

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148969.8 - phase: 0
(219 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
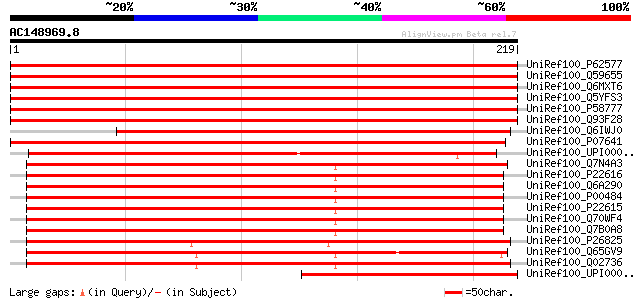
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_P62577 Chloramphenicol acetyltransferase [Escherichia ... 469 e-131
UniRef100_Q59655 PP-CAT [Pasteurella piscicida)] 468 e-131
UniRef100_Q6MXT6 Chloramphenicol acetyltransferase [Serratia mar... 466 e-130
UniRef100_Q5YFS3 Chloramphenicol acetyltransferase [Alcaligenes ... 466 e-130
UniRef100_P58777 Chloramphenicol acetyltransferase [Klebsiella sp] 466 e-130
UniRef100_Q93F28 Chloramphenicol acetyltransferase [Shigella fle... 459 e-128
UniRef100_Q6IWJ0 Chloramphenicol acetyltransferase [Salmonella p... 369 e-101
UniRef100_P07641 Chloramphenicol acetyltransferase [Proteus mira... 357 9e-98
UniRef100_UPI00004382DB UPI00004382DB UniRef100 entry 280 2e-74
UniRef100_Q7N4A3 Chloramphenicol acetyltransferase [Photorhabdus... 238 6e-62
UniRef100_P22616 Chloramphenicol acetyltransferase II [Haemophil... 220 2e-56
UniRef100_Q6A290 Chloramphenicol acetyltransferase II [Haemophil... 218 1e-55
UniRef100_P00484 Chloramphenicol acetyltransferase III [Escheric... 215 6e-55
UniRef100_P22615 Chloramphenicol acetyltransferase II [Escherich... 215 8e-55
UniRef100_Q70WF4 Chloramphenicol aminotransferase [Aeromonas sal... 214 1e-54
UniRef100_Q7B0A8 Chloramphenicol acetyl transferase [uncultured ... 213 2e-54
UniRef100_P26825 Chloramphenicol acetyltransferase [Clostridium ... 206 3e-52
UniRef100_Q65GV9 Hypothetical protein [Bacillus licheniformis] 204 1e-51
UniRef100_Q02736 Chloramphenicol acetyltransferase [Clostridium ... 204 2e-51
UniRef100_UPI000026EBA5 UPI000026EBA5 UniRef100 entry 200 3e-50
>UniRef100_P62577 Chloramphenicol acetyltransferase [Escherichia coli]
Length = 219
Score = 469 bits (1206), Expect = e-131
Identities = 219/219 (100%), Positives = 219/219 (100%)
Query: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI
Sbjct: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
Query: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY
Sbjct: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
Query: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG
Sbjct: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
Query: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA
Sbjct: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
>UniRef100_Q59655 PP-CAT [Pasteurella piscicida)]
Length = 219
Score = 468 bits (1205), Expect = e-131
Identities = 218/219 (99%), Positives = 219/219 (99%)
Query: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI
Sbjct: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
Query: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY
Sbjct: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
Query: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
SQD+ACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG
Sbjct: 121 SQDIACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
Query: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA
Sbjct: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
>UniRef100_Q6MXT6 Chloramphenicol acetyltransferase [Serratia marcescens]
Length = 219
Score = 466 bits (1200), Expect = e-130
Identities = 217/219 (99%), Positives = 219/219 (99%)
Query: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI
Sbjct: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
Query: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
HILARLMNAHPEFRMAMKDGELVIWDS+HPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY
Sbjct: 61 HILARLMNAHPEFRMAMKDGELVIWDSIHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
Query: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG
Sbjct: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
Query: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYC+EWQGGA
Sbjct: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCNEWQGGA 219
>UniRef100_Q5YFS3 Chloramphenicol acetyltransferase [Alcaligenes xylosoxydans
xylosoxydans]
Length = 219
Score = 466 bits (1200), Expect = e-130
Identities = 218/219 (99%), Positives = 218/219 (99%)
Query: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI
Sbjct: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
Query: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
HILARLMNAHPE RMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY
Sbjct: 61 HILARLMNAHPELRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
Query: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG
Sbjct: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
Query: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA
Sbjct: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
>UniRef100_P58777 Chloramphenicol acetyltransferase [Klebsiella sp]
Length = 219
Score = 466 bits (1198), Expect = e-130
Identities = 218/219 (99%), Positives = 218/219 (99%)
Query: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI
Sbjct: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
Query: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY
Sbjct: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
Query: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVA MDNFFAPVFTMGKYYTQG
Sbjct: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVAAMDNFFAPVFTMGKYYTQG 180
Query: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA
Sbjct: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
>UniRef100_Q93F28 Chloramphenicol acetyltransferase [Shigella flexneri 2a]
Length = 219
Score = 459 bits (1181), Expect = e-128
Identities = 215/219 (98%), Positives = 215/219 (98%)
Query: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI
Sbjct: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
Query: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY
Sbjct: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
Query: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
S DVACYGENLAYFPK F ENM FVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG
Sbjct: 121 SXDVACYGENLAYFPKXFXENMXFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
Query: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA
Sbjct: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
>UniRef100_Q6IWJ0 Chloramphenicol acetyltransferase [Salmonella pullorum]
Length = 170
Score = 369 bits (946), Expect = e-101
Identities = 170/170 (100%), Positives = 170/170 (100%)
Query: 47 TVKKNKHKFYPAFIHILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLW 106
TVKKNKHKFYPAFIHILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLW
Sbjct: 1 TVKKNKHKFYPAFIHILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLW 60
Query: 107 SEYHDDFRQFLHIYSQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNF 166
SEYHDDFRQFLHIYSQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNF
Sbjct: 61 SEYHDDFRQFLHIYSQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNF 120
Query: 167 FAPVFTMGKYYTQGDKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQ 216
FAPVFTMGKYYTQGDKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQ
Sbjct: 121 FAPVFTMGKYYTQGDKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQ 170
>UniRef100_P07641 Chloramphenicol acetyltransferase [Proteus mirabilis]
Length = 217
Score = 357 bits (917), Expect = 9e-98
Identities = 165/214 (77%), Positives = 190/214 (88%)
Query: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
M+ K G VD+SQW RKEHFEAFQS AQCT++QTVQLDIT+ LKTVK+N +KFYP FI
Sbjct: 1 MDTKRVGILVVDLSQWGRKEHFEAFQSFAQCTFSQTVQLDITSLLKTVKQNGYKFYPTFI 60
Query: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
+I++ L+N H EFRMAMKDGELVIWDSV+P Y +FHEQTETFSSLWS YH D +FL Y
Sbjct: 61 YIISLLVNKHAEFRMAMKDGELVIWDSVNPGYNIFHEQTETFSSLWSYYHKDINRFLKTY 120
Query: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
S+D+A YG++LAYFPK FIENMFFVSANPWVSFTSF+LN+AN++NFFAPVFT+GKYYTQG
Sbjct: 121 SEDIAQYGDDLAYFPKEFIENMFFVSANPWVSFTSFNLNMANINNFFAPVFTIGKYYTQG 180
Query: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDE 214
DKVLMPLAIQVHHAVCDGFHVGR+LNE+QQYCDE
Sbjct: 181 DKVLMPLAIQVHHAVCDGFHVGRLLNEIQQYCDE 214
>UniRef100_UPI00004382DB UPI00004382DB UniRef100 entry
Length = 202
Score = 280 bits (715), Expect = 2e-74
Identities = 138/203 (67%), Positives = 158/203 (76%), Gaps = 2/203 (0%)
Query: 9 TTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLMN 68
TT DISQWHRK FEAFQSV CTYN +VQ++ITAFLK+V ++KHK+ A I I+ L+N
Sbjct: 1 TTSDISQWHRKGLFEAFQSVTPCTYNNSVQVNITAFLKSVSRDKHKYVAAIIGIVVGLIN 60
Query: 69 AHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVACYG 128
AHP+F M MK G LVIW SVHPC++VFHE TFSSLWSEYH D RQFL IY+QDV CYG
Sbjct: 61 AHPKFPMTMKAGSLVIWHSVHPCHSVFHELPATFSSLWSEYHADLRQFLRIYTQDV-CYG 119
Query: 129 ENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLMPLA 188
EN+AY KGF +MFFVSAN WVSFTSFDLNV NMD F APVF +GKYY QGDKVL+P
Sbjct: 120 ENMAYIRKGFSHHMFFVSANLWVSFTSFDLNVHNMDCFIAPVFNIGKYYAQGDKVLIPAG 179
Query: 189 IQVH-HAVCDGFHVGRMLNELQQ 210
VC GFHVGRM+N+L+Q
Sbjct: 180 NSGSIIPVCKGFHVGRMVNKLRQ 202
>UniRef100_Q7N4A3 Chloramphenicol acetyltransferase [Photorhabdus luminescens]
Length = 212
Score = 238 bits (608), Expect = 6e-62
Identities = 100/209 (47%), Positives = 158/209 (74%), Gaps = 1/209 (0%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
Y+ VDI W RKEHF +++V QC ++ T ++DIT L ++ + ++KFYP I++++ ++
Sbjct: 3 YSKVDIDLWDRKEHFLHYRNVVQCGFSLTAKIDITHLLSSLVEKQYKFYPTMIYLISTVV 62
Query: 68 NAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVACY 127
N++ EFRMA+KD EL++WD V+P YT+FH++TETFS++W+E++ D +F+ YS D Y
Sbjct: 63 NSYSEFRMAIKDEELIVWDGVNPAYTIFHKETETFSAIWTEFNSDLAEFMKNYSADYETY 122
Query: 128 GENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLMP 186
++L +F K + EN F +S+ PWVSF F+LN+A++ ++F P+FTMGK+Y G++ +P
Sbjct: 123 KDDLCFFSKPELPENHFHISSVPWVSFDGFNLNMASVMDYFPPIFTMGKFYQNGNQTQLP 182
Query: 187 LAIQVHHAVCDGFHVGRMLNELQQYCDEW 215
LAIQVHHA CDGFHVGR++N LQ+ C+++
Sbjct: 183 LAIQVHHATCDGFHVGRVINNLQELCNDF 211
>UniRef100_P22616 Chloramphenicol acetyltransferase II [Haemophilus influenzae]
Length = 213
Score = 220 bits (560), Expect = 2e-56
Identities = 96/207 (46%), Positives = 144/207 (69%), Gaps = 1/207 (0%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+T +D++ W+R+EHF ++ +C ++ T +LDITAF + + +KFYP I++++R++
Sbjct: 3 FTRIDLNTWNRREHFALYRQQIKCGFSLTTKLDITAFRTALAETDYKFYPVMIYLISRVV 62
Query: 68 NAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVACY 127
N PEFRMAMKD L+ WD P +TVFH++TETFS+L+ Y D +F+ Y+ +A Y
Sbjct: 63 NQFPEFRMAMKDNALIYWDQTDPVFTVFHKETETFSALFCRYCPDISEFMAGYNAVMAEY 122
Query: 128 GENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLMP 186
N A FP+G + EN +S+ PWVSF F+LN+ D++FAPVFTM K+ + ++VL+P
Sbjct: 123 QHNTALFPQGALPENHLNISSLPWVSFDGFNLNITGNDDYFAPVFTMAKFQQEDNRVLLP 182
Query: 187 LAIQVHHAVCDGFHVGRMLNELQQYCD 213
+++QVHHAVCDGFH R +N LQ CD
Sbjct: 183 VSVQVHHAVCDGFHAARFINTLQMMCD 209
>UniRef100_Q6A290 Chloramphenicol acetyltransferase II [Haemophilus influenzae]
Length = 213
Score = 218 bits (554), Expect = 1e-55
Identities = 95/207 (45%), Positives = 143/207 (68%), Gaps = 1/207 (0%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+T +D++ W+R+EHF ++ +C ++ T +LDITA + + +KFYP I++++R++
Sbjct: 3 FTRIDLNTWNRREHFALYRQQIKCGFSLTTKLDITALRTALAETDYKFYPVMIYLISRVV 62
Query: 68 NAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVACY 127
N PEFRMAMKD L+ WD P +TVFH++TETFS+L+ Y D +F+ Y+ +A Y
Sbjct: 63 NQFPEFRMAMKDNALIYWDQTDPVFTVFHKETETFSALFCRYCPDISEFMAGYNAVMAEY 122
Query: 128 GENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLMP 186
N A FP+G + EN +S+ PWVSF F+LN+ D++FAPVFTM K+ + ++VL+P
Sbjct: 123 QHNTALFPQGALPENHLNISSLPWVSFDGFNLNITGNDDYFAPVFTMAKFQQEDNRVLLP 182
Query: 187 LAIQVHHAVCDGFHVGRMLNELQQYCD 213
+++QVHHAVCDGFH R +N LQ CD
Sbjct: 183 VSVQVHHAVCDGFHAARFINTLQMMCD 209
>UniRef100_P00484 Chloramphenicol acetyltransferase III [Escherichia coli]
Length = 213
Score = 215 bits (548), Expect = 6e-55
Identities = 98/207 (47%), Positives = 141/207 (67%), Gaps = 1/207 (0%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
YT D+ W R+EHFE ++ C ++ T ++DIT K++ + +KFYP I+++A+ +
Sbjct: 3 YTKFDVKNWVRREHFEFYRHRLPCGFSLTSKIDITTLKKSLDDSAYKFYPVMIYLIAQAV 62
Query: 68 NAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVACY 127
N E RMA+KD EL++WDSV P +TVFH++TETFS+L Y D QF+ Y + Y
Sbjct: 63 NQFDELRMAIKDDELIVWDSVDPQFTVFHQETETFSALSCPYSSDIDQFMVNYLSVMERY 122
Query: 128 GENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLMP 186
+ FP+G EN +SA PWV+F SF+LNVAN ++FAP+ TM KY +GD++L+P
Sbjct: 123 KSDTKLFPQGVTPENHLNISALPWVNFDSFNLNVANFTDYFAPIITMAKYQQEGDRLLLP 182
Query: 187 LAIQVHHAVCDGFHVGRMLNELQQYCD 213
L++QVHHAVCDGFHV R +N LQ+ C+
Sbjct: 183 LSVQVHHAVCDGFHVARFINRLQELCN 209
>UniRef100_P22615 Chloramphenicol acetyltransferase II [Escherichia coli]
Length = 213
Score = 215 bits (547), Expect = 8e-55
Identities = 95/207 (45%), Positives = 141/207 (67%), Gaps = 1/207 (0%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+T +D++ W+R+EHF ++ +C ++ T +LDITA + + +KFYP I++++R +
Sbjct: 3 FTRIDLNTWNRREHFALYRQQIKCGFSLTTKLDITALRTALAETGYKFYPLMIYLISRAV 62
Query: 68 NAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVACY 127
N PEFRMA+KD EL+ WD P +TVFH++TETFS+L Y D +F+ Y+ A Y
Sbjct: 63 NQFPEFRMALKDNELIYWDQSDPVFTVFHKETETFSALSCRYFPDLSEFMAGYNAVTAEY 122
Query: 128 GENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLMP 186
+ FP+G + EN +S+ PWVSF F+LN+ D++FAPVFTM K+ +GD+VL+P
Sbjct: 123 QHDTRLFPQGNLPENHLNISSLPWVSFDGFNLNITGNDDYFAPVFTMAKFQQEGDRVLLP 182
Query: 187 LAIQVHHAVCDGFHVGRMLNELQQYCD 213
+++QVHHAVCDGFH R +N LQ CD
Sbjct: 183 VSVQVHHAVCDGFHAARFINTLQLMCD 209
>UniRef100_Q70WF4 Chloramphenicol aminotransferase [Aeromonas salmonicida]
Length = 213
Score = 214 bits (546), Expect = 1e-54
Identities = 95/207 (45%), Positives = 141/207 (67%), Gaps = 1/207 (0%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+T +D++ W+R+EHF ++ +C ++ T +LDITA + + +KFYP I++++R +
Sbjct: 3 FTRIDLNTWNRREHFAFYRQQIKCGFSLTTKLDITALRTALAETGYKFYPLMIYLISRAV 62
Query: 68 NAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVACY 127
N PEFRMA+KD EL+ WD P +TVFH++TETFS+L Y D +F+ Y+ A Y
Sbjct: 63 NQFPEFRMALKDNELIYWDQSDPVFTVFHKETETFSALSCRYFPDLSEFMAGYNAVTAEY 122
Query: 128 GENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLMP 186
+ FP+G + EN +S+ PWVSF F+LN+ D++FAPVFTM K+ +GD+VL+P
Sbjct: 123 QHDTRLFPQGNLPENHLNISSLPWVSFDGFNLNITGNDDYFAPVFTMAKFQQEGDRVLLP 182
Query: 187 LAIQVHHAVCDGFHVGRMLNELQQYCD 213
+++QVHHAVCDGFH R +N LQ CD
Sbjct: 183 VSVQVHHAVCDGFHAARFINTLQLMCD 209
>UniRef100_Q7B0A8 Chloramphenicol acetyl transferase [uncultured eubacterium]
Length = 213
Score = 213 bits (543), Expect = 2e-54
Identities = 97/207 (46%), Positives = 141/207 (67%), Gaps = 1/207 (0%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
YT D+ W R+EHFE ++ C ++ T ++DIT K++ + +KFYP I+++A+ +
Sbjct: 3 YTKFDVKNWVRREHFEFYRHRLPCGFSLTSKIDITTLKKSLDDSAYKFYPVMIYLIAQAV 62
Query: 68 NAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVACY 127
N E RMA+KD EL++WDSV P +TVFH++TETFS+L Y D QF+ Y + Y
Sbjct: 63 NQFDELRMAIKDDELIVWDSVDPQFTVFHQETETFSALSCPYSSDIDQFMVNYLSVMERY 122
Query: 128 GENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLMP 186
+ FP+G EN +SA PWV+F SF+LNVAN ++FAP+ TM KY +GD++L+P
Sbjct: 123 KSDTKLFPQGVTPENHLNISALPWVNFDSFNLNVANFTDYFAPIITMAKYQQEGDRLLLP 182
Query: 187 LAIQVHHAVCDGFHVGRMLNELQQYCD 213
L++QVHHAVCDGFHV R ++ LQ+ C+
Sbjct: 183 LSVQVHHAVCDGFHVARFISRLQELCN 209
>UniRef100_P26825 Chloramphenicol acetyltransferase [Clostridium perfringens]
Length = 219
Score = 206 bits (524), Expect = 3e-52
Identities = 89/211 (42%), Positives = 139/211 (65%), Gaps = 2/211 (0%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+ +DI W+RK +FE + + +CTY+ T ++IT L+ +K K YP I+I+ ++
Sbjct: 3 FNLIDIEDWNRKPYFEHYLNAVRCTYSMTANIEITGLLREIKLKGLKLYPTLIYIITTVV 62
Query: 68 NAHPEFRMAM-KDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVAC 126
N H EFR + G+L WDS++P YTVFH+ ETFSS+W+EY ++F +F + Y +D+
Sbjct: 63 NRHKEFRTCFDQKGKLGYWDSMNPSYTVFHKDNETFSSIWTEYDENFPRFYYNYLEDIRN 122
Query: 127 YGENLAYFPK-GFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLM 185
Y + L + PK G N VS+ PWV+FT F+LN+ N + P+FT+GKY+ Q +K+L+
Sbjct: 123 YSDVLNFMPKTGEPANTINVSSIPWVNFTGFNLNIYNDATYLIPIFTLGKYFQQDNKILL 182
Query: 186 PLAIQVHHAVCDGFHVGRMLNELQQYCDEWQ 216
P+++QVHHAVCDG+H+ R NE Q+ ++
Sbjct: 183 PMSVQVHHAVCDGYHISRFFNEAQELASNYE 213
>UniRef100_Q65GV9 Hypothetical protein [Bacillus licheniformis]
Length = 216
Score = 204 bits (520), Expect = 1e-51
Identities = 89/213 (41%), Positives = 144/213 (66%), Gaps = 6/213 (2%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+ T+++ W+RK +F+ + A+C+++ T +++T L +KK K K YPAFI+I++R++
Sbjct: 3 FQTIELDTWYRKSYFDHYMKEAKCSFSITANVNVTNLLAVLKKKKLKLYPAFIYIVSRVI 62
Query: 68 NAHPEFRMAMKD-GELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVAC 126
++ PEFR D G+L W+ +HPCY +FH+ +TFS+LW+EY DDF QF H Y D
Sbjct: 63 HSRPEFRTTFDDKGQLGYWEQMHPCYAIFHQDDQTFSALWTEYSDDFSQFYHQYLLDAER 122
Query: 127 YGENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLM 185
+G+ + K I N F VS+ PWV F++F+LN+ N ++ P+ T GKY+++G + +
Sbjct: 123 FGDKRGLWAKPDIPPNTFSVSSIPWVRFSNFNLNLDNSEHLL-PIITNGKYFSEGRETFL 181
Query: 186 PLAIQVHHAVCDGFHVGRMLNELQQY---CDEW 215
P+++QVHHAVCDG+H G +NEL++ C+EW
Sbjct: 182 PVSLQVHHAVCDGYHAGAFINELERLAADCEEW 214
>UniRef100_Q02736 Chloramphenicol acetyltransferase [Clostridium butyricum]
Length = 219
Score = 204 bits (518), Expect = 2e-51
Identities = 91/211 (43%), Positives = 137/211 (64%), Gaps = 2/211 (0%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+ +DI+ W RK +FE + + +CTY+ T ++IT L +K KFYP I+++A ++
Sbjct: 3 FNLIDINHWSRKPYFEHYLNNVKCTYSMTANIEITDLLYEIKLKNIKFYPTLIYMIATVV 62
Query: 68 NAHPEFRMAMKD-GELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVAC 126
N H EFR+ G L WDS++P YT+FH++ ETFSS+W+EY+ F +F Y D+
Sbjct: 63 NNHKEFRICFDHKGSLGYWDSMNPSYTIFHKENETFSSIWTEYNKSFLRFYSDYLDDIKN 122
Query: 127 YGENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLM 185
YG + + PK +N F VS+ PWVSFT F+LNV N + P+FT GKY+ Q +K+ +
Sbjct: 123 YGNIMKFTPKSNEPDNTFSVSSIPWVSFTGFNLNVYNEGTYLIPIFTAGKYFKQENKIFI 182
Query: 186 PLAIQVHHAVCDGFHVGRMLNELQQYCDEWQ 216
P++IQVHHA+CDG+H R +NE+Q+ +Q
Sbjct: 183 PISIQVHHAICDGYHASRFINEMQELAFSFQ 213
>UniRef100_UPI000026EBA5 UPI000026EBA5 UniRef100 entry
Length = 93
Score = 200 bits (508), Expect = 3e-50
Identities = 91/93 (97%), Positives = 92/93 (98%)
Query: 127 YGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLMP 186
YGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYY QGDKV+MP
Sbjct: 1 YGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYMQGDKVMMP 60
Query: 187 LAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
LAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA
Sbjct: 61 LAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 93
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.137 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 382,889,185
Number of Sequences: 2790947
Number of extensions: 15150095
Number of successful extensions: 39722
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 68
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 39528
Number of HSP's gapped (non-prelim): 77
length of query: 219
length of database: 848,049,833
effective HSP length: 122
effective length of query: 97
effective length of database: 507,554,299
effective search space: 49232767003
effective search space used: 49232767003
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 72 (32.3 bits)
Medicago: description of AC148969.8


