

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148969.14 + phase: 0
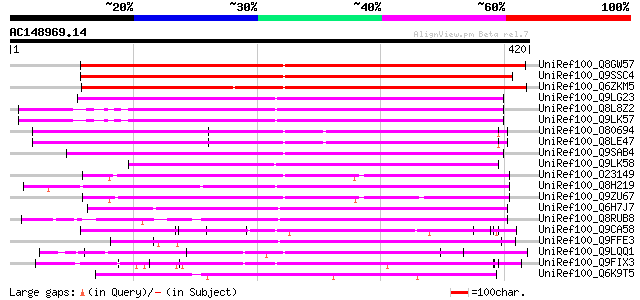
(420 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8GW57 Hypothetical protein At1g80150/F18B13_23 [Arabi... 419 e-116
UniRef100_Q9SSC4 F18B13.23 protein [Arabidopsis thaliana] 416 e-115
UniRef100_Q6ZKM5 Putative drought-inducible protein 1OS [Oryza s... 358 2e-97
UniRef100_Q9LG23 F14J16.14 [Arabidopsis thaliana] 204 3e-51
UniRef100_Q8L8Z2 Hypothetical protein [Arabidopsis thaliana] 202 2e-50
UniRef100_Q9LK57 Arabidopsis thaliana genomic DNA, chromosome 3,... 201 3e-50
UniRef100_O80694 Hypothetical protein At1g61870; F8K4.8 [Arabido... 191 3e-47
UniRef100_Q8LE47 Putative membrane-associated salt-inducible pro... 191 3e-47
UniRef100_Q9SAB4 F25C20.22 protein [Arabidopsis thaliana] 184 5e-45
UniRef100_Q9LK58 Arabidopsis thaliana genomic DNA, chromosome 3,... 169 1e-40
UniRef100_O23149 Membrane-associated salt-inducible protein like... 167 7e-40
UniRef100_Q8H219 Drought-inducible protein 1OS [Oryza sativa] 158 3e-37
UniRef100_Q9ZU67 Expressed protein [Arabidopsis thaliana] 153 8e-36
UniRef100_Q6H7J7 Membrane-associated salt-inducible protein like... 142 1e-32
UniRef100_Q8RUB8 Putative drought-inducible protein 1OS [Oryza s... 138 3e-31
UniRef100_Q9CA58 Hypothetical protein F1M20.26 [Arabidopsis thal... 133 8e-30
UniRef100_Q9FFE3 Salt-inducible protein-like [Arabidopsis thaliana] 121 3e-26
UniRef100_Q9LQQ1 F24B9.15 protein [Arabidopsis thaliana] 116 1e-24
UniRef100_Q9FIX3 Gb|AAD30619.1 [Arabidopsis thaliana] 115 2e-24
UniRef100_Q6K9T5 Membrane-associated salt-inducible protein-like... 114 5e-24
>UniRef100_Q8GW57 Hypothetical protein At1g80150/F18B13_23 [Arabidopsis thaliana]
Length = 397
Score = 419 bits (1077), Expect = e-116
Identities = 213/360 (59%), Positives = 264/360 (73%), Gaps = 1/360 (0%)
Query: 58 KPALEALKSEWDPHKLFHIFKSNATNPIVVENRFAFYDTVSRLAGAKRFDYIEQLLEQQK 117
+PAL LKSE DP KL+++FK+NATN +V+ENRFAF DTVSRLAGA R D+IE LLE QK
Sbjct: 39 EPALVKLKSERDPEKLYNLFKANATNRLVIENRFAFEDTVSRLAGAGRLDFIEDLLEHQK 98
Query: 118 KLPQSRREGFVVRIITLYGNAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFD 177
LPQ RREGF+VRII LYG AGM + AL+TF+ M + C R+VKSFNA L VL+ + D
Sbjct: 99 TLPQGRREGFMVRIIMLYGKAGMTKQALDTFFNMDLYGCKRSVKSFNAALQVLSFNPDLH 158
Query: 178 EISRFLNEVPRRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTT 237
I FL++ P ++ I +D S NIA+K+FCE L AY+ M + + G+ PDVVTYTT
Sbjct: 159 TIWEFLHDAPSKYGIDIDAVSFNIAIKSFCELGILDGAYMAMRE-MEKSGLTPDVVTYTT 217
Query: 238 LISAFYDHKRWEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNG 297
LISA Y H+R IGNGLWN +VLKG PNL TFNVRIQFLV RR WDAN L+ LM +
Sbjct: 218 LISALYKHERCVIGNGLWNLIVLKGCKPNLTTFNVRIQFLVNRRRAWDANDLLLLMPKLQ 277
Query: 298 VTPDEVTLVLVIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAY 357
V PD +T +VIKGFF A +P+MA RVY+A+H KGYK + IYQTMIH LCK G+F AY
Sbjct: 278 VEPDSITYNMVIKGFFLARFPDMAERVYTAMHGKGYKPNLKIYQTMIHYLCKAGNFDLAY 337
Query: 358 TLCKDSMRKNWFPNVDTIFMLLEGLKKSGKINKAKVIVALAEGRKPPFSFSYLASMQSIL 417
T+CKD MRK W+PN+DT+ MLL+GL K G++++AK I+ L R PPF L S++SIL
Sbjct: 338 TMCKDCMRKKWYPNLDTVEMLLKGLVKKGQLDQAKSIMELVHRRVPPFRSKQLLSLKSIL 397
>UniRef100_Q9SSC4 F18B13.23 protein [Arabidopsis thaliana]
Length = 419
Score = 416 bits (1070), Expect = e-115
Identities = 210/350 (60%), Positives = 259/350 (74%), Gaps = 1/350 (0%)
Query: 58 KPALEALKSEWDPHKLFHIFKSNATNPIVVENRFAFYDTVSRLAGAKRFDYIEQLLEQQK 117
+PAL LKSE DP KL+++FK+NATN +V+ENRFAF DTVSRLAGA R D+IE LLE QK
Sbjct: 39 EPALVKLKSERDPEKLYNLFKANATNRLVIENRFAFEDTVSRLAGAGRLDFIEDLLEHQK 98
Query: 118 KLPQSRREGFVVRIITLYGNAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFD 177
LPQ RREGF+VRII LYG AGM + AL+TF+ M + C R+VKSFNA L VL+ + D
Sbjct: 99 TLPQGRREGFIVRIIMLYGKAGMTKQALDTFFNMDLYGCKRSVKSFNAALQVLSFNPDLH 158
Query: 178 EISRFLNEVPRRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTT 237
I FL++ P ++ I +D S NIA+K+FCE L AY+ M + + G+ PDVVTYTT
Sbjct: 159 TIWEFLHDAPSKYGIDIDAVSFNIAIKSFCELGILDGAYMAMRE-MEKSGLTPDVVTYTT 217
Query: 238 LISAFYDHKRWEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNG 297
LISA Y H+R IGNGLWN MVLKG PNL TFNVRIQFLV RR WDAN L+ LM +
Sbjct: 218 LISALYKHERCVIGNGLWNLMVLKGCKPNLTTFNVRIQFLVNRRRAWDANDLLLLMPKLQ 277
Query: 298 VTPDEVTLVLVIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAY 357
V PD +T +VIKGFF A +P+MA RVY+A+H KGYK + IYQTMIH LCK G+F AY
Sbjct: 278 VEPDSITYNMVIKGFFLARFPDMAERVYTAMHGKGYKPNLKIYQTMIHYLCKAGNFDLAY 337
Query: 358 TLCKDSMRKNWFPNVDTIFMLLEGLKKSGKINKAKVIVALAEGRKPPFSF 407
T+CKD MRK W+PN+DT+ MLL+GL K G++++AK I+ L R PPFS+
Sbjct: 338 TMCKDCMRKKWYPNLDTVEMLLKGLVKKGQLDQAKSIMELVHRRVPPFSW 387
>UniRef100_Q6ZKM5 Putative drought-inducible protein 1OS [Oryza sativa]
Length = 443
Score = 358 bits (919), Expect = 2e-97
Identities = 186/360 (51%), Positives = 246/360 (67%), Gaps = 2/360 (0%)
Query: 59 PALEALKSEWDPHKLFHIFKSNATNPIVVENRFAFYDTVSRLAGAKRFDYIEQLLEQQKK 118
P L +K+E DP +L+ +F++NA N +++ENRFAF D V+RLAGA+R D +E++LEQ K
Sbjct: 80 PVLVRIKNERDPVRLYELFRANAHNRLLIENRFAFEDAVARLAGARRNDLVEEILEQHKA 139
Query: 119 LPQSRREGFVVRIITLYGNAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFDE 178
LPQ RREGFVVRII LYG A M HAL+TF +MG + C RT KS NAT+ VL ++R FDE
Sbjct: 140 LPQGRREGFVVRIIGLYGKARMPDHALSTFREMGLYGCPRTAKSLNATMKVLLRARLFDE 199
Query: 179 ISRFLNEVPRRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTL 238
+ + L E + + LD S N VK C+ +L+ A+ M + + GV+PDV+TYTTL
Sbjct: 200 VLQ-LFESSETYGVELDDISYNTVVKMMCDLGELRAAFRVMQE-MEKAGVRPDVITYTTL 257
Query: 239 ISAFYDHKRWEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGV 298
+ AFY + E+G+GLWN M L+G MP L ++NVRIQFLV RR W AN L+ M +G+
Sbjct: 258 MDAFYKCGQREVGDGLWNLMRLRGCMPTLASYNVRIQFLVNRRRGWQANDLVRKMYASGL 317
Query: 299 TPDEVTLVLVIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYT 358
PDE+T LVIKGFF G EMA V+ A+H +G K +A +YQTM+H LC+R +F A+
Sbjct: 318 RPDEITYNLVIKGFFMMGEHEMAKTVFGAMHGRGCKPNAKVYQTMVHYLCERREFDLAFR 377
Query: 359 LCKDSMRKNWFPNVDTIFMLLEGLKKSGKINKAKVIVALAEGRKPPFSFSYLASMQSILS 418
LCKDSM KNWFP+VDTI LL+GL K A+ I+ L GRKP +S + + Q ILS
Sbjct: 378 LCKDSMEKNWFPSVDTINQLLKGLISISKDRNAREIMKLVIGRKPSYSNDEVKTFQDILS 437
>UniRef100_Q9LG23 F14J16.14 [Arabidopsis thaliana]
Length = 398
Score = 204 bits (520), Expect = 3e-51
Identities = 114/344 (33%), Positives = 183/344 (53%), Gaps = 1/344 (0%)
Query: 56 PSKPALEALKSEWDPHKLFHIFKSNATNPIVVENRFAFYDTVSRLAGAKRFDYIEQLLEQ 115
P K + E +P ++ FK + N + TV RL AKR Y+E++LE+
Sbjct: 40 PQKSLTSLVNGERNPKRIVEKFKKACESERFRTNIAVYDRTVRRLVAAKRLHYVEEILEE 99
Query: 116 QKKLPQSRREGFVVRIITLYGNAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRD 175
QKK +EGF RII+LYG AGM ++A F +M + C R+V SFNA L+ S+
Sbjct: 100 QKKYRDMSKEGFAARIISLYGKAGMFENAQKVFEEMPNRDCKRSVLSFNALLSAYRLSKK 159
Query: 176 FDEISRFLNEVPRRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTY 235
FD + NE+P + I+ D+ S N +KA CE + L EA + +LD + NKG+KPD+VT+
Sbjct: 160 FDVVEELFNELPGKLSIKPDIVSYNTLIKALCEKDSLPEA-VALLDEIENKGLKPDIVTF 218
Query: 236 TTLISAFYDHKRWEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQR 295
TL+ + Y ++E+G +W +MV K V ++ T+N R+ L + + L ++
Sbjct: 219 NTLLLSSYLKGQFELGEEIWAKMVEKNVAIDIRTYNARLLGLANEAKSKELVNLFGELKA 278
Query: 296 NGVTPDEVTLVLVIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQ 355
+G+ PD + +I+G G + A Y + GY+ + ++ +CK GDF
Sbjct: 279 SGLKPDVFSFNAMIRGSINEGKMDEAEAWYKEIVKHGYRPDKATFALLLPAMCKAGDFES 338
Query: 356 AYTLCKDSMRKNWFPNVDTIFMLLEGLKKSGKINKAKVIVALAE 399
A L K++ K + T+ L++ L K K +A+ IV +A+
Sbjct: 339 AIELFKETFSKRYLVGQTTLQQLVDELVKGSKREEAEEIVKIAK 382
>UniRef100_Q8L8Z2 Hypothetical protein [Arabidopsis thaliana]
Length = 394
Score = 202 bits (513), Expect = 2e-50
Identities = 127/392 (32%), Positives = 196/392 (49%), Gaps = 28/392 (7%)
Query: 8 STRTLLKIFAPSSTAIKTLKPSKPALEVLKSDLEPSKPALEVLKSEWVPSKPALEALKSE 67
ST T + F+ + A T P KP+L L +D K E K +A ++E
Sbjct: 16 STPTNRRFFSAVTAAAATPSPPKPSLITLVNDERDPKFITEKFK----------KACQAE 65
Query: 68 WDPHKLFHIFKSNATNPIVVENRFAFYDTVSRLAGAKRFDYIEQLLEQQKKLPQSRREGF 127
W F+ N I V R TV RLA AK+F+++E++LE+Q K P +EGF
Sbjct: 66 W--------FRKN----IAVYER-----TVRRLAAAKKFEWVEEILEEQNKYPNMSKEGF 108
Query: 128 VVRIITLYGNAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFDEISRFLNEVP 187
V RII LYG GM ++A F +M + C RTV SFNA LN S+ FD + E+P
Sbjct: 109 VARIINLYGRVGMFENAQKVFDEMPERNCKRTVLSFNALLNACVNSKKFDLVEGIFEELP 168
Query: 188 RRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFYDHKR 247
+ I DV S N +K C EA + ++D + NKG+KPD +T+ L+ Y +
Sbjct: 169 GKLSIEPDVASYNTLIKGLCGKGSFTEA-VALIDEIENKGLKPDHITFNILLHESYTKGK 227
Query: 248 WEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTLVL 307
+E G +W +MV K V ++ ++N R+ L + + L ++ N + PD T
Sbjct: 228 FEEGEQIWARMVEKNVKRDIRSYNARLLGLAMENKSEEMVSLFDKLKGNELKPDVFTFTA 287
Query: 308 VIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYTLCKDSMRKN 367
+IKGF G + A+ Y + G + ++ +++ +CK GD AY LCK+ K
Sbjct: 288 MIKGFVSEGKLDEAITWYKEIEKNGCRPLKFVFNSLLPAICKAGDLESAYELCKEIFAKR 347
Query: 368 WFPNVDTIFMLLEGLKKSGKINKAKVIVALAE 399
+ + +++ L K K ++A+ IV LA+
Sbjct: 348 LLVDEAVLQEVVDALVKGSKQDEAEEIVELAK 379
>UniRef100_Q9LK57 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone:MJG19
[Arabidopsis thaliana]
Length = 394
Score = 201 bits (511), Expect = 3e-50
Identities = 126/392 (32%), Positives = 195/392 (49%), Gaps = 28/392 (7%)
Query: 8 STRTLLKIFAPSSTAIKTLKPSKPALEVLKSDLEPSKPALEVLKSEWVPSKPALEALKSE 67
ST T + F+ + A T P KP+L L +D K E K +A ++E
Sbjct: 16 STHTNRRFFSAVTAAAATPSPPKPSLITLVNDERDPKFITEKFK----------KACQAE 65
Query: 68 WDPHKLFHIFKSNATNPIVVENRFAFYDTVSRLAGAKRFDYIEQLLEQQKKLPQSRREGF 127
W F+ N I V R TV RLA AK+F+++E++LE+Q K P +EGF
Sbjct: 66 W--------FRKN----IAVYER-----TVRRLAAAKKFEWVEEILEEQNKYPNMSKEGF 108
Query: 128 VVRIITLYGNAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFDEISRFLNEVP 187
V RII LYG GM ++A F +M + C RT SFNA LN S+ FD + E+P
Sbjct: 109 VARIINLYGRVGMFENAQKVFDEMPERNCKRTALSFNALLNACVNSKKFDLVEGIFKELP 168
Query: 188 RRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFYDHKR 247
+ I DV S N +K C EA + ++D + NKG+KPD +T+ L+ Y +
Sbjct: 169 GKLSIEPDVASYNTLIKGLCGKGSFTEA-VALIDEIENKGLKPDHITFNILLHESYTKGK 227
Query: 248 WEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTLVL 307
+E G +W +MV K V ++ ++N R+ L + + L ++ N + PD T
Sbjct: 228 FEEGEQIWARMVEKNVKRDIRSYNARLLGLAMENKSEEMVSLFDKLKGNELKPDVFTFTA 287
Query: 308 VIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYTLCKDSMRKN 367
+IKGF G + A+ Y + G + ++ +++ +CK GD AY LCK+ K
Sbjct: 288 MIKGFVSEGKLDEAITWYKEIEKNGCRPLKFVFNSLLPAICKAGDLESAYELCKEIFAKR 347
Query: 368 WFPNVDTIFMLLEGLKKSGKINKAKVIVALAE 399
+ + +++ L K K ++A+ IV LA+
Sbjct: 348 LLVDEAVLQEVVDALVKGSKQDEAEEIVELAK 379
>UniRef100_O80694 Hypothetical protein At1g61870; F8K4.8 [Arabidopsis thaliana]
Length = 408
Score = 191 bits (485), Expect = 3e-47
Identities = 123/379 (32%), Positives = 192/379 (50%), Gaps = 4/379 (1%)
Query: 19 SSTAIKTLKPSKPALEVLKSDLEPSKPALEVLKSEWVPSKPALEALKSEWDPHKLFHIFK 78
SST++ + P + L S P + + SK AL LKSE DP ++ I +
Sbjct: 9 SSTSLFRHLNASPQIRSLSSASTILSPDSKTPLTSKEKSKAALSLLKSEKDPDRILEICR 68
Query: 79 SNATNPIVVENRFAFYDTVSRLAGAKRFDYIEQLLEQQ-KKLPQSRREGFVVRIITLYGN 137
+ + P +R AF V LA K F + LL+ + P + E F I LY
Sbjct: 69 AASLTPDCRIDRIAFSAAVENLAEKKHFSAVSNLLDGFIENRPDLKSERFAAHAIVLYAQ 128
Query: 138 AGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFDEISRFLNEVPRRFDIRLDVY 197
A M+ H+L F + F RTVKS NA L ++D+ E R E+P+ + I D+
Sbjct: 129 ANMLDHSLRVFRDLEKFEISRTVKSLNALLFACLVAKDYKEAKRVYIEMPKMYGIEPDLE 188
Query: 198 SVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFY-DHKRWEIGNGLWN 256
+ N +K FCE+ +Y + + + KG+KP+ ++ +IS FY + K E+G L
Sbjct: 189 TYNRMIKVFCESGSASSSYSIVAE-MERKGIKPNSSSFGLMISGFYAEDKSDEVGKVL-A 246
Query: 257 QMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTLVLVIKGFFRAG 316
M +GV + T+N+RIQ L ++ +A L+ M G+ P+ VT +I GF
Sbjct: 247 MMKDRGVNIGVSTYNIRIQSLCKRKKSKEAKALLDGMLSAGMKPNTVTYSHLIHGFCNED 306
Query: 317 YPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYTLCKDSMRKNWFPNVDTIF 376
E A +++ + ++G K + Y T+I+ LCK GDF A +LCK+SM KNW P+ +
Sbjct: 307 DFEEAKKLFKIMVNRGCKPDSECYFTLIYYLCKGGDFETALSLCKESMEKNWVPSFSIMK 366
Query: 377 MLLEGLKKSGKINKAKVIV 395
L+ GL K K+ +AK ++
Sbjct: 367 SLVNGLAKDSKVEEAKELI 385
Score = 69.3 bits (168), Expect = 2e-10
Identities = 55/246 (22%), Positives = 110/246 (44%), Gaps = 6/246 (2%)
Query: 162 SFNATLNVLAKSRDFDEISRFLNE-VPRRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFML 220
+F+A + LA+ + F +S L+ + R D++ + ++ + A+ + +A L +
Sbjct: 82 AFSAAVENLAEKKHFSAVSNLLDGFIENRPDLKSERFAAH-AIVLYAQANMLDHSLRVFR 140
Query: 221 DCVNNKGVKPDVVTYTTLISAFYDHKRWEIGNGLWNQMV-LKGVMPNLHTFNVRIQFLVT 279
D + + V + L+ A K ++ ++ +M + G+ P+L T+N I+
Sbjct: 141 D-LEKFEISRTVKSLNALLFACLVAKDYKEAKRVYIEMPKMYGIEPDLETYNRMIKVFCE 199
Query: 280 VRRVWDANKLMALMQRNGVTPDEVTLVLVIKGFFRAGYPEMAMRVYSALHDKGYKISANI 339
+ ++A M+R G+ P+ + L+I GF+ + +V + + D+G I +
Sbjct: 200 SGSASSSYSIVAEMERKGIKPNSSSFGLMISGFYAEDKSDEVGKVLAMMKDRGVNIGVST 259
Query: 340 YQTMIHNLCKRGDFSQAYTLCKDSMRKNWFPNVDTIFMLLEGLKKSGKINKAKVI--VAL 397
Y I +LCKR +A L + PN T L+ G +AK + + +
Sbjct: 260 YNIRIQSLCKRKKSKEAKALLDGMLSAGMKPNTVTYSHLIHGFCNEDDFEEAKKLFKIMV 319
Query: 398 AEGRKP 403
G KP
Sbjct: 320 NRGCKP 325
>UniRef100_Q8LE47 Putative membrane-associated salt-inducible protein [Arabidopsis
thaliana]
Length = 407
Score = 191 bits (485), Expect = 3e-47
Identities = 121/378 (32%), Positives = 190/378 (50%), Gaps = 3/378 (0%)
Query: 19 SSTAIKTLKPSKPALEVLKSDLEPSKPALEVLKSEWVPSKPALEALKSEWDPHKLFHIFK 78
SST++ + P + L S P + + SK AL LKSE DP ++ I +
Sbjct: 9 SSTSLFRYLNASPQIRSLSSASTILAPDSKTPLTSKEKSKAALSLLKSEKDPDRILEICR 68
Query: 79 SNATNPIVVENRFAFYDTVSRLAGAKRFDYIEQLLEQQKKLPQSRREGFVVRIITLYGNA 138
+ + P +R AF V LA F + LL+ + + E F I LY A
Sbjct: 69 AASLTPDCHIDRIAFSAAVENLAEKNHFSAVSNLLDGFIENRHLKSERFAAHAIVLYAQA 128
Query: 139 GMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFDEISRFLNEVPRRFDIRLDVYS 198
M+ H+L F + F RTVKS NA L ++D+ E R E+P+ + I D+ +
Sbjct: 129 NMLDHSLRVFRDLEKFEISRTVKSLNALLFACLVAKDYKEAKRVYIEMPKMYGIEPDLET 188
Query: 199 VNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFY-DHKRWEIGNGLWNQ 257
N +K FCE+ +Y + + + KG+KP+ ++ +IS FY + K E+G L
Sbjct: 189 YNRMIKVFCESGSASSSYSIVAE-MERKGIKPNSSSFGLMISGFYAEDKSDEVGKVL-AM 246
Query: 258 MVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTLVLVIKGFFRAGY 317
M +GV + T+N+RIQ L ++ +A L+ M G+ P+ VT +I GF
Sbjct: 247 MKARGVNIGVSTYNIRIQSLCKKKKSKEAKALLDGMLSAGMKPNTVTYSHLIHGFCNEDD 306
Query: 318 PEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYTLCKDSMRKNWFPNVDTIFM 377
E A +++ + ++G K + Y T+I+ LCK GDF A +LCK+SM KNW P+ +
Sbjct: 307 FEEAKKLFKVMVNRGCKPDSECYFTLIYYLCKGGDFETALSLCKESMEKNWVPSFSIMKS 366
Query: 378 LLEGLKKSGKINKAKVIV 395
L+ GL K K+ +AK ++
Sbjct: 367 LVNGLAKDSKVEEAKELI 384
Score = 62.8 bits (151), Expect = 2e-08
Identities = 52/245 (21%), Positives = 105/245 (42%), Gaps = 5/245 (2%)
Query: 162 SFNATLNVLAKSRDFDEISRFLNEVPRRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLD 221
+F+A + LA+ F +S L+ ++ + ++ + A+ + +A L + D
Sbjct: 82 AFSAAVENLAEKNHFSAVSNLLDGFIENRHLKSERFAAH-AIVLYAQANMLDHSLRVFRD 140
Query: 222 CVNNKGVKPDVVTYTTLISAFYDHKRWEIGNGLWNQMV-LKGVMPNLHTFNVRIQFLVTV 280
+ + V + L+ A K ++ ++ +M + G+ P+L T+N I+
Sbjct: 141 -LEKFEISRTVKSLNALLFACLVAKDYKEAKRVYIEMPKMYGIEPDLETYNRMIKVFCES 199
Query: 281 RRVWDANKLMALMQRNGVTPDEVTLVLVIKGFFRAGYPEMAMRVYSALHDKGYKISANIY 340
+ ++A M+R G+ P+ + L+I GF+ + +V + + +G I + Y
Sbjct: 200 GSASSSYSIVAEMERKGIKPNSSSFGLMISGFYAEDKSDEVGKVLAMMKARGVNIGVSTY 259
Query: 341 QTMIHNLCKRGDFSQAYTLCKDSMRKNWFPNVDTIFMLLEGLKKSGKINKAKVI--VALA 398
I +LCK+ +A L + PN T L+ G +AK + V +
Sbjct: 260 NIRIQSLCKKKKSKEAKALLDGMLSAGMKPNTVTYSHLIHGFCNEDDFEEAKKLFKVMVN 319
Query: 399 EGRKP 403
G KP
Sbjct: 320 RGCKP 324
>UniRef100_Q9SAB4 F25C20.22 protein [Arabidopsis thaliana]
Length = 405
Score = 184 bits (466), Expect = 5e-45
Identities = 109/354 (30%), Positives = 182/354 (50%), Gaps = 2/354 (0%)
Query: 47 LEVLKSEWVPSKPALEALKSEWDPHKLFHIFKSNATNPIVVENRFAFYDTVSRLAGAKRF 106
L+ L S+ S+ L LKSE +P ++ I +S + +P +R F V LA K F
Sbjct: 32 LKSLTSKQKKSRDTLSLLKSENNPDRILEICRSTSLSPDYHVDRIIFSVAVVTLAREKHF 91
Query: 107 DYIEQLLEQQ-KKLPQSRREGFVVRIITLYGNAGMIQHALNTFYQMGDFRCVRTVKSFNA 165
+ QLL+ + P + E F VR I LYG A M+ ++ TF + + RTVKS NA
Sbjct: 92 VAVSQLLDGFIQNQPDPKSESFAVRAIILYGRANMLDRSIQTFRNLEQYEIPRTVKSLNA 151
Query: 166 TLNVLAKSRDFDEISRFLNEVPRRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNN 225
L ++D+ E +R E+P+ + I D+ + N ++ CE+ +Y + + +
Sbjct: 152 LLFACLMAKDYKEANRVYLEMPKMYGIEPDLETYNRMIRVLCESGSTSSSYSIVAE-MER 210
Query: 226 KGVKPDVVTYTTLISAFYDHKRWEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWD 285
K +KP ++ +I FY ++++ + M GV + T+N+ IQ L ++ +
Sbjct: 211 KWIKPTAASFGLMIDGFYKEEKFDEVRKVMRMMDEFGVHVGVATYNIMIQCLCKRKKSAE 270
Query: 286 ANKLMALMQRNGVTPDEVTLVLVIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIH 345
A L+ + + P+ VT L+I GF + AM ++ + GYK + Y T+IH
Sbjct: 271 AKALIDGVMSCRMRPNSVTYSLLIHGFCSEENLDEAMNLFEVMVCNGYKPDSECYFTLIH 330
Query: 346 NLCKRGDFSQAYTLCKDSMRKNWFPNVDTIFMLLEGLKKSGKINKAKVIVALAE 399
LCK GDF A LC++SM KNW P+ + L+ GL K+++AK ++A+ +
Sbjct: 331 CLCKGGDFETALILCRESMEKNWVPSFSVMKWLVNGLASRSKVDEAKELIAVVK 384
>UniRef100_Q9LK58 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone:MJG19
[Arabidopsis thaliana]
Length = 551
Score = 169 bits (429), Expect = 1e-40
Identities = 91/299 (30%), Positives = 163/299 (54%), Gaps = 1/299 (0%)
Query: 97 VSRLAGAKRFDYIEQLLEQQKKLPQSRREGFVVRIITLYGNAGMIQHALNTFYQMGDFRC 156
+ RL AK+F I+++L+ QKK + E FV+RI+ LYG +GM +HA F +M + C
Sbjct: 94 IRRLREAKKFSTIDEVLQYQKKFDDIKSEDFVIRIMLLYGYSGMAEHAHKLFDEMPELNC 153
Query: 157 VRTVKSFNATLNVLAKSRDFDEISRFLNEVPRRFDIRLDVYSVNIAVKAFCEAEKLKEAY 216
RTVKSFNA L+ S+ DE + E+P + I D+ + N +KA C + +
Sbjct: 154 ERTVKSFNALLSAYVNSKKLDEAMKTFKELPEKLGITPDLVTYNTMIKALCRKGSMDD-I 212
Query: 217 LFMLDCVNNKGVKPDVVTYTTLISAFYDHKRWEIGNGLWNQMVLKGVMPNLHTFNVRIQF 276
L + + + G +PD++++ TL+ FY + + G+ +W+ M K + PN+ ++N R++
Sbjct: 213 LSIFEELEKNGFEPDLISFNTLLEEFYRRELFVEGDRIWDLMKSKNLSPNIRSYNSRVRG 272
Query: 277 LVTVRRVWDANKLMALMQRNGVTPDEVTLVLVIKGFFRAGYPEMAMRVYSALHDKGYKIS 336
L ++ DA L+ +M+ G++PD T +I + E M+ Y+ + +KG
Sbjct: 273 LTRNKKFTDALNLIDVMKTEGISPDVHTYNALITAYRVDNNLEEVMKCYNEMKEKGLTPD 332
Query: 337 ANIYQTMIHNLCKRGDFSQAYTLCKDSMRKNWFPNVDTIFMLLEGLKKSGKINKAKVIV 395
Y +I LCK+GD +A + +++++ + ++E L +GKI++A +V
Sbjct: 333 TVTYCMLIPLLCKKGDLDRAVEVSEEAIKHKLLSRPNMYKPVVERLMGAGKIDEATQLV 391
>UniRef100_O23149 Membrane-associated salt-inducible protein like [Arabidopsis
thaliana]
Length = 428
Score = 167 bits (422), Expect = 7e-40
Identities = 107/351 (30%), Positives = 184/351 (51%), Gaps = 12/351 (3%)
Query: 60 ALEALKSEWDPHKLFHIFKS---NATNPIVVENRFAFYDTVSRLAGAKRFDYIEQLLEQQ 116
A L+ E DP K I+ + ++ +P V +R+A TV RLA +RF IE L+E
Sbjct: 52 AKSTLRKEHDPDKALKIYANVSDHSASP--VSSRYAQELTVRRLAKCRRFSDIETLIESH 109
Query: 117 KKLPQSRREGFVVRIITLYGNAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDF 176
K P+ + E F +I YG A M HA+ TF QM + R+ SFNA LN S++F
Sbjct: 110 KNDPKIKEEPFYSTLIRSYGQASMFNHAMRTFEQMDQYGTPRSAVSFNALLNACLHSKNF 169
Query: 177 DEISRFLNEVPRRFD-IRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTY 235
D++ + +E+P+R++ I D S I +K++C++ ++A M + KG++ + +
Sbjct: 170 DKVPQLFDEIPQRYNKIIPDKISYGILIKSYCDSGTPEKAIEIMRQ-MQGKGMEVTTIAF 228
Query: 236 TTLISAFYDHKRWEIGNGLWNQMVLKGVMPNLHTFNVRIQFL--VTVRRVWDANKLMALM 293
TT++S+ Y E+ + LWN+MV KG + +NVRI + RV +L+ M
Sbjct: 229 TTILSSLYKKGELEVADNLWNEMVKKGCELDNAAYNVRIMSAQKESPERV---KELIEEM 285
Query: 294 QRNGVTPDEVTLVLVIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDF 353
G+ PD ++ ++ + G + A +VY L +A ++T+I +LC +
Sbjct: 286 SSMGLKPDTISYNYLMTAYCERGMLDEAKKVYEGLEGNNCAPNAATFRTLIFHLCYSRLY 345
Query: 354 SQAYTLCKDSMRKNWFPNVDTIFMLLEGLKKSGKINKAKVIVALAEGRKPP 404
Q Y + K S+ + P+ +T+ L+ GL ++ K + AK ++ + + PP
Sbjct: 346 EQGYAIFKKSVYMHKIPDFNTLKHLVVGLVENKKRDDAKGLIRTVKKKFPP 396
>UniRef100_Q8H219 Drought-inducible protein 1OS [Oryza sativa]
Length = 434
Score = 158 bits (399), Expect = 3e-37
Identities = 107/399 (26%), Positives = 194/399 (47%), Gaps = 11/399 (2%)
Query: 12 LLKIFAPSSTAIKTLKPSK----PALEVLKSDLEPSKPALEVLKSEWVPSKPALEALKSE 67
L +IF+ SS +++ KP + P K P A L+ + ++ L E
Sbjct: 13 LSRIFSSSSPSVRPPKPGQVKEAPKPAPTKKAPAPGAEANPNLRRNAIDD--IIKGLLRE 70
Query: 68 WDPHKLFHIFKSNATNPIVVENRFAFYDT-VSRLAGAKRFDYIEQLLEQQKKLPQSRREG 126
DP KL F + ++ R YD VSRLA R D +E +++ QK ++ +EG
Sbjct: 71 RDPDKLVSGFIAASSTHPRFRARHRVYDVAVSRLATFGRLDGVEAIIDAQKPFLETSKEG 130
Query: 127 FVVRIITLYGNAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFDEISRFLNEV 186
F R+I LYG+A M HA TF+ + +++ +FN+ L ++ +F+ ++ E+
Sbjct: 131 FAARLIRLYGHASMASHAAATFHDLPPQ--LKSTMTFNSLLAAYVEAGEFEALAAAFKEI 188
Query: 187 P-RRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFYDH 245
P + VYS NI ++A C+ L A L + + G+ PD+VT+ TL++ FY+H
Sbjct: 189 PVSNPSVVPSVYSYNILLQALCKVPDLSAA-LDTMTLMEKSGISPDLVTFNTLLNGFYNH 247
Query: 246 KRWEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTL 305
+ +W + + ++P+ +N +++ LV R+ DA ++ M+++G PD ++
Sbjct: 248 GDMDGAEKVWEMITERNMVPDAKNYNAKLRGLVAQGRIEDAVAVVEKMEKDGPKPDTISY 307
Query: 306 VLVIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYTLCKDSMR 365
+I+G+ + G E A +++ + + GY + Y T+I L K G+ A C +
Sbjct: 308 NELIRGYCKDGRLEEAKKLFEDMAENGYVANRGTYHTLIPCLVKAGELDYALKCCHEIYG 367
Query: 366 KNWFPNVDTIFMLLEGLKKSGKINKAKVIVALAEGRKPP 404
KN + + ++ L + ++ A IV L P
Sbjct: 368 KNLRVDCFVLQEVVTALVTASRVEDATKIVELGWNNSYP 406
>UniRef100_Q9ZU67 Expressed protein [Arabidopsis thaliana]
Length = 418
Score = 153 bits (387), Expect = 8e-36
Identities = 108/351 (30%), Positives = 178/351 (49%), Gaps = 16/351 (4%)
Query: 60 ALEALKSEWDPHKLFHIFKS---NATNPIVVENRFAFYDTVSRLAGAKRFDYIEQLLEQQ 116
A L+ DP K I+KS N+T+P+ +R+A TV RLA ++RF IE L+E
Sbjct: 36 AKSKLRKVQDPDKALAIYKSVSNNSTSPL--SSRYAMELTVQRLAKSQRFSDIEALIESH 93
Query: 117 KKLPQSRREGFVVRIITLYGNAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDF 176
K P+ + E F+ +I YG A M HA+ F +M RTV SFNA L S F
Sbjct: 94 KNNPKIKTETFLSTLIRSYGRASMFDHAMKMFEEMDKLGTPRTVVSFNALLAACLHSDLF 153
Query: 177 DEISRFLNEVPRRF-DIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTY 235
+ + + +E P+R+ +I D S + +K++C++ K ++A M D + KGV+ ++ +
Sbjct: 154 ERVPQLFDEFPQRYNNITPDKISYGMLIKSYCDSGKPEKAMEIMRD-MEVKGVEVTIIAF 212
Query: 236 TTLISAFYDHKRWEIGNGLWNQMVLKGVMPNLHTFNVRIQFLV--TVRRVWDANKLMALM 293
TT++ + Y + + LW +MV KG + +NVR+ + RV +LM M
Sbjct: 213 TTILGSLYKNGLVDEAESLWIEMVNKGCDLDNTVYNVRLMNAAKESPERV---KELMEEM 269
Query: 294 QRNGVTPDEVTLVLVIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDF 353
G+ PD V+ ++ + G A +VY L +A ++T+I +LC G +
Sbjct: 270 SSVGLKPDTVSYNYLMTAYCVKGMMSEAKKVYEGLEQP----NAATFRTLIFHLCINGLY 325
Query: 354 SQAYTLCKDSMRKNWFPNVDTIFMLLEGLKKSGKINKAKVIVALAEGRKPP 404
Q T+ K S + P+ T L EGL K+ ++ A+ + + + + PP
Sbjct: 326 DQGLTVFKKSAIVHKIPDFKTCKHLTEGLVKNNRMEDARGVARIVKKKFPP 376
>UniRef100_Q6H7J7 Membrane-associated salt-inducible protein like [Oryza sativa]
Length = 455
Score = 142 bits (359), Expect = 1e-32
Identities = 100/342 (29%), Positives = 164/342 (47%), Gaps = 5/342 (1%)
Query: 64 LKSEWDPHKLFHIFKS-NATNPIVVENRFAFYDTVSRLAGAKRFDYIEQLLEQQKKLPQS 122
L+ E DP ++ +F++ + + R A RL+ ++RF E LL +P S
Sbjct: 44 LRREHDPDRVVSLFEAIDDASLSASSTRHALSLAARRLSRSRRFADAEALLSSH--IPAS 101
Query: 123 RREGFVVRIITLYGNAGMIQHALNTFYQMG-DFRCVRTVKSFNATLNVLAKSRDFDEISR 181
E + ++ Y A + + AL F + FNA L+V + R +
Sbjct: 102 PTEPQLAAVLCSYAAASLPEKALAAFRSAAPSLPSPISPLPFNAVLSVFLRCRRHRRVPV 161
Query: 182 FLNEVPRRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISA 241
+E+ + F I D S I VKA+C K +A+ +LD + +G P YTT+I +
Sbjct: 162 LFDELSKEFSITPDASSYGILVKAYCMLSKDAKAHE-VLDQMRGQGFTPTNSIYTTMIDS 220
Query: 242 FYDHKRWEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPD 301
Y K+ E LW QM+ G P+ +N +I + D +++A M+ GV PD
Sbjct: 221 MYKQKKMEQAERLWKQMLESGRKPDQAVYNTKIMHHSLHGKTEDVLEVIAEMEEAGVKPD 280
Query: 302 EVTLVLVIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYTLCK 361
+T ++ + + G E A +Y +L +KG +A Y+ M+ LC GD A + K
Sbjct: 281 TITYNFLMTSYCKHGKMETAKELYRSLGEKGCSANAATYKHMMAQLCAHGDLDGALVIFK 340
Query: 362 DSMRKNWFPNVDTIFMLLEGLKKSGKINKAKVIVALAEGRKP 403
+S R N P+ T+ L+EGL K+G++ +AK IVA + + P
Sbjct: 341 ESYRSNKVPDFRTMSGLVEGLTKAGRVAEAKNIVAKMKKKFP 382
>UniRef100_Q8RUB8 Putative drought-inducible protein 1OS [Oryza sativa]
Length = 390
Score = 138 bits (347), Expect = 3e-31
Identities = 112/396 (28%), Positives = 181/396 (45%), Gaps = 33/396 (8%)
Query: 10 RTLLKIFAPSSTAIKTLKPSKPALEVLKSDLEPSKPALEVLKSEWVPSKPALEALKSEWD 69
R L AP T L PS P+ + + + S+ A+ +LKS P P
Sbjct: 11 RGLSTATAPPPTPGSILNPSSPSTPL--TSRQKSRLAISLLKS--TPPPP---------- 56
Query: 70 PHKLFHIFKSNATNPIVVENRFAFYDTVSRLAGAKRF--DYIEQLLEQQKKLPQSRREGF 127
P ++ I ++ A +P +R A S+L+ A D LL
Sbjct: 57 PDQILSICRAAALSPDSHLDRVALSLAASKLSSAPDSVRDLASSLLTPHH---------- 106
Query: 128 VVRIITLYGNAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFDEISRFLNEVP 187
I L+G AG++ A++TF + + +S NA L S + E +R P
Sbjct: 107 APHAIALFGQAGLLPDAVSTF------KSSPSTRSLNALLFACLVSGNHAEAARVFQTFP 160
Query: 188 RRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFYDHKR 247
+ ++ + + N +K+F E+ + Y +LD + KGVKP+ T+TT I+ FY +R
Sbjct: 161 DAYSVKPNTDTFNAIIKSFAESGTTRSFYS-VLDEMCQKGVKPNATTFTTAIAGFYKEER 219
Query: 248 WEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTLVL 307
++ + M G +L FNVR+Q L + R DA L+ M + G P +T
Sbjct: 220 FDDVGKVIELMKKHGCGESLPVFNVRVQGLCKLGRSGDAKALLNEMVKKGTKPSWLTYNH 279
Query: 308 VIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYTLCKDSMRKN 367
+I GF + G E A R+Y + KG ++ Y +I+ LCK GDF A + + +N
Sbjct: 280 LIHGFCKEGDLEEAKRLYKEMAKKGLVGDSSFYYMLIYYLCKGGDFDTAVGVYNEIAARN 339
Query: 368 WFPNVDTIFMLLEGLKKSGKINKAKVIVALAEGRKP 403
W P T+ ML+ GL S ++++AK I+ + + P
Sbjct: 340 WVPCFSTMKMLVNGLAGSSRVDEAKGIIEKMKEKFP 375
>UniRef100_Q9CA58 Hypothetical protein F1M20.26 [Arabidopsis thaliana]
Length = 763
Score = 133 bits (335), Expect = 8e-30
Identities = 95/339 (28%), Positives = 162/339 (47%), Gaps = 3/339 (0%)
Query: 58 KPALEALKSEWDPHKLFHIFKSNATNPIVVENRFAFYDTVSRLAGAKRFDYIEQLL-EQQ 116
K +K + DP K +F S + + +L +F+ +E++L + +
Sbjct: 8 KHVTAVIKCQKDPMKALEMFNSMRKEVGFKHTLSTYRSVIEKLGYYGKFEAMEEVLVDMR 67
Query: 117 KKLPQSRREGFVVRIITLYGNAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDF 176
+ + EG V + YG G +Q A+N F +M + C TV S+NA ++VL S F
Sbjct: 68 ENVGNHMLEGVYVGAMKNYGRKGKVQEAVNVFERMDFYDCEPTVFSYNAIMSVLVDSGYF 127
Query: 177 DEISRFLNEVPRRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYT 236
D+ + + R I DVYS I +K+FC+ + A L +L+ ++++G + +VV Y
Sbjct: 128 DQAHKVYMRMRDR-GITPDVYSFTIRMKSFCKTSR-PHAALRLLNNMSSQGCEMNVVAYC 185
Query: 237 TLISAFYDHKRWEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRN 296
T++ FY+ G L+ +M+ GV L TFN ++ L V + KL+ + +
Sbjct: 186 TVVGGFYEENFKAEGYELFGKMLASGVSLCLSTFNKLLRVLCKKGDVKECEKLLDKVIKR 245
Query: 297 GVTPDEVTLVLVIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQA 356
GV P+ T L I+G + G + A+R+ L ++G K Y +I+ LCK F +A
Sbjct: 246 GVLPNLFTYNLFIQGLCQRGELDGAVRMVGCLIEQGPKPDVITYNNLIYGLCKNSKFQEA 305
Query: 357 YTLCKDSMRKNWFPNVDTIFMLLEGLKKSGKINKAKVIV 395
+ + P+ T L+ G K G + A+ IV
Sbjct: 306 EVYLGKMVNEGLEPDSYTYNTLIAGYCKGGMVQLAERIV 344
Score = 102 bits (253), Expect = 3e-20
Identities = 71/254 (27%), Positives = 126/254 (48%), Gaps = 5/254 (1%)
Query: 160 VKSFNATLNVLAKSRDFDEISRFLNEVPRRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFM 219
+ +FN L VL K D E + L++V +R + ++++ N+ ++ C+ +L A + M
Sbjct: 216 LSTFNKLLRVLCKKGDVKECEKLLDKVIKR-GVLPNLFTYNLFIQGLCQRGELDGA-VRM 273
Query: 220 LDCVNNKGVKPDVVTYTTLISAFYDHKRWEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVT 279
+ C+ +G KPDV+TY LI + +++ +MV +G+ P+ +T+N I
Sbjct: 274 VGCLIEQGPKPDVITYNNLIYGLCKNSKFQEAEVYLGKMVNEGLEPDSYTYNTLIAGYCK 333
Query: 280 VRRVWDANKLMALMQRNGVTPDEVTLVLVIKGFFRAGYPEMAMRVYSALHDKGYKISANI 339
V A +++ NG PD+ T +I G G A+ +++ KG K + +
Sbjct: 334 GGMVQLAERIVGDAVFNGFVPDQFTYRSLIDGLCHEGETNRALALFNEALGKGIKPNVIL 393
Query: 340 YQTMIHNLCKRGDFSQAYTLCKDSMRKNWFPNVDTIFMLLEGLKKSGKINKAK--VIVAL 397
Y T+I L +G +A L + K P V T +L+ GL K G ++ A V V +
Sbjct: 394 YNTLIKGLSNQGMILEAAQLANEMSEKGLIPEVQTFNILVNGLCKMGCVSDADGLVKVMI 453
Query: 398 AEGRKPP-FSFSYL 410
++G P F+F+ L
Sbjct: 454 SKGYFPDIFTFNIL 467
Score = 95.9 bits (237), Expect = 2e-18
Identities = 68/255 (26%), Positives = 129/255 (49%), Gaps = 5/255 (1%)
Query: 137 NAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFDEISRFLNEVPRRFDIRLDV 196
N GMI A +M + + V++FN +N L K + + + + D+
Sbjct: 403 NQGMILEAAQLANEMSEKGLIPEVQTFNILVNGLCKMGCVSDADGLVKVMISKGYFP-DI 461
Query: 197 YSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFYDHKRWEIGNGLWN 256
++ NI + + K++ A L +LD + + GV PDV TY +L++ ++E +
Sbjct: 462 FTFNILIHGYSTQLKMENA-LEILDVMLDNGVDPDVYTYNSLLNGLCKTSKFEDVMETYK 520
Query: 257 QMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTLVLVIKGFFRAG 316
MV KG PNL TFN+ ++ L R++ +A L+ M+ V PD VT +I GF + G
Sbjct: 521 TMVEKGCAPNLFTFNILLESLCRYRKLDEALGLLEEMKNKSVNPDAVTFGTLIDGFCKNG 580
Query: 317 YPEMAMRVYSALHDKGYKISAN--IYQTMIHNLCKRGDFSQAYTLCKDSMRKNWFPNVDT 374
+ A ++ + ++ YK+S++ Y +IH ++ + + A L ++ + + P+ T
Sbjct: 581 DLDGAYTLFRKM-EEAYKVSSSTPTYNIIIHAFTEKLNVTMAEKLFQEMVDRCLGPDGYT 639
Query: 375 IFMLLEGLKKSGKIN 389
++++G K+G +N
Sbjct: 640 YRLMVDGFCKTGNVN 654
Score = 84.3 bits (207), Expect = 6e-15
Identities = 65/266 (24%), Positives = 111/266 (41%), Gaps = 35/266 (13%)
Query: 160 VKSFNATLNVLAKSRDFDEISRFLNEVPRRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFM 219
V ++N + L K+ F E +L ++ + D Y+ N + +C+ ++ A +
Sbjct: 286 VITYNNLIYGLCKNSKFQEAEVYLGKMVNE-GLEPDSYTYNTLIAGYCKGGMVQLAERIV 344
Query: 220 LDCVNN----------------------------------KGVKPDVVTYTTLISAFYDH 245
D V N KG+KP+V+ Y TLI +
Sbjct: 345 GDAVFNGFVPDQFTYRSLIDGLCHEGETNRALALFNEALGKGIKPNVILYNTLIKGLSNQ 404
Query: 246 KRWEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTL 305
L N+M KG++P + TFN+ + L + V DA+ L+ +M G PD T
Sbjct: 405 GMILEAAQLANEMSEKGLIPEVQTFNILVNGLCKMGCVSDADGLVKVMISKGYFPDIFTF 464
Query: 306 VLVIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYTLCKDSMR 365
++I G+ E A+ + + D G Y ++++ LCK F K +
Sbjct: 465 NILIHGYSTQLKMENALEILDVMLDNGVDPDVYTYNSLLNGLCKTSKFEDVMETYKTMVE 524
Query: 366 KNWFPNVDTIFMLLEGLKKSGKINKA 391
K PN+ T +LLE L + K+++A
Sbjct: 525 KGCAPNLFTFNILLESLCRYRKLDEA 550
Score = 73.2 bits (178), Expect = 1e-11
Identities = 49/200 (24%), Positives = 94/200 (46%), Gaps = 1/200 (0%)
Query: 192 IRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFYDHKRWEIG 251
I+ +V N +K + EA + ++ KG+ P+V T+ L++
Sbjct: 387 IKPNVILYNTLIKGLSNQGMILEAAQLANE-MSEKGLIPEVQTFNILVNGLCKMGCVSDA 445
Query: 252 NGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTLVLVIKG 311
+GL M+ KG P++ TFN+ I T ++ +A +++ +M NGV PD T ++ G
Sbjct: 446 DGLVKVMISKGYFPDIFTFNILIHGYSTQLKMENALEILDVMLDNGVDPDVYTYNSLLNG 505
Query: 312 FFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYTLCKDSMRKNWFPN 371
+ E M Y + +KG + + ++ +LC+ +A L ++ K+ P+
Sbjct: 506 LCKTSKFEDVMETYKTMVEKGCAPNLFTFNILLESLCRYRKLDEALGLLEEMKNKSVNPD 565
Query: 372 VDTIFMLLEGLKKSGKINKA 391
T L++G K+G ++ A
Sbjct: 566 AVTFGTLIDGFCKNGDLDGA 585
Score = 65.9 bits (159), Expect = 2e-09
Identities = 61/243 (25%), Positives = 107/243 (43%), Gaps = 4/243 (1%)
Query: 135 YGNAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFDEISRFLNEVPRRFDIRL 194
Y +++AL M D V ++N+ LN L K+ F+++ + +
Sbjct: 471 YSTQLKMENALEILDVMLDNGVDPDVYTYNSLLNGLCKTSKFEDVMETYKTMVEK-GCAP 529
Query: 195 DVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFYDHKRWEIGNGL 254
++++ NI +++ C KL EA L +L+ + NK V PD VT+ TLI F + + L
Sbjct: 530 NLFTFNILLESLCRYRKLDEA-LGLLEEMKNKSVNPDAVTFGTLIDGFCKNGDLDGAYTL 588
Query: 255 WNQMV-LKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTLVLVIKGFF 313
+ +M V + T+N+ I V A KL M + PD T L++ GF
Sbjct: 589 FRKMEEAYKVSSSTPTYNIIIHAFTEKLNVTMAEKLFQEMVDRCLGPDGYTYRLMVDGFC 648
Query: 314 RAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYTLCKDSMRKNWFPN-V 372
+ G + + + + G+ S +I+ LC +A + ++K P V
Sbjct: 649 KTGNVNLGYKFLLEMMENGFIPSLTTLGRVINCLCVEDRVYEAAGIIHRMVQKGLVPEAV 708
Query: 373 DTI 375
+TI
Sbjct: 709 NTI 711
>UniRef100_Q9FFE3 Salt-inducible protein-like [Arabidopsis thaliana]
Length = 535
Score = 121 bits (304), Expect = 3e-26
Identities = 77/330 (23%), Positives = 161/330 (48%), Gaps = 3/330 (0%)
Query: 82 TNPIVVENRFAFYDTVSRLAGAKRFDYIEQLLEQQKKL--PQSRREGFVVRIITLYGNAG 139
++P N ++ + +L+ A+ FD +E L+ + P E + ++ YG AG
Sbjct: 75 SHPGFTHNYDTYHSILFKLSRARAFDPVESLMADLRNSYPPIKCGENLFIDLLRNYGLAG 134
Query: 140 MIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFDEISRFLNEVPRRFDIRLDVYSV 199
+ ++ F ++ DF R+V+S N LNVL +++ FD + F I ++++
Sbjct: 135 RYESSMRIFLRIPDFGVKRSVRSLNTLLNVLIQNQRFDLVHAMFKNSKESFGITPNIFTC 194
Query: 200 NIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFYDHKRWEIGNGLWNQMV 259
N+ VKA C+ ++ AY +LD + + G+ P++VTYTT++ + E + +M+
Sbjct: 195 NLLVKALCKKNDIESAYK-VLDEIPSMGLVPNLVTYTTILGGYVARGDMESAKRVLEEML 253
Query: 260 LKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTLVLVIKGFFRAGYPE 319
+G P+ T+ V + + R +A +M M++N + P+EVT ++I+ +
Sbjct: 254 DRGWYPDATTYTVLMDGYCKLGRFSEAATVMDDMEKNEIEPNEVTYGVMIRALCKEKKSG 313
Query: 320 MAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYTLCKDSMRKNWFPNVDTIFMLL 379
A ++ + ++ + +++ +I LC+ +A L + ++ N P+ + L+
Sbjct: 314 EARNMFDEMLERSFMPDSSLCCKVIDALCEDHKVDEACGLWRKMLKNNCMPDNALLSTLI 373
Query: 380 EGLKKSGKINKAKVIVALAEGRKPPFSFSY 409
L K G++ +A+ + E P +Y
Sbjct: 374 HWLCKEGRVTEARKLFDEFEKGSIPSLLTY 403
Score = 81.3 bits (199), Expect = 5e-14
Identities = 67/288 (23%), Positives = 126/288 (43%), Gaps = 9/288 (3%)
Query: 117 KKLPQSRREGFVVRIITL------YGNAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVL 170
K L + G V ++T Y G ++ A +M D ++ ++
Sbjct: 212 KVLDEIPSMGLVPNLVTYTTILGGYVARGDMESAKRVLEEMLDRGWYPDATTYTVLMDGY 271
Query: 171 AKSRDFDEISRFLNEVPRRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKP 230
K F E + ++++ + +I + + + ++A C+ +K EA M D + + P
Sbjct: 272 CKLGRFSEAATVMDDMEKN-EIEPNEVTYGVMIRALCKEKKSGEARN-MFDEMLERSFMP 329
Query: 231 DVVTYTTLISAFYDHKRWEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLM 290
D +I A + + + GLW +M+ MP+ + I +L RV +A KL
Sbjct: 330 DSSLCCKVIDALCEDHKVDEACGLWRKMLKNNCMPDNALLSTLIHWLCKEGRVTEARKLF 389
Query: 291 ALMQRNGVTPDEVTLVLVIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKR 350
++ + P +T +I G G A R++ ++++ K +A Y +I L K
Sbjct: 390 DEFEKGSI-PSLLTYNTLIAGMCEKGELTEAGRLWDDMYERKCKPNAFTYNVLIEGLSKN 448
Query: 351 GDFSQAYTLCKDSMRKNWFPNVDTIFMLLEGLKKSGKINKAKVIVALA 398
G+ + + ++ + FPN T +L EGL+K GK A IV++A
Sbjct: 449 GNVKEGVRVLEEMLEIGCFPNKTTFLILFEGLQKLGKEEDAMKIVSMA 496
Score = 40.0 bits (92), Expect = 0.12
Identities = 57/286 (19%), Positives = 103/286 (35%), Gaps = 16/286 (5%)
Query: 35 VLKSDLEPSKPALE-VLKSEWVPSKPALEALKSEW-------DPHKLFHIFKSNATNPIV 86
V + D+E +K LE +L W P L + + + + N P
Sbjct: 237 VARGDMESAKRVLEEMLDRGWYPDATTYTVLMDGYCKLGRFSEAATVMDDMEKNEIEP-- 294
Query: 87 VENRFAFYDTVSRLAGAKRFDYIEQLLEQQKKLPQSRREGFVVRIITLYGNAGMIQHALN 146
N + + L K+ + ++ + ++I + A
Sbjct: 295 --NEVTYGVMIRALCKEKKSGEARNMFDEMLERSFMPDSSLCCKVIDALCEDHKVDEACG 352
Query: 147 TFYQMGDFRCVRTVKSFNATLNVLAKSRDFDEISRFLNEVPRRFDIRLDVYSVNIAVKAF 206
+ +M C+ + ++ L K E + +E + L Y+ IA
Sbjct: 353 LWRKMLKNNCMPDNALLSTLIHWLCKEGRVTEARKLFDEFEKGSIPSLLTYNTLIA--GM 410
Query: 207 CEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFYDHKRWEIGNGLWNQMVLKGVMPN 266
CE +L EA D K KP+ TY LI + + G + +M+ G PN
Sbjct: 411 CEKGELTEAGRLWDDMYERK-CKPNAFTYNVLIEGLSKNGNVKEGVRVLEEMLEIGCFPN 469
Query: 267 LHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTLVLVIKGF 312
TF + + L + + DA K++++ NG D+ + L +K F
Sbjct: 470 KTTFLILFEGLQKLGKEEDAMKIVSMAVMNGKV-DKESWELFLKKF 514
>UniRef100_Q9LQQ1 F24B9.15 protein [Arabidopsis thaliana]
Length = 459
Score = 116 bits (290), Expect = 1e-24
Identities = 94/400 (23%), Positives = 176/400 (43%), Gaps = 18/400 (4%)
Query: 25 TLKPSKPALEVLKSDLEPSKPALEVLKSEWVPSKPALEALKSEWDPHKLFHIFKSNATNP 84
T +P KP + + P + + W P L LK DP + +F +
Sbjct: 23 TSRPEKPTKKASSHE-----PTHKFTRKPW-EEVPFLTDLKEIEDPEEALSLF--HQYQE 74
Query: 85 IVVENRFAFYDT-VSRLAGAKRFDYIEQLLEQQKKLPQSRREGFVVRIITLYGNAGMIQH 143
+ + + Y + + +LA ++ FD ++Q+L + RE + +I YG AG +
Sbjct: 75 MGFRHDYPSYSSLIYKLAKSRNFDAVDQILRLVRYRNVRCRESLFMGLIQHYGKAGSVDK 134
Query: 144 ALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFDEISRFLNEVPRRFDIRLDVYSVNIAV 203
A++ F+++ F CVRT++S N +NVL + + ++ F + + +R + S NI +
Sbjct: 135 AIDVFHKITSFDCVRTIQSLNTLINVLVDNGELEKAKSFFDGA-KDMRLRPNSVSFNILI 193
Query: 204 KAF---CEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFYDHKRWEIGNGLWNQMVL 260
K F C+ E + + ML+ V+P VVTY +LI + L M+
Sbjct: 194 KGFLDKCDWEAACKVFDEMLEME----VQPSVVTYNSLIGFLCRNDDMGKAKSLLEDMIK 249
Query: 261 KGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTLVLVIKGFFRAGYPEM 320
K + PN TF + ++ L +A KLM M+ G P V +++ + G +
Sbjct: 250 KRIRPNAVTFGLLMKGLCCKGEYNEAKKLMFDMEYRGCKPGLVNYGILMSDLGKRGRIDE 309
Query: 321 AMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYTLCKDSMRKNWFPNVDTIFMLLE 380
A + + + K IY ++++LC +AY + + K PN T M+++
Sbjct: 310 AKLLLGEMKKRRIKPDVVIYNILVNHLCTECRVPEAYRVLTEMQMKGCKPNAATYRMMID 369
Query: 381 GLKKSGKINKA-KVIVALAEGRKPPFSFSYLASMQSILSG 419
G + + V+ A+ R P +++ + ++ G
Sbjct: 370 GFCRIEDFDSGLNVLNAMLASRHCPTPATFVCMVAGLIKG 409
Score = 66.6 bits (161), Expect = 1e-09
Identities = 51/224 (22%), Positives = 102/224 (44%), Gaps = 2/224 (0%)
Query: 144 ALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFDEISRFLNEVPRRFDIRLDVYSVNIAV 203
A F +M + +V ++N+ + L ++ D + L ++ ++ IR + + + +
Sbjct: 205 ACKVFDEMLEMEVQPSVVTYNSLIGFLCRNDDMGKAKSLLEDMIKK-RIRPNAVTFGLLM 263
Query: 204 KAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFYDHKRWEIGNGLWNQMVLKGV 263
K C + EA M D + +G KP +V Y L+S R + L +M + +
Sbjct: 264 KGLCCKGEYNEAKKLMFD-MEYRGCKPGLVNYGILMSDLGKRGRIDEAKLLLGEMKKRRI 322
Query: 264 MPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTLVLVIKGFFRAGYPEMAMR 323
P++ +N+ + L T RV +A +++ MQ G P+ T ++I GF R + +
Sbjct: 323 KPDVVIYNILVNHLCTECRVPEAYRVLTEMQMKGCKPNAATYRMMIDGFCRIEDFDSGLN 382
Query: 324 VYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYTLCKDSMRKN 367
V +A+ + + + M+ L K G+ A + + +KN
Sbjct: 383 VLNAMLASRHCPTPATFVCMVAGLIKGGNLDHACFVLEVMGKKN 426
Score = 49.7 bits (117), Expect = 2e-04
Identities = 54/288 (18%), Positives = 114/288 (38%), Gaps = 12/288 (4%)
Query: 61 LEALKSEWDPHKLFHIFKSNATNPIVVENRFAFYDTVSRLAGAKRFDYIEQLLEQQKKLP 120
LE KS +D K + ++ + I+++ F D A K FD + ++ Q +
Sbjct: 167 LEKAKSFFDGAKDMRLRPNSVSFNILIKG---FLDKCDWEAACKVFDEMLEMEVQPSVVT 223
Query: 121 QSRREGFVVRIITLYGNAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFDEIS 180
+ GF+ R + +++ + R +F + L +++E
Sbjct: 224 YNSLIGFLCRNDDMGKAKSLLEDMIKK-------RIRPNAVTFGLLMKGLCCKGEYNEAK 276
Query: 181 RFLNEVPRRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLIS 240
+ + ++ R + + + I + + ++ EA L +L + + +KPDVV Y L++
Sbjct: 277 KLMFDMEYR-GCKPGLVNYGILMSDLGKRGRIDEAKL-LLGEMKKRRIKPDVVIYNILVN 334
Query: 241 AFYDHKRWEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTP 300
R + +M +KG PN T+ + I + ++ M + P
Sbjct: 335 HLCTECRVPEAYRVLTEMQMKGCKPNAATYRMMIDGFCRIEDFDSGLNVLNAMLASRHCP 394
Query: 301 DEVTLVLVIKGFFRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLC 348
T V ++ G + G + A V + K + +Q ++ +LC
Sbjct: 395 TPATFVCMVAGLIKGGNLDHACFVLEVMGKKNLSFGSGAWQNLLSDLC 442
>UniRef100_Q9FIX3 Gb|AAD30619.1 [Arabidopsis thaliana]
Length = 747
Score = 115 bits (288), Expect = 2e-24
Identities = 101/379 (26%), Positives = 175/379 (45%), Gaps = 16/379 (4%)
Query: 22 AIKTLKPSKPALEVLKSDLEPSKPALEVLKSEWVPSKPALEALKSEW-DPHKLFHIFKSN 80
A+ LK L L ++ P + +LKS+ + AL W +PH+ F +
Sbjct: 28 ALTFLKRHPYQLHHLSANFTPEAASNLLLKSQ---NDQALILKFLNWANPHQFFTLRCKC 84
Query: 81 ATNPIVVENRFAFYDTVSRLA---GAKRFD-----YIEQLLEQQKKLPQSRREGFVVRII 132
T I+ +F Y T LA AK D + + L++ L S F + ++
Sbjct: 85 ITLHILT--KFKLYKTAQILAEDVAAKTLDDEYASLVFKSLQETYDLCYSTSSVFDL-VV 141
Query: 133 TLYGNAGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFDEISRFLNEVPRRFDI 192
Y +I AL+ + + V S+NA L+ +S+ + + + +
Sbjct: 142 KSYSRLSLIDKALSIVHLAQAHGFMPGVLSYNAVLDATIRSKRNISFAENVFKEMLESQV 201
Query: 193 RLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFYDHKRWEIGN 252
+V++ NI ++ FC A + A L + D + KG P+VVTY TLI + ++ + G
Sbjct: 202 SPNVFTYNILIRGFCFAGNIDVA-LTLFDKMETKGCLPNVVTYNTLIDGYCKLRKIDDGF 260
Query: 253 GLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTLVLVIKGF 312
L M LKG+ PNL ++NV I L R+ + + ++ M R G + DEVT +IKG+
Sbjct: 261 KLLRSMALKGLEPNLISYNVVINGLCREGRMKEVSFVLTEMNRRGYSLDEVTYNTLIKGY 320
Query: 313 FRAGYPEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYTLCKDSMRKNWFPNV 372
+ G A+ +++ + G S Y ++IH++CK G+ ++A + PN
Sbjct: 321 CKEGNFHQALVMHAEMLRHGLTPSVITYTSLIHSMCKAGNMNRAMEFLDQMRVRGLCPNE 380
Query: 373 DTIFMLLEGLKKSGKINKA 391
T L++G + G +N+A
Sbjct: 381 RTYTTLVDGFSQKGYMNEA 399
Score = 112 bits (279), Expect = 3e-23
Identities = 83/303 (27%), Positives = 143/303 (46%), Gaps = 23/303 (7%)
Query: 114 EQQKKLPQSRREGFVVRIIT----LYGNA--GMIQHALNTFYQMGDFRCVRTVKSFNATL 167
E + L + GF ++T + G+ G ++ A+ M + V S++ L
Sbjct: 398 EAYRVLREMNDNGFSPSVVTYNALINGHCVTGKMEDAIAVLEDMKEKGLSPDVVSYSTVL 457
Query: 168 NVLAKSRDFDEISRFLNEVPRRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKG 227
+ +S D DE R E+ + I+ D + + ++ FCE + KEA + + + G
Sbjct: 458 SGFCRSYDVDEALRVKREMVEK-GIKPDTITYSSLIQGFCEQRRTKEA-CDLYEEMLRVG 515
Query: 228 VKPDVVTYTTLISAFYDHKRWEIGNGLWNQMVLKGVMPNLHTFNVRIQFLVTVRRVWDAN 287
+ PD TYT LI+A+ E L N+MV KGV+P++ T++V I L R +A
Sbjct: 516 LPPDEFTYTALINAYCMEGDLEKALQLHNEMVEKGVLPDVVTYSVLINGLNKQSRTREAK 575
Query: 288 KLMALMQRNGVTPDEVT---------------LVLVIKGFFRAGYPEMAMRVYSALHDKG 332
+L+ + P +VT +V +IKGF G A +V+ ++ K
Sbjct: 576 RLLLKLFYEESVPSDVTYHTLIENCSNIEFKSVVSLIKGFCMKGMMTEADQVFESMLGKN 635
Query: 333 YKISANIYQTMIHNLCKRGDFSQAYTLCKDSMRKNWFPNVDTIFMLLEGLKKSGKINKAK 392
+K Y MIH C+ GD +AYTL K+ ++ + + T+ L++ L K GK+N+
Sbjct: 636 HKPDGTAYNIMIHGHCRAGDIRKAYTLYKEMVKSGFLLHTVTVIALVKALHKEGKVNELN 695
Query: 393 VIV 395
++
Sbjct: 696 SVI 698
Score = 96.7 bits (239), Expect = 1e-18
Identities = 69/278 (24%), Positives = 128/278 (45%), Gaps = 3/278 (1%)
Query: 138 AGMIQHALNTFYQMGDFRCVRTVKSFNATLNVLAKSRDFDEISRFLNEVPRRFDIRLDVY 197
AG I AL F +M C+ V ++N ++ K R D+ + L + + + ++
Sbjct: 218 AGNIDVALTLFDKMETKGCLPNVVTYNTLIDGYCKLRKIDDGFKLLRSMALK-GLEPNLI 276
Query: 198 SVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFYDHKRWEIGNGLWNQ 257
S N+ + C ++KE F+L +N +G D VTY TLI + + + +
Sbjct: 277 SYNVVINGLCREGRMKEVS-FVLTEMNRRGYSLDEVTYNTLIKGYCKEGNFHQALVMHAE 335
Query: 258 MVLKGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTLVLVIKGFFRAGY 317
M+ G+ P++ T+ I + + A + + M+ G+ P+E T ++ GF + GY
Sbjct: 336 MLRHGLTPSVITYTSLIHSMCKAGNMNRAMEFLDQMRVRGLCPNERTYTTLVDGFSQKGY 395
Query: 318 PEMAMRVYSALHDKGYKISANIYQTMIHNLCKRGDFSQAYTLCKDSMRKNWFPNVDTIFM 377
A RV ++D G+ S Y +I+ C G A + +D K P+V +
Sbjct: 396 MNEAYRVLREMNDNGFSPSVVTYNALINGHCVTGKMEDAIAVLEDMKEKGLSPDVVSYST 455
Query: 378 LLEGLKKSGKINKA-KVIVALAEGRKPPFSFSYLASMQ 414
+L G +S +++A +V + E P + +Y + +Q
Sbjct: 456 VLSGFCRSYDVDEALRVKREMVEKGIKPDTITYSSLIQ 493
Score = 77.0 bits (188), Expect = 9e-13
Identities = 66/304 (21%), Positives = 128/304 (41%), Gaps = 2/304 (0%)
Query: 89 NRFAFYDTVSRLAGAKRFDYIEQLLEQQKKLPQSRREGFVVRIITLYGNAGMIQHALNTF 148
N ++ ++ L R + +L + + S E +I Y G AL
Sbjct: 274 NLISYNVVINGLCREGRMKEVSFVLTEMNRRGYSLDEVTYNTLIKGYCKEGNFHQALVMH 333
Query: 149 YQMGDFRCVRTVKSFNATLNVLAKSRDFDEISRFLNEVPRRFDIRLDVYSVNIAVKAFCE 208
+M +V ++ + ++ + K+ + + FL+++ R + + + V F +
Sbjct: 334 AEMLRHGLTPSVITYTSLIHSMCKAGNMNRAMEFLDQMRVR-GLCPNERTYTTLVDGFSQ 392
Query: 209 AEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFYDHKRWEIGNGLWNQMVLKGVMPNLH 268
+ EAY + + +N+ G P VVTY LI+ + E + M KG+ P++
Sbjct: 393 KGYMNEAYRVLRE-MNDNGFSPSVVTYNALINGHCVTGKMEDAIAVLEDMKEKGLSPDVV 451
Query: 269 TFNVRIQFLVTVRRVWDANKLMALMQRNGVTPDEVTLVLVIKGFFRAGYPEMAMRVYSAL 328
+++ + V +A ++ M G+ PD +T +I+GF + A +Y +
Sbjct: 452 SYSTVLSGFCRSYDVDEALRVKREMVEKGIKPDTITYSSLIQGFCEQRRTKEACDLYEEM 511
Query: 329 HDKGYKISANIYQTMIHNLCKRGDFSQAYTLCKDSMRKNWFPNVDTIFMLLEGLKKSGKI 388
G Y +I+ C GD +A L + + K P+V T +L+ GL K +
Sbjct: 512 LRVGLPPDEFTYTALINAYCMEGDLEKALQLHNEMVEKGVLPDVVTYSVLINGLNKQSRT 571
Query: 389 NKAK 392
+AK
Sbjct: 572 REAK 575
>UniRef100_Q6K9T5 Membrane-associated salt-inducible protein-like [Oryza sativa]
Length = 423
Score = 114 bits (285), Expect = 5e-24
Identities = 86/340 (25%), Positives = 160/340 (46%), Gaps = 21/340 (6%)
Query: 70 PHKLFHIFKSNATNPIVVENRFAFYDTVSRLAGAKRFDYIEQLLEQQ-KKLPQSR-REGF 127
P L +F S ++P + +R F +V RLA A R D + +L LP EGF
Sbjct: 58 PEALADLFISGLSHPAFLADRPIFTLSVHRLASAGRRDLVASILSSSLTSLPAPHPSEGF 117
Query: 128 VVRIITLYGNAGMIQHALNTFYQMGDFRCVR--TVKSFNATLNVLAKSRDFDEISRFLNE 185
++R+I+LY AGM H+L+TF R V + ++ +A L+ +R +D +
Sbjct: 118 LIRLISLYSAAGMPDHSLSTF------RIVTPPSDRALSALLSAYHDNRLYDRAIQAFRT 171
Query: 186 VPRRFDIRLDVYSVNIAVKAFCEAEKLKEAYLFMLDCVNNKGVKPDVVTYTTLISAFYDH 245
+P I+ V S N+ +K+F + L A + + V+PD+V+ ++ + +
Sbjct: 172 LPAELGIKPSVVSHNVLLKSFVASGDLASARALFDEMPSKADVEPDIVSCNEILKGYLNA 231
Query: 246 KRWEIGNGLWNQMVL-----KGVMPNLHTFNVRIQFLVTVRRVWDANKLMALMQRNGVTP 300
+ + + + PN+ T+N+R+ L + R ++A +L+ M+ GV P
Sbjct: 232 ADYAAFDQFLKDNTTAAGGKRRLKPNVSTYNLRMASLCSKGRSFEAAELLDAMEAKGVPP 291
Query: 301 DEVTLVLVIKGFFRAGYPEMAMRVYSAL------HDKGYKISANIYQTMIHNLCKRGDFS 354
+ + VI+G + G A+ ++ + + KG ++ Y ++ L +G F+
Sbjct: 292 NRGSFNTVIQGLCKEGEVGAAVAIFKRMPEVPRPNGKGVLPNSETYIMLLEGLVNKGVFA 351
Query: 355 QAYTLCKDSMRKNWFPNVDTIFMLLEGLKKSGKINKAKVI 394
A + K+ ++ W P + L++GL KS K AK +
Sbjct: 352 PALEVFKECLQNKWAPPFQAVQGLIKGLLKSRKAKHAKEV 391
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 668,071,145
Number of Sequences: 2790947
Number of extensions: 27255557
Number of successful extensions: 76802
Number of sequences better than 10.0: 823
Number of HSP's better than 10.0 without gapping: 533
Number of HSP's successfully gapped in prelim test: 297
Number of HSP's that attempted gapping in prelim test: 69176
Number of HSP's gapped (non-prelim): 3885
length of query: 420
length of database: 848,049,833
effective HSP length: 130
effective length of query: 290
effective length of database: 485,226,723
effective search space: 140715749670
effective search space used: 140715749670
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 76 (33.9 bits)
Medicago: description of AC148969.14


