

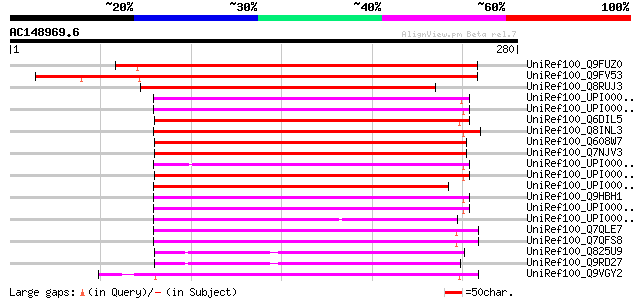
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148969.6 + phase: 0
(280 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FUZ0 Peptide deformylase, mitochondrial precursor [L... 243 3e-63
UniRef100_Q9FV53 Peptide deformylase, mitochondrial precursor [A... 242 9e-63
UniRef100_Q8RUJ3 Peptide deformylase-like protein [Oryza sativa] 202 1e-50
UniRef100_UPI0000017B99 UPI0000017B99 UniRef100 entry 148 1e-34
UniRef100_UPI00003374D4 UPI00003374D4 UniRef100 entry 148 1e-34
UniRef100_Q6DIL5 MGC89413 protein [Xenopus tropicalis] 148 2e-34
UniRef100_Q8INL3 CG31373-PA [Drosophila melanogaster] 144 3e-33
UniRef100_Q608W7 Polypeptide deformylase [Methylococcus capsulatus] 143 4e-33
UniRef100_Q7NJV3 Peptide deformylase 1 [Gloeobacter violaceus] 143 4e-33
UniRef100_UPI0000436F74 UPI0000436F74 UniRef100 entry 140 5e-32
UniRef100_UPI00001D1446 UPI00001D1446 UniRef100 entry 140 5e-32
UniRef100_UPI0000026615 UPI0000026615 UniRef100 entry 139 6e-32
UniRef100_Q9HBH1 Peptide deformylase, mitochondrial precursor [H... 133 4e-30
UniRef100_UPI000036A88F UPI000036A88F UniRef100 entry 131 2e-29
UniRef100_UPI00002484BE UPI00002484BE UniRef100 entry 130 5e-29
UniRef100_Q7QLE7 ENSANGP00000012713 [Anopheles gambiae str. PEST] 128 2e-28
UniRef100_Q7QFS8 ENSANGP00000017891 [Anopheles gambiae str. PEST] 128 2e-28
UniRef100_Q825U9 Peptide deformylase 3 [Streptomyces avermitilis] 121 2e-26
UniRef100_Q9RD27 Peptide deformylase 1 [Streptomyces coelicolor] 120 4e-26
UniRef100_Q9VGY2 CG31278-PA [Drosophila melanogaster] 120 5e-26
>UniRef100_Q9FUZ0 Peptide deformylase, mitochondrial precursor [Lycopersicon
esculentum]
Length = 277
Score = 243 bits (621), Expect = 3e-63
Identities = 122/203 (60%), Positives = 158/203 (77%), Gaps = 3/203 (1%)
Query: 59 SFSSASSETAF---LSKTLKSELPHIVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMIL 115
++SSA++ + L + K +P IV+AGDPVLHEP++++ EI S++IQKII+ M+
Sbjct: 66 NYSSATARAGWFLGLGEKKKQAMPDIVKAGDPVLHEPSQDIPLEEIGSERIQKIIEEMVK 125
Query: 116 VMRNAPGISLSAQKIGIPLRIIVLEEPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIK 175
VMRNAPG+ L+A +IGIPL+IIVLE+ E + ++ K DRRPF LLVI+NPKLK K
Sbjct: 126 VMRNAPGVGLAAPQIGIPLKIIVLEDTNEYISYAPKDETKAQDRRPFGLLVIINPKLKKK 185
Query: 176 SNKTFLFFEGCLSVHGFQAVVERYLDVEVEGFDRYGEPIKINASGWHARILQHECDHLDG 235
NKT LFFEGCLSV GF+AVVER+L+VEV G DR G+ IK++ASGW ARILQHE DHLDG
Sbjct: 186 GNKTALFFEGCLSVDGFRAVVERHLEVEVTGLDRNGKAIKVDASGWQARILQHEYDHLDG 245
Query: 236 TLYVDKMVPRTFRSWENINMSIA 258
TLYVDKM PRTFR+ EN+++ +A
Sbjct: 246 TLYVDKMAPRTFRTVENLDLPLA 268
>UniRef100_Q9FV53 Peptide deformylase, mitochondrial precursor [Arabidopsis thaliana]
Length = 259
Score = 242 bits (617), Expect = 9e-63
Identities = 136/250 (54%), Positives = 171/250 (68%), Gaps = 6/250 (2%)
Query: 15 MEAARRLASRLRVVPMMLVFSNAT--VSSSSSSCNTETPPSNTKLSSFSSASSETAFL-- 70
ME R++ RL V + + VS SS N SS SS S++ +L
Sbjct: 1 METLFRVSLRLLPVSAAVTCRSIRFPVSRPGSSHLLNRKLYNLPTSSSSSLSTKAGWLLG 60
Query: 71 --SKTLKSELPHIVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQ 128
K K +LP IV +GDPVLHE AREVD EI S++IQKIID MI VMR APG+ L+A
Sbjct: 61 LGEKKKKVDLPEIVASGDPVLHEKAREVDPGEIGSERIQKIIDDMIKVMRLAPGVGLAAP 120
Query: 129 KIGIPLRIIVLEEPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLS 188
+IG+PLRIIVLE+ KE + +E +RR FDL+V++NP LK +SNK LFFEGCLS
Sbjct: 121 QIGVPLRIIVLEDTKEYISYAPKEEILAQERRHFDLMVMVNPVLKERSNKKALFFEGCLS 180
Query: 189 VHGFQAVVERYLDVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTFR 248
V GF+A VERYL+V V G+DR G+ I++NASGW ARILQHECDHLDG LYVDKMVPRTFR
Sbjct: 181 VDGFRAAVERYLEVVVTGYDRQGKRIEVNASGWQARILQHECDHLDGNLYVDKMVPRTFR 240
Query: 249 SWENINMSIA 258
+ +N+++ +A
Sbjct: 241 TVDNLDLPLA 250
>UniRef100_Q8RUJ3 Peptide deformylase-like protein [Oryza sativa]
Length = 246
Score = 202 bits (513), Expect = 1e-50
Identities = 98/163 (60%), Positives = 126/163 (77%)
Query: 73 TLKSELPHIVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGI 132
T + P V+AGDPVLHEPA++V +I S+K+Q +ID M+ VMR APG+ L+A +IG+
Sbjct: 66 TAMTVTPGTVKAGDPVLHEPAQDVAPGDIPSEKVQGVIDRMVAVMRKAPGVGLAAPQIGV 125
Query: 133 PLRIIVLEEPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGF 192
PL+IIVLE+ +E + ++ + DRRPFDLLVI+NPKLK S +T LFFEGCLSV G+
Sbjct: 126 PLKIIVLEDTQEYISYAPKKDIEAQDRRPFDLLVIINPKLKTTSKRTALFFEGCLSVDGY 185
Query: 193 QAVVERYLDVEVEGFDRYGEPIKINASGWHARILQHECDHLDG 235
+A+VER+LDVEV G DR G PIK+ ASGW ARILQHECDHL+G
Sbjct: 186 RALVERHLDVEVSGLDRNGRPIKVEASGWQARILQHECDHLEG 228
>UniRef100_UPI0000017B99 UPI0000017B99 UniRef100 entry
Length = 198
Score = 148 bits (374), Expect = 1e-34
Identities = 79/177 (44%), Positives = 105/177 (58%), Gaps = 2/177 (1%)
Query: 80 HIVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVL 139
H+ Q GDPVL A VD + I +IQK I+ ++ VMR + LSA +IG+PLRI+ L
Sbjct: 21 HVCQVGDPVLRSRAAAVDPAAIRGAEIQKTINTLVKVMRKLDCVGLSAPQIGVPLRILAL 80
Query: 140 EEPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERY 199
E P++ L + + L + +NP L++ +T LF E C S+ G+ A V RY
Sbjct: 81 EYPEKMLEESSPASREARGLSAQPLRIFVNPHLRVLDGRTVLFQEACESISGYSATVPRY 140
Query: 200 LDVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTFR--SWENIN 254
L VEV G + GE + ASGW ARILQHE DHLDG LYVD+M RTF +W+ N
Sbjct: 141 LSVEVSGLNEKGEDVTWEASGWPARILQHEMDHLDGVLYVDRMDSRTFLNINWQAFN 197
>UniRef100_UPI00003374D4 UPI00003374D4 UniRef100 entry
Length = 198
Score = 148 bits (374), Expect = 1e-34
Identities = 75/177 (42%), Positives = 105/177 (58%), Gaps = 2/177 (1%)
Query: 80 HIVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVL 139
H+ Q GDPVL A VD + ++QK++ ++ VMR + LSA +IG+PLRI+ L
Sbjct: 21 HVCQVGDPVLRSRAAPVDPGAVGGAEVQKVVHTLVKVMRELDCVGLSAPQIGVPLRILAL 80
Query: 140 EEPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERY 199
E P++ L + + L + +NP+L++ +T LF E C S+ GF A V RY
Sbjct: 81 EYPEKMLEESSPASREARGLSAQPLRIFVNPQLRVLDGRTALFQEACESISGFSATVPRY 140
Query: 200 LDVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTFRS--WENIN 254
L VEV G + GE ++ A GW ARILQHE DHLDG LY+D+M RTF + W+ N
Sbjct: 141 LSVEVSGLNENGEEVRWQARGWPARILQHEMDHLDGVLYIDRMDSRTFTNIHWQAFN 197
>UniRef100_Q6DIL5 MGC89413 protein [Xenopus tropicalis]
Length = 239
Score = 148 bits (373), Expect = 2e-34
Identities = 76/176 (43%), Positives = 107/176 (60%), Gaps = 2/176 (1%)
Query: 81 IVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVLE 140
+ Q GDPVL A V ++I+ Q +++ M+ V+R + LSA +IG+PLRI+ +
Sbjct: 63 VTQTGDPVLRCTAARVPCAQISHPDTQAVVNQMVRVLRAGCCVGLSAPQIGVPLRILAVA 122
Query: 141 EPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERYL 200
P++ EV + PF L + +NP+++I ++T F EGC SV GF AVV RY
Sbjct: 123 FPQQMYQAVPPEVRNAREMSPFPLQIFINPEMRIVDSRTLSFPEGCSSVQGFSAVVPRYY 182
Query: 201 DVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTF--RSWENIN 254
VE++G + GE + A GW ARI+QHE DHLDG LY+DKM PRTF SW +N
Sbjct: 183 AVELQGMNPKGEHVTWQAQGWAARIIQHEMDHLDGVLYIDKMDPRTFVNISWMEVN 238
>UniRef100_Q8INL3 CG31373-PA [Drosophila melanogaster]
Length = 196
Score = 144 bits (363), Expect = 3e-33
Identities = 76/183 (41%), Positives = 112/183 (60%), Gaps = 2/183 (1%)
Query: 80 HIVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVL 139
H Q GDPVL + A EV +I+S +I +IIDGM+ V+R+ + ++A ++GIPLRIIV+
Sbjct: 8 HFTQIGDPVLRQRAEEVPPEDIDSREINQIIDGMVKVLRHYDCVGVAAPQVGIPLRIIVM 67
Query: 140 EEPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERY 199
E + + E+ + L V +NP+L+I S++ EGC+SV G+ A VERY
Sbjct: 68 EFREGKQEQFKPEIYEERKMSILPLAVFINPELEIISSQVNKHPEGCMSVRGYSAEVERY 127
Query: 200 LDVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTFRS--WENINMSI 257
V + G + G P ++ GW+ARI QHE DHL+GT+Y+D+M TF WE IN +
Sbjct: 128 DKVRIRGIGKLGTPSEMELEGWNARIAQHEVDHLNGTIYMDRMDLSTFNCILWEQINAAE 187
Query: 258 ARA 260
R+
Sbjct: 188 GRS 190
>UniRef100_Q608W7 Polypeptide deformylase [Methylococcus capsulatus]
Length = 191
Score = 143 bits (361), Expect = 4e-33
Identities = 79/172 (45%), Positives = 106/172 (60%)
Query: 81 IVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVLE 140
IVQAG+PVL + AR + EI S +Q +I M MR+APG+ L+A +IG L++ V+E
Sbjct: 10 IVQAGEPVLRQRARPLSPEEIRSAAVQALIGHMRETMRDAPGVGLAAPQIGQGLQLAVIE 69
Query: 141 EPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERYL 200
+ + + E R P VI+NP++ +S +T +F EGCLS+ GF A VER
Sbjct: 70 DRADYHRGLSAEELAARGREPVPFHVIVNPEIVARSEETDVFHEGCLSLAGFSARVERAR 129
Query: 201 DVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTFRSWEN 252
V V D GEP I ASGW+ARILQHE DHL G LY+D+M PR+F + N
Sbjct: 130 WVRVSCLDHRGEPQTIEASGWYARILQHEIDHLHGRLYIDRMDPRSFTTQPN 181
>UniRef100_Q7NJV3 Peptide deformylase 1 [Gloeobacter violaceus]
Length = 227
Score = 143 bits (361), Expect = 4e-33
Identities = 71/172 (41%), Positives = 109/172 (63%)
Query: 81 IVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVLE 140
IV+ GDPVL A+ ++ EI S+ IQ++I M MR APG+ L+A ++G+ ++++V+E
Sbjct: 48 IVKTGDPVLRLTAKPLNSDEIQSEAIQQLIAAMAERMREAPGVGLAAPQVGVSVQLVVIE 107
Query: 141 EPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERYL 200
+ E + + + +R P V++NP L ++ ++ +FFEGCLS+ G+Q +V R
Sbjct: 108 DRPEYIERLSGAERREREREPVPFHVLINPVLSVEGEESAVFFEGCLSIPGYQGLVARAR 167
Query: 201 DVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTFRSWEN 252
V VE D P+ I A GW+ARILQHE DHL+G L VD+M +TF + EN
Sbjct: 168 VVRVEALDERAAPVVIRAHGWYARILQHEIDHLNGLLCVDRMDLQTFSTLEN 219
>UniRef100_UPI0000436F74 UPI0000436F74 UniRef100 entry
Length = 198
Score = 140 bits (352), Expect = 5e-32
Identities = 74/177 (41%), Positives = 106/177 (59%), Gaps = 3/177 (1%)
Query: 80 HIVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVL 139
H+ Q GDPVL A EV+ + I ++QK+I ++ VMR + LSA +IG+PLRI+ L
Sbjct: 22 HVCQVGDPVLRSHAAEVEGA-IQGPEVQKVIKTLVKVMRKLECVGLSAPQIGVPLRILAL 80
Query: 140 EEPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERY 199
E PK+ L + + L++ +NP+L++ +T +F E C S+ G+ A V RY
Sbjct: 81 EYPKKMLEESSTASVEARGLVAVPLMIFINPQLRVLDGRTVIFQEACESISGYSASVPRY 140
Query: 200 LDVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTFRS--WENIN 254
+ VEV G + E + ASGW ARILQHE DHL+G LY+D M +TF + WE N
Sbjct: 141 VSVEVSGLNEKAEEVSWKASGWPARILQHEMDHLNGVLYIDHMDSKTFINVKWEEHN 197
>UniRef100_UPI00001D1446 UPI00001D1446 UniRef100 entry
Length = 231
Score = 140 bits (352), Expect = 5e-32
Identities = 68/176 (38%), Positives = 108/176 (60%), Gaps = 2/176 (1%)
Query: 81 IVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVLE 140
+ Q GDPVL A V+ ++ ++Q++++ ++ VMR + LSA ++G+PL+++VLE
Sbjct: 55 VCQVGDPVLRTVAAPVEPKQLAGPELQRLVEQLVQVMRRRGCVGLSAPQLGVPLQVLVLE 114
Query: 141 EPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERYL 200
P ++ + ++ PF L V++NP L++ ++ F EGC SV GF A V R+
Sbjct: 115 FPDRLFRAFSPRLRELRQMEPFPLRVLVNPSLRVLDSRLVTFPEGCESVAGFLACVPRFQ 174
Query: 201 DVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTFRS--WENIN 254
V++ G D GEP+ +ASGW ARI+QHE DHL G L++DKM TF + W +N
Sbjct: 175 AVQISGLDPKGEPVVWSASGWTARIIQHEMDHLHGCLFIDKMDSGTFTNLHWMEVN 230
>UniRef100_UPI0000026615 UPI0000026615 UniRef100 entry
Length = 175
Score = 139 bits (351), Expect = 6e-32
Identities = 66/163 (40%), Positives = 103/163 (62%)
Query: 80 HIVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVL 139
H+ Q GDPVL A V+ ++ ++Q+++ M+ VMR + LSA ++G+PL+++ L
Sbjct: 12 HVCQVGDPVLRVVAAPVEPEQLAGPELQRLVGRMVQVMRRRGCVGLSAPQLGVPLQVLAL 71
Query: 140 EEPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERY 199
E P + L ++ + ++ PF L V++NP L++ ++ F EGC SV GF A V R+
Sbjct: 72 EFPDKLLRAFSPRLRELRQMEPFPLRVLVNPSLRVLDSRLVTFPEGCESVAGFLACVPRF 131
Query: 200 LDVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKM 242
V++ G D GEP+ +ASGW ARI+QHE DHL G L++DKM
Sbjct: 132 QAVQISGLDPKGEPVVWSASGWTARIIQHEMDHLQGCLFIDKM 174
>UniRef100_Q9HBH1 Peptide deformylase, mitochondrial precursor [Homo sapiens]
Length = 243
Score = 133 bits (335), Expect = 4e-30
Identities = 67/177 (37%), Positives = 101/177 (56%), Gaps = 2/177 (1%)
Query: 80 HIVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVL 139
H+ Q GDPVL A V+ +++ ++Q++ ++ VMR + LSA ++G+P +++ L
Sbjct: 66 HVCQVGDPVLRGVAAPVERAQLGGPELQRLTQRLVQVMRRRRCVGLSAPQLGVPRQVLAL 125
Query: 140 EEPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERY 199
E P+ + PF L V +NP L++ ++ F EGC SV GF A V R+
Sbjct: 126 ELPEALCRECPPRQRALRQMEPFPLRVFVNPSLRVLDSRLVTFPEGCESVAGFLACVPRF 185
Query: 200 LDVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTFRS--WENIN 254
V++ G D GE + ASGW ARI+QHE DHL G L++DKM RTF + W +N
Sbjct: 186 QAVQISGLDPNGEQVVWQASGWAARIIQHEMDHLQGCLFIDKMDSRTFTNVYWMKVN 242
>UniRef100_UPI000036A88F UPI000036A88F UniRef100 entry
Length = 260
Score = 131 bits (330), Expect = 2e-29
Identities = 66/177 (37%), Positives = 100/177 (56%), Gaps = 2/177 (1%)
Query: 80 HIVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVL 139
H+ Q GDPVL A V+ +++ ++Q++ ++ VMR + LSA ++G+P +++ L
Sbjct: 83 HVCQVGDPVLRGVAAPVERAQLGGPELQRLTQRLVQVMRRRRCVGLSAPQLGVPRQVLAL 142
Query: 140 EEPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERY 199
E P+ + PF L V +NP L++ ++ F EGC SV GF A V R+
Sbjct: 143 ELPEALCRECPPRQRALRQMEPFPLRVFVNPSLRVLDSRLVTFPEGCESVAGFLACVPRF 202
Query: 200 LDVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTFRS--WENIN 254
V++ G D GE + SGW ARI+QHE DHL G L++DKM RTF + W +N
Sbjct: 203 QAVQISGLDPNGEQVVWQPSGWAARIIQHEMDHLQGCLFIDKMDSRTFTNVYWMKVN 259
>UniRef100_UPI00002484BE UPI00002484BE UniRef100 entry
Length = 194
Score = 130 bits (326), Expect = 5e-29
Identities = 69/168 (41%), Positives = 98/168 (58%), Gaps = 1/168 (0%)
Query: 80 HIVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVL 139
H+ Q GDPVL A +V+ I ++QK+I ++ VMR + LSA +IG+PLRI+ L
Sbjct: 21 HVCQVGDPVLRSHAAKVEPGAIQGPEVQKVIKTLVKVMRKLECVGLSAPQIGVPLRILAL 80
Query: 140 EEPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERY 199
E PK+ L + + L++ +NP+L + +T L E C S+ G+ A V RY
Sbjct: 81 EYPKKMLEESSTASVEARGLLAVPLMIFINPQLPVLDRRTVLP-EACESISGYSASVPRY 139
Query: 200 LDVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTF 247
+ VE+ + E I ASGW ARILQHE DHL+G LY+D M +TF
Sbjct: 140 VSVEISCLNEKAEAISWKASGWPARILQHEMDHLNGVLYIDHMDSKTF 187
>UniRef100_Q7QLE7 ENSANGP00000012713 [Anopheles gambiae str. PEST]
Length = 221
Score = 128 bits (321), Expect = 2e-28
Identities = 68/182 (37%), Positives = 104/182 (56%), Gaps = 2/182 (1%)
Query: 80 HIVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVL 139
HIVQ GDPVL PA + E+ S ++Q + + VMR + L+A ++G+ LR V+
Sbjct: 33 HIVQLGDPVLRVPANAIPEKELQSAEVQYLARHLTKVMRAYRCVGLAAPQLGLSLRAFVM 92
Query: 140 EEPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERY 199
E E YT+ K+ + P L ++LNP+LK+ + + + E C SV G++A V RY
Sbjct: 93 EFKDELRDQYTKADYKLREMEPLPLTILLNPELKVLNYEKVIHTEACESVRGYRADVPRY 152
Query: 200 LDVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPR--TFRSWENINMSI 257
++ ++GFD G ++ SGW+ARI QHE DHL+G +Y D M + T W+ +N
Sbjct: 153 REILLQGFDATGNRQELRLSGWNARIAQHEMDHLNGIVYTDIMNRKSLTCTCWQAVNAKS 212
Query: 258 AR 259
R
Sbjct: 213 GR 214
>UniRef100_Q7QFS8 ENSANGP00000017891 [Anopheles gambiae str. PEST]
Length = 240
Score = 128 bits (321), Expect = 2e-28
Identities = 68/182 (37%), Positives = 104/182 (56%), Gaps = 2/182 (1%)
Query: 80 HIVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVL 139
HIVQ GDPVL PA + E+ S ++Q + + VMR + L+A ++G+ LR V+
Sbjct: 52 HIVQLGDPVLRVPANAIPEKELQSAEVQYLARHLTKVMRAYRCVGLAAPQLGLSLRAFVM 111
Query: 140 EEPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERY 199
E E YT+ K+ + P L ++LNP+LK+ + + + E C SV G++A V RY
Sbjct: 112 EFKDELRDQYTKADYKLREMEPLPLTILLNPELKVLNYEKVIHTEACESVRGYRADVPRY 171
Query: 200 LDVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPR--TFRSWENINMSI 257
++ ++GFD G ++ SGW+ARI QHE DHL+G +Y D M + T W+ +N
Sbjct: 172 REILLQGFDATGNRQELRLSGWNARIAQHEMDHLNGIVYTDIMNRKSLTCTCWQAVNAKS 231
Query: 258 AR 259
R
Sbjct: 232 GR 233
>UniRef100_Q825U9 Peptide deformylase 3 [Streptomyces avermitilis]
Length = 224
Score = 121 bits (304), Expect = 2e-26
Identities = 68/171 (39%), Positives = 96/171 (55%), Gaps = 5/171 (2%)
Query: 81 IVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVLE 140
IV AGDPVL A D ++ + + ++ + L M APG+ L+A ++G+ LRI V+E
Sbjct: 35 IVAAGDPVLRRGAEPYD-GQLGPGLLARFVEALRLTMHAAPGVGLAAPQVGVGLRIAVIE 93
Query: 141 EPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERYL 200
+P EEV + R P V++NP + + FFEGCLSV G+QAVV R
Sbjct: 94 DPAP----VPEEVGAVRGRVPQPFRVLVNPSYEAVGSDRAAFFEGCLSVPGWQAVVARPA 149
Query: 201 DVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTFRSWE 251
V + D +G + +GW ARI+QHE DHLDG LY+D+ R+ S E
Sbjct: 150 RVRLTALDEHGRAVDEEFTGWPARIVQHETDHLDGMLYLDRAELRSLSSNE 200
>UniRef100_Q9RD27 Peptide deformylase 1 [Streptomyces coelicolor]
Length = 218
Score = 120 bits (301), Expect = 4e-26
Identities = 66/169 (39%), Positives = 96/169 (56%), Gaps = 5/169 (2%)
Query: 81 IVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVLE 140
IV AGDPVL A D ++ ++ ++ + L M APG+ L+A ++G+ LR+ V+E
Sbjct: 26 IVAAGDPVLRRAAEPYD-GQVAPALFERFVEALRLTMHAAPGVGLAAPQVGVGLRVAVIE 84
Query: 141 EPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERYL 200
+P +EV R P V++NP + FFEGCLSV G+QAVV R+
Sbjct: 85 DPAP----VPDEVRVARGRVPQPFRVLVNPSYEPAGAGRAAFFEGCLSVPGWQAVVARHA 140
Query: 201 DVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTFRS 249
+V + D +G + +GW ARI+QHE DHLDGTLY+D+ R+ S
Sbjct: 141 EVRLRAHDEHGRAVDEVFAGWPARIVQHETDHLDGTLYLDRAELRSLAS 189
>UniRef100_Q9VGY2 CG31278-PA [Drosophila melanogaster]
Length = 238
Score = 120 bits (300), Expect = 5e-26
Identities = 73/215 (33%), Positives = 108/215 (49%), Gaps = 11/215 (5%)
Query: 50 TPPSNTKLSSFSSASSETAFLSKTLKSELP---HIVQAGDPVLHEPAREVDHSEINSDKI 106
+PP+N + L T ++ LP H Q GDPVL + A V + S +I
Sbjct: 23 SPPANQSFRKWYQ------HLWTTERTNLPPYNHFTQIGDPVLRQQAALVPKEHMASPEI 76
Query: 107 QKIIDGMILVMRNAPGISLSAQKIGIPLRIIVLEEPKENLYNYTEEVNKIIDRRPFDLLV 166
+ I++ M+ V+R + ++A +IG+ LRII +E E V + L +
Sbjct: 77 KAIVERMVKVLRKFDCVGIAAPQIGVSLRIIAMEFKGRIRKELPEAVYQARQMSELPLTI 136
Query: 167 ILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERYLDVEVEGFDRYGEPIKINASGWHARIL 226
+NP L + + EGC+SV G+ A VER+ V++ G D+ G ++ SGW+ARI
Sbjct: 137 FINPVLTVTNYAKLKHPEGCMSVRGYSAEVERFEGVKLTGLDQLGNQSELALSGWNARIA 196
Query: 227 QHECDHLDGTLYVDKMVPRTF--RSWENINMSIAR 259
QHE DHL+G LY D M TF WE +N R
Sbjct: 197 QHEMDHLEGKLYTDHMDRSTFACTCWEAVNTKSGR 231
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.133 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 429,268,987
Number of Sequences: 2790947
Number of extensions: 17263379
Number of successful extensions: 73822
Number of sequences better than 10.0: 832
Number of HSP's better than 10.0 without gapping: 707
Number of HSP's successfully gapped in prelim test: 130
Number of HSP's that attempted gapping in prelim test: 71883
Number of HSP's gapped (non-prelim): 1081
length of query: 280
length of database: 848,049,833
effective HSP length: 126
effective length of query: 154
effective length of database: 496,390,511
effective search space: 76444138694
effective search space used: 76444138694
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC148969.6


