 4e-05
UniRef100_Q7X975 Putative Mutator protein [Oryza sativa] 50 8e-05
UniRef100_Q69S26 Serine/threonine phosphatase PP7-related-like [... 50 8e-05
UniRef100_Q8S0N3 Putative mutator-like transposase [Oryza sativa] 50 1e-04
UniRef100_Q8H3F8 Serine/threonine phosphatase PP7-related-like p... 47 0.001
UniRef100_Q6Z2C7 Calcineurin-like phosphoesterase-like [Oryza sa... 42 0.037
>UniRef100_Q9AS80 P0028E10.12 protein [Oryza sativa]
Length = 792
Score = 65.1 bits (157), Expect = 3e-09
Identities = 55/161 (34%), Positives = 74/161 (45%), Gaps = 24/161 (14%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
MP GE+T+TL DVA + LPI G + P++E L+ YLG A
Sbjct: 278 MPCGEITITLQDVAMILGLPIAGHAVTVNPTEPQNE---LVERYLGKAPPPDR------- 327
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRS 120
P LR + RA ED +E E ++ + R Y+L L+ LLF D S
Sbjct: 328 ----PRPGLRVSWV----RAEFNNCPEDADE----ETIKQH-ARAYILSLISGLLFPDAS 374
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACR 161
+AD + +YSWG+ TLAYLY + DACR
Sbjct: 375 GDLYTFYPFPLIAD-LENIGSYSWGSATLAYLYRAMCDACR 414
>UniRef100_Q9LMT7 F2H15.15 protein [Arabidopsis thaliana]
Length = 478
Score = 63.2 bits (152), Expect = 1e-08
Identities = 49/181 (27%), Positives = 85/181 (46%), Gaps = 28/181 (15%)
Query: 2 PMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEYG 61
P GEMT+TLD+V+ + L ++G+ + G K + + + + LG K+ E
Sbjct: 84 PCGEMTITLDEVSLILGLAVDGKPVV-GVKEKDEDPSQVCLRLLG-------KLPKGELS 135
Query: 62 GY-ISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRS 120
G ++ L++ + A + +E+E Y R YL+Y+V +F
Sbjct: 136 GNRVTAKWLKESFAECPKGATM-------KEIE-------YHTRAYLIYIVGSTIFATTD 181
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHRVLGGSVTLLTLDDVC 180
+I + YL D + Y+WGA LA+LY ++ +A + ++GG +TLL C
Sbjct: 182 PSKISVDYLILFED-FEKAGEYAWGAAALAFLYRQIGNASQRSQSIIGGCLTLLQ----C 236
Query: 181 W 181
W
Sbjct: 237 W 237
>UniRef100_Q8LEM2 Hypothetical protein [Arabidopsis thaliana]
Length = 478
Score = 63.2 bits (152), Expect = 1e-08
Identities = 49/181 (27%), Positives = 85/181 (46%), Gaps = 28/181 (15%)
Query: 2 PMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEYG 61
P GEMT+TLD+V+ + L ++G+ + G K + + + + LG K+ E
Sbjct: 84 PCGEMTITLDEVSLILGLAVDGKPVV-GVKEKDEDPSQVCLKLLG-------KLPKGELS 135
Query: 62 GY-ISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRS 120
G ++ L++ + A + +E+E Y R YL+Y+V +F
Sbjct: 136 GNRVTAKWLKESFAECPKGATM-------KEIE-------YHTRAYLIYIVGSTIFATTD 181
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHRVLGGSVTLLTLDDVC 180
+I + YL D + Y+WGA LA+LY ++ +A + ++GG +TLL C
Sbjct: 182 PSKISVDYLILFED-FEKAGEYAWGAAALAFLYRQIGNASQRSQSIIGGCLTLLQ----C 236
Query: 181 W 181
W
Sbjct: 237 W 237
>UniRef100_Q9LD66 Similar to maize transposon MuDR mudrA protein [Oryza sativa]
Length = 1230
Score = 62.4 bits (150), Expect = 2e-08
Identities = 55/176 (31%), Positives = 79/176 (44%), Gaps = 19/176 (10%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
+P GE+TVTL+DVA + LPI G+ + + YLG+ A Q
Sbjct: 592 LPCGELTVTLEDVAMILGLPIRGQAVTGDTASGNWRER--VEEYLGLEPPVAPDGQRQTK 649
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRS 120
+ LR + + P E +E V+ YC R Y+LY+ +LF D
Sbjct: 650 TSGVPLSWLRANFG------------QCPAEADEAT-VQRYC-RAYVLYIFGSILFPDSG 695
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACR--PGHRVLGGSVTLL 174
++L +AD + +SWG+ LA+LY +L DACR G L G V LL
Sbjct: 696 GDMASWMWLPLLAD-WDEAGTFSWGSAALAWLYRQLCDACRRQGGDANLAGCVWLL 750
>UniRef100_Q60D76 Putative polyprotein [Oryza sativa]
Length = 1754
Score = 62.4 bits (150), Expect = 2e-08
Identities = 55/176 (31%), Positives = 79/176 (44%), Gaps = 19/176 (10%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
+P GE+TVTL+DVA + LPI G+ + + YLG+ A Q
Sbjct: 974 LPCGELTVTLEDVAMILGLPIRGQAVTGDTASGNWRER--VEEYLGLEPPVAPDGQRQTK 1031
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRS 120
+ LR + + P E +E V+ YC R Y+LY+ +LF D
Sbjct: 1032 TSGVPLSWLRANFG------------QCPAEADEAT-VQRYC-RAYVLYIFGSILFPDSG 1077
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACR--PGHRVLGGSVTLL 174
++L +AD + +SWG+ LA+LY +L DACR G L G V LL
Sbjct: 1078 GDMASWMWLPLLAD-WDEAGTFSWGSAALAWLYRQLCDACRRQGGDANLAGCVWLL 1132
>UniRef100_Q69L69 Calcineurin-like phosphoesterase-like protein [Oryza sativa]
Length = 766
Score = 58.5 bits (140), Expect = 3e-07
Identities = 58/175 (33%), Positives = 89/175 (50%), Gaps = 26/175 (14%)
Query: 4 GEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEYGGY 63
GEMTVTL DV+ L LPI+GR + H+ A +++ LG H+ + + +
Sbjct: 153 GEMTVTLQDVSMLLALPIDGRPV---CSTTDHDYAQMVIDCLG---HD-PRGPSMPGKSF 205
Query: 64 ISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRSNKR 123
+ Y L+ + + GT+D + +ER VR Y+L L+ +LF D + R
Sbjct: 206 LHYKWLKKHF------YELPEGTDD----QTVER----HVRAYILSLLCGVLFPDGTG-R 250
Query: 124 IELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHRV--LGGSVTLLTL 176
+ LIYL +AD + + YSWG+ LA+LY L H + +GGS+ LL L
Sbjct: 251 MSLIYLPLIAD-LSLVGTYSWGSAVLAFLYRSLCSVA-SSHNIKNIGGSLLLLQL 303
>UniRef100_Q94JI4 P0638D12.5 protein [Oryza sativa]
Length = 761
Score = 57.0 bits (136), Expect = 9e-07
Identities = 57/175 (32%), Positives = 84/175 (47%), Gaps = 26/175 (14%)
Query: 4 GEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEYGGY 63
GEM VTL DVA L LPI+GR + H+ A +++ LG H+ + + +
Sbjct: 170 GEMAVTLQDVAMLLALPIDGRPV---CSTTDHDYAQMVIDCLG---HD-PRGPSMPGKSF 222
Query: 64 ISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRSNKR 123
+ Y L+ + L D + VE VR Y+L L+ +LF D + R
Sbjct: 223 LHYKWLKKHFYE-------LPEGADDQTVER-------HVRAYILSLLCGVLFPDGTG-R 267
Query: 124 IELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHRV--LGGSVTLLTL 176
+ LIYL +AD + + YSWG+ LA+LY L H + +GGS+ LL L
Sbjct: 268 MSLIYLPLIAD-LSRVGTYSWGSAVLAFLYRSLCSVA-SSHNIKNIGGSLLLLQL 320
>UniRef100_Q9SK32 Expressed protein [Arabidopsis thaliana]
Length = 509
Score = 57.0 bits (136), Expect = 9e-07
Identities = 52/181 (28%), Positives = 83/181 (45%), Gaps = 23/181 (12%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
+P+GEMT+TLD+VA + L I+G + K G + M G + N+E
Sbjct: 92 LPLGEMTITLDEVALVLGLEIDGDPIVGSK-----VGDEVAMDMCGRLLGKLPSAANKE- 145
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRS 120
++ R++ ++L R +E PE+ + V+ + R YLLYL+ +F
Sbjct: 146 ---VNCSRVK---LNWLKR----TFSECPEDA-SFDVVKCH-TRAYLLYLIGSTIFATTD 193
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHRVLGGSVTLLTLDDVC 180
++ + YL D + Y+WGA LA LY L +A + G +TLL C
Sbjct: 194 GDKVSVKYLPLFED-FDQAGRYAWGAAALACLYRALGNASLKSQSNICGCLTLLQ----C 248
Query: 181 W 181
W
Sbjct: 249 W 249
>UniRef100_Q8LQS9 P0702H08.15 protein [Oryza sativa]
Length = 718
Score = 57.0 bits (136), Expect = 9e-07
Identities = 57/175 (32%), Positives = 84/175 (47%), Gaps = 26/175 (14%)
Query: 4 GEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEYGGY 63
GEM VTL DVA L LPI+GR + H+ A +++ LG H+ + + +
Sbjct: 184 GEMAVTLQDVAMLLALPIDGRPV---CSTTDHDYAQMVIDCLG---HD-PRGPSMPGKSF 236
Query: 64 ISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRSNKR 123
+ Y L+ + L D + VE VR Y+L L+ +LF D + R
Sbjct: 237 LHYKWLKKHFYE-------LPEGADDQTVER-------HVRAYILSLLCGVLFPDGTG-R 281
Query: 124 IELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHRV--LGGSVTLLTL 176
+ LIYL +AD + + YSWG+ LA+LY L H + +GGS+ LL L
Sbjct: 282 MSLIYLPLIAD-LSRVGTYSWGSAVLAFLYRSLCSVA-SSHNIKNIGGSLLLLQL 334
>UniRef100_Q8LFH9 Hypothetical protein [Arabidopsis thaliana]
Length = 509
Score = 57.0 bits (136), Expect = 9e-07
Identities = 52/181 (28%), Positives = 79/181 (42%), Gaps = 23/181 (12%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
+P+GEMT+TLD+VA + L I+G + G K+ + LG A K N
Sbjct: 92 LPLGEMTITLDEVALVLGLEIDGDPI-VGSKVDDEVAMDMCGRLLGKLPSAANKEVN--- 147
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRS 120
S +L ++ +E PE+ + V+ + R YLLYL+ +F
Sbjct: 148 ---CSRVKLNWLKRTF---------SECPEDA-SFDVVKCH-TRAYLLYLIGSTIFATTD 193
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHRVLGGSVTLLTLDDVC 180
++ + YL D + Y+WGA LA LY L +A + G +TLL C
Sbjct: 194 GDKVSVKYLPLFED-FDQAGRYAWGAAALACLYRALGNASLKSQSNICGCLTLLQ----C 248
Query: 181 W 181
W
Sbjct: 249 W 249
>UniRef100_Q7XHQ1 Calcineurin-like phosphoesterase-like [Oryza sativa]
Length = 327
Score = 55.8 bits (133), Expect = 2e-06
Identities = 46/180 (25%), Positives = 77/180 (42%), Gaps = 15/180 (8%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHE----AEKIC 56
+P+GEMT+TL DV+CL LPI GR + ++ LG+ E +K
Sbjct: 87 LPVGEMTITLQDVSCLWGLPIHGRPI---TGQADGSWVDMIERLLGIPMEEQHMKQKKRK 143
Query: 57 NQEYGGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLF 116
++ +SY R + R V+ E+ + R +L ++ ++F
Sbjct: 144 KEDDMTMVSYSRYSISLSKLRDRFRVMPKNATEREI-------NWYTRALVLDIIGSMVF 196
Query: 117 GDRSNKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHRVLGGSVTLLTL 176
D S + +YL M + + Y+WGA L+ LY +L+ A + + LL L
Sbjct: 197 TDTSGDGVPAMYLQFMMN-LSEQTEYNWGAAALSMLYRQLSIASEKERAEISRPLLLLQL 255
>UniRef100_Q75LY8 Putative MuDR family transposase [Oryza sativa]
Length = 1403
Score = 54.7 bits (130), Expect = 4e-06
Identities = 63/246 (25%), Positives = 102/246 (40%), Gaps = 37/246 (15%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMP--KHEGAALLMTYLGVAQHEAEKICNQ 58
+P GEMT+TL DVA + LP+ G + + P + + L L + Q + K
Sbjct: 888 LPSGEMTITLQDVAMILALPLRGHAVTGRTETPGWRAQVEQLFGITLNIEQGQGGK---- 943
Query: 59 EYGGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGD 118
+ S+L + N +D E RV Y R Y+L+L+ +LF +
Sbjct: 944 --------KKQNGIPLSWLSQ-NFSHLHDDAEPW----RVECYA-RAYILHLLGGVLFPN 989
Query: 119 RSNKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADAC--RPGHRVLGGSVTLLTL 176
I++ +A+ + +SWG+ LA+ Y +L +AC + + G V L+ L
Sbjct: 990 AGGDIASAIWIPLVAN-LGDLGRFSWGSAVLAWTYRQLCEACCRQAPSSNMSGCVLLIQL 1048
Query: 177 -----DDVCWRPYEEHRE--------IQDFEEVFWYSGWIMCGVRRVYRHLSERVLRQYG 223
+ W PY + + F Y ++C V HL RV+RQ+G
Sbjct: 1049 WMWLRLPIEWLPYHTNEASSLTLNSMCNRDSDYFMYQCPLIC-FWAVEYHLPHRVMRQFG 1107
Query: 224 YVQTIP 229
Q P
Sbjct: 1108 KKQDWP 1113
>UniRef100_Q9LNG5 F21D18.16 [Arabidopsis thaliana]
Length = 1340
Score = 54.3 bits (129), Expect = 6e-06
Identities = 51/177 (28%), Positives = 79/177 (43%), Gaps = 25/177 (14%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
+P GE+TVTL DV L L ++G + K + A L LG + +
Sbjct: 101 LPAGEITVTLQDVNILLGLRVDGPAVTGSTK---YNWADLCEDLLGHRPGPKDL-----H 152
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYC-VRCYLLYLVRCLLFGDR 119
G ++S LR+ + + DP+EV C R ++L L+ L+GD+
Sbjct: 153 GSHVSLAWLRENFRNL---------PADPDEVT------LKCHTRAFVLALMSGFLYGDK 197
Query: 120 SNKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHRVLGGSVTLLTL 176
S + L +L + D + + SWG+ TLA LY EL A + + G + LL L
Sbjct: 198 SKHDVALTFLPLLRD-FDEVAKLSWGSATLALLYRELCRASKRTVSTICGPLVLLQL 253
>UniRef100_Q7XRQ6 OSJNBb0096E05.18 protein [Oryza sativa]
Length = 567
Score = 54.3 bits (129), Expect = 6e-06
Identities = 81/337 (24%), Positives = 122/337 (36%), Gaps = 59/337 (17%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
+P GE+TVTL+DVA + LPI G+ + + YLG+ A Q
Sbjct: 30 LPCGELTVTLEDVAMILGLPIRGQAVTGDTASGNWRER--VEEYLGLEPPVAPDGQRQTK 87
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRS 120
+ LR + + P E +E V+ YC R Y+LY+ +LF D
Sbjct: 88 TSGVPLSWLRANFG------------QCPAEADEAT-VQRYC-RAYVLYIFGSILFPDSG 133
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHRVLGGSVTLLTLDDVC 180
++L +AD + +SWG+ LA+ L L V
Sbjct: 134 GDMASWMWLPLLAD-WDEAGTFSWGSAALAWSL----------------QCVLFLLLQVV 176
Query: 181 WRPYEEHREIQ-------DFEEVFWYSGWIMCGVRRVYRHLSERVLRQYGYVQTIPR--- 230
W PY+ ++ E+ + V + RV+RQ+G +QTIP
Sbjct: 177 WMPYQAQEVLELELNPMCHIEDALKTLRCPLICFYAVEFQMCHRVMRQFGRLQTIPHRFS 236
Query: 231 -----HPTDVR-DLPPPSIVQMFVDFRTHTLKADARG-EQAGEDTWRVADGYVLWYTRVS 283
H D+R + D T K + G G+ D Y+ W R
Sbjct: 237 TSIDLHKVDLRKNKKVTDWAYYHQDHITQWEKFEENGVPDQGQHNGTEFDLYLAWLHRTY 296
Query: 284 HPQILPPIPGDLLRPANEEQIIAEQWQRYEARNSPDT 320
+LRPA IA+ + E +N DT
Sbjct: 297 RL---------VLRPAWTLADIADDPEDVEEQNEYDT 324
>UniRef100_Q84JZ9 Mutator-like transposase [Oryza sativa]
Length = 1381
Score = 51.6 bits (122), Expect = 4e-05
Identities = 34/88 (38%), Positives = 47/88 (52%), Gaps = 5/88 (5%)
Query: 89 PEEVEELERVRTYCVRCYLLYLVRCLLFGDRSNKRIELIYLTTMADGYAGMRNYSWGAMT 148
P E +E V+ YC R Y+LY+ +LF D ++L +AD + +SWG+
Sbjct: 804 PAEADEAT-VQRYC-RAYVLYIFGSILFPDSGGDMASWMWLPLLAD-WDEAGTFSWGSAA 860
Query: 149 LAYLYGELADACR--PGHRVLGGSVTLL 174
LA+LY +L DACR G L G V LL
Sbjct: 861 LAWLYRQLCDACRRQGGDANLAGCVWLL 888
>UniRef100_Q7X975 Putative Mutator protein [Oryza sativa]
Length = 1456
Score = 50.4 bits (119), Expect = 8e-05
Identities = 61/248 (24%), Positives = 95/248 (37%), Gaps = 46/248 (18%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
+P GE+TVTL+DVA + LPI G+ + + YLG+ A Q
Sbjct: 913 LPCGELTVTLEDVAMILGLPIRGQAVTGDTASGNWRER--VEEYLGLEPPVAPDGQRQTK 970
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRS 120
+ LR + + P E +E V+ YC R Y+LY+ +LF D
Sbjct: 971 TSGVPLSWLRANFG------------QCPAEADEAT-VQRYC-RAYVLYIFGSILFPDSG 1016
Query: 121 NKRIELIYLTTMAD---GYAGMRNYSWGAMTLAYLYGELADACRPGHRVLGGSVTLLTLD 177
++L +AD G RN ++ Y Y + D +P H
Sbjct: 1017 GDMASWMWLPLLADWDEAVVGRRNLAY------YHYTKEMDYLQPEH------------- 1057
Query: 178 DVCWRPYEEHREIQ-------DFEEVFWYSGWIMCGVRRVYRHLSERVLRQYGYVQTIPR 230
V W PY+ ++ E+ + V + RV++Q+G +QTIP
Sbjct: 1058 -VVWMPYQAQEALELELNPMCHIEDALKTLRCPLICFYAVEFQMCHRVMQQFGRLQTIPH 1116
Query: 231 HPTDVRDL 238
+ DL
Sbjct: 1117 RFSTSIDL 1124
>UniRef100_Q69S26 Serine/threonine phosphatase PP7-related-like [Oryza sativa]
Length = 619
Score = 50.4 bits (119), Expect = 8e-05
Identities = 50/171 (29%), Positives = 77/171 (44%), Gaps = 22/171 (12%)
Query: 5 EMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEYGGYI 64
EM +L DV+ + +P+ G ++ + + + + +LG + E++C G I
Sbjct: 96 EMAPSLRDVSYILGIPVTGHVVT-AEPIGDEAVRRMCLHFLGESPGNGEQLC-----GLI 149
Query: 65 SYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRSNKRI 124
RL Y + E+P + E+ Y R YLLYLV LF D +
Sbjct: 150 ---RLTWLYRKFHQLP------ENPT-INEI----AYSTRAYLLYLVGSTLFPDTMRGFV 195
Query: 125 ELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHRV-LGGSVTLL 174
YL +AD + +R Y+WG+ LA+LY L+ A P GS TLL
Sbjct: 196 SPRYLPLLAD-FRKIREYAWGSAALAHLYRGLSVAVTPNATTQFLGSATLL 245
>UniRef100_Q8S0N3 Putative mutator-like transposase [Oryza sativa]
Length = 1556
Score = 50.1 bits (118), Expect = 1e-04
Identities = 62/248 (25%), Positives = 94/248 (37%), Gaps = 46/248 (18%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
+P GE+TVTL+DVA + LPI G+ + + YLG+ A Q
Sbjct: 982 LPCGELTVTLEDVAMILGLPIRGQAVTGDTASGNWRER--VEEYLGLEPPVAPDGQRQTK 1039
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRS 120
+ LR + + P E +E V+ YC R Y+LY+ +LF D
Sbjct: 1040 TSGVPLSWLRANFG------------QCPAEADEAI-VQRYC-RAYVLYIFGSILFPDSG 1085
Query: 121 NKRIELIYLTTMAD---GYAGMRNYSWGAMTLAYLYGELADACRPGHRVLGGSVTLLTLD 177
++L +AD G RN ++ Y Y D +P H
Sbjct: 1086 GNMASWMWLPLLADWDEAVVGRRNLAY------YHYTNEMDYLQPEH------------- 1126
Query: 178 DVCWRPYEEHREIQ-------DFEEVFWYSGWIMCGVRRVYRHLSERVLRQYGYVQTIPR 230
V W PY+ ++ E+ + V + RV+RQ+G +QTIP
Sbjct: 1127 -VVWMPYQAQEVLELELNPMCHIEDALKTLRCPLICFYAVEFQMWHRVMRQFGRLQTIPH 1185
Query: 231 HPTDVRDL 238
+ DL
Sbjct: 1186 RFSTSIDL 1193
>UniRef100_Q8H3F8 Serine/threonine phosphatase PP7-related-like protein [Oryza
sativa]
Length = 589
Score = 46.6 bits (109), Expect = 0.001
Identities = 45/171 (26%), Positives = 72/171 (41%), Gaps = 22/171 (12%)
Query: 5 EMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEYGGYI 64
EM +L D A + +P+ GR++ G + K L YLG C G ++
Sbjct: 83 EMAPSLRDAAYILGIPVTGRVVTTGAVLNKSV-EDLCFQYLGQVPD-----CRDCRGSHV 136
Query: 65 SYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRSNKRI 124
L+ ++ E P + + Y R YLL+L+ L +R +
Sbjct: 137 KLSWLQSKFSRI---------PERPTNDQTM-----YGTRAYLLFLIGSALLPERDRGYV 182
Query: 125 ELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADA-CRPGHRVLGGSVTLL 174
YL ++D + ++ Y+WGA LA+LY L+ A + L GS LL
Sbjct: 183 SPKYLPLLSD-FDKVQEYAWGAAALAHLYKALSIAVAHSARKRLFGSAALL 232
>UniRef100_Q6Z2C7 Calcineurin-like phosphoesterase-like [Oryza sativa]
Length = 432
Score = 41.6 bits (96), Expect = 0.037
Identities = 38/146 (26%), Positives = 56/146 (38%), Gaps = 18/146 (12%)
Query: 104 RCYLLYLVRCLLFGDRSNKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPG 163
R +L ++ ++F D S + +YL M D A Y+WG LA LY +L +
Sbjct: 15 RALVLEILGSIIFTDSSGDSVPAMYLQFM-DNLATRTEYNWGGAVLAMLYRQLNNGAEKA 73
Query: 164 HRVLGGSVTLLTLDDVCWRPY--------------EEHREIQDFEEVF---WYSGWIMCG 206
+ G + LL L P E+ ++ D +F W S
Sbjct: 74 RSEIFGPLVLLQLWSWSRLPLGRPKNIIQKTDEVEEQEKKESDGYPIFGAKWCSYHEFPT 133
Query: 207 VRRVYRHLSERVLRQYGYVQTIPRHP 232
H ERV+RQ+G Q I P
Sbjct: 134 PHNCEFHYPERVMRQFGRKQLITPPP 159
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 670,344,259
Number of Sequences: 2790947
Number of extensions: 28324450
Number of successful extensions: 54917
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 49
Number of HSP's that attempted gapping in prelim test: 54847
Number of HSP's gapped (non-prelim): 116
length of query: 382
length of database: 848,049,833
effective HSP length: 129
effective length of query: 253
effective length of database: 488,017,670
effective search space: 123468470510
effective search space used: 123468470510
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Medicago: description of AC148970.11


